
Feasibility and clinical value of circulating tumor DNA testing in patients with gastric adenocarcinomas
Introduction
Gastric cancer is prevalent in various parts of the world, with an average of 1 million patients diagnosed annually. Despite recent advances, the 5-year survival rate is less than 30–40% in patients with advanced gastric cancer (1). Therefore development of biomarkers that can assist in timely detection, prognosis and in assessment of treatment response is of great importance. Circulating tumor DNA (ctDNA) has shown potential in various studies for further development as a biomarker with predictive and prognostic implications. This is supported by studies showing ctDNA levels to correlate with the tumor size, tumor stage and depth of invasion in patients with gastric, breast, lung, ovarian and pancreatic cancers (2,3). Further, studies have shown the level of ctDNA to be 2–3 times higher in the plasma of cancer patients (4,5). Correlation of ctDNA with treatment response has also been noted, implicating a possible role in monitoring treatment response (2). Over 80% of patients with advanced gastroesophageal cancers have been shown to have genetic aberrations detected by ctDNA (6) However, limited literature is available elucidating the feasibility of ctDNA testing in a real-time clinical setting in patients with gastric cancer including those with gastroesophageal junction cancer. Herein, we describe a cohort of such patients characterized using a clinically available ctDNA assay.
Methods
During the period extending from January 2015 to June 2018, a total of 46 patients with gastric cancer including those with gastroesophageal junction malignancies underwent ctDNA testing by Guardant Health (Redwood City, CA, USA). All patients were seen at Mayo Clinic Cancer Center. Of the 46 patients, 55 samples were obtained, with 7 patients having serial testing. Demographic information and date of blood collection were available for all patients. The analysis of data from patients in this cohort was approved by the Mayo Clinic institutional review board.
Cell-free DNA (ctDNA) isolation
ctDNA was isolated as previously described (7,8). Briefly, blood samples were collected in StreckTM tubes. Samples were shipped at room temperature overnight to Guardant Health (Redwood City, CA, USA). On receipt, 10 mL of blood was processed by centrifugation by 1,600 g for 10 minutes at 4 °C to isolate plasma. ctDNA was then extracted from 1mL aliquots of plasma using the QIAamp circulating nucleic acid kit (Qiagen), concentrated and size selected using Agencourt Ampure XP beads (Beckman Coulter), and quantified by Qubit fluorometer (Life Technologies, Carlsbad, CA, USA).
ctDNA sequencing
The G360 panel is a CLIA-certified, College of American Pathologists (CAP)-accredited, New York State Department of Health (NYSDOH)-approved test that detects single nucleotide variants (SNV) in 73 genes as well as copy number amplifications (CNAs) in 18 genes, fusions in 6 genes, and insertions or deletions (indels) in 23 genes. Following ctDNA isolation, 5–30 ng of DNA underwent oligonucleotide barcoding for digital sequencing library preparation. This library was amplified and enriched for the target genes using biotinylated custom baits. Each of the cancer-related genes was pair-end sequenced on an Illumina HiSeq 2500. Each base pair had a 15,000× average coverage depth. After sequencing, algorithmic reconstruction of the digitized sequencing signals was used to reconstruct the ctDNA fragments. Analytic and clinical validation has been previously reported (7).
The Illumina sequencing reads were mapped to the hg19/GRCh37, and ctDNA genomic alterations were identified from the sequencing data by Guardant Health’s proprietary bioinformatics algorithms. The absolute number of unique DNA fragments at a given nucleotide position is quantified, enabling a quantitative measurement of ctDNA as a percentage of the total ctDNA. The variant allele frequency (VAF) for a given somatic alteration is calculated as the fraction of ctDNA molecules harboring the variant of interest divided by the total number of unique ctDNA molecules mapping to the variant position. The reportable range for SNV, indels, fusions, and CNAs in ctDNA by the G360 assay is cs in cportable range for SNV, y the total numctively. Plasma copy number of is reported by centiles with 2+ being between the 50th to 90th percentile in the Guardant Health database and 3+ being greater than the 90th percentile.
Over the course of the study, the gene panel composition expanded from 54 to 68 to 70 to 73 genes. The currently utilized 73 gene panel includes the addition of 5 genes to and removal of 2 genes from the prior list. The vast majority of samples in this study were tested under the 73-gene panel.
Results
Data from a total of 46 patients was included in this study. A total of 55 samples were obtained from these patients including variants of undetermined significance (VUS) with six patients undergoing testing twice and one patient undergoing testing four times. Eleven samples were noted to have no alterations. After excluding VUS we had a total of 43 samples from 34 patients. Eighteen patients were diagnosed with gastric adenocarcinoma (53%) and 16 patients were diagnosed as gastroesophageal junction adenocarcinoma (47%). Nine patients were female (26%) and twenty five were male (73%). The median age at the time of first test was at 65 years with a minimum age of 33 years and a maximum of 98 years in our cohort of patients.
Among all samples the median number of alterations, excluding VUS, per sample was 1 (range, 1–52) with an average of 4.39. The median minor allele frequency (MAF), excluding VUS, per sample was 1.2 (range, 0.03–52) with an average of 3.7.
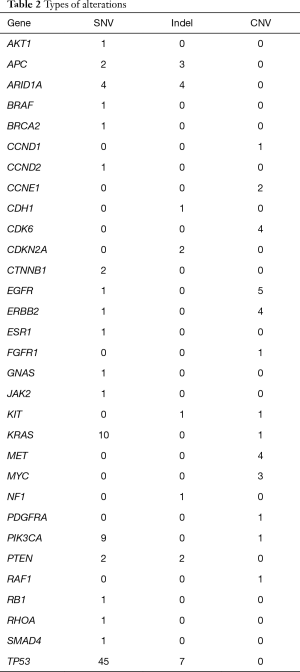
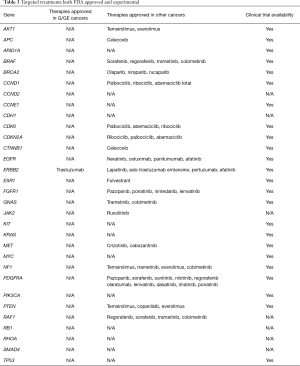
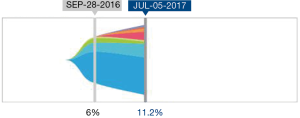
A total of 136 genetic alterations excluding VUS or synonymous alterations were seen. The profile of genetic alterations observed was broad as shown in Table 1. The top 11 genes altered in this cohort of patients, excluding VUS or synonymous alterations, were TP53 (38.2%), KRAS (8.1%), PIK3CA (7.4%), ARID1A (5.96%), EGFR (4.4%), APC (3.7%), ERBB2/HER2 (3.7%), CDK6 (2.9%), MET (2.9%), PTEN (2.9%) and MYC (2.2%). This illustrates that only a few genes are altered in the majority of the patients. Regarding the type of alterations, the majority (63.2%) were single nucleotide variations followed by copy number variations (31%) and indels (20%) (Table 2). Currently the only approved treatment by FDA that is targeted against a genetic aberration in patients with gastric cancer/GE-junction adenocarcinoma and HER-2 positivity is trastuzumab however there are approved treatments for multiple targets in various other malignancies along with the availability of experimental therapeutics (Table 3). HER2/Neu amplification/alteration was only detected in 5 out of the 46 patients included in this study. Data regarding HER2/Neu testing on tissue samples was available in 3 out of 5 patients at the time of consultation. All three of these patients were found to be positive for HER2/Neu via IHC testing on tissue samples. Serial testing done in patients at the time of disease progression correlated with increase in somatic mutation burden. This is shown in Figure 1 which shows ctDNA tumor response in a patient with metastatic GE-junction adenocarcinoma who underwent serial testing. Somatic mutation burden was at 6% upon initial consultation; subsequently patient underwent repeat testing upon radiographic disease progression and was found to have increase in somatic mutation burden along with emergence of new alterations.

Full table
Full table
Full table
Since this analysis we have identified two patients with GE junction cancer who where noted to be have a BRCA mutation via ctDNA testing. BRCA mutation is uncommon in patients with GE junction cancer. One of the patients was identified as germ line BRCA mutation while the other patient was identified as somatic BRCA mutation. Interestingly the patient who was found to have somatic BRCA mutation was also positive for HE2/Neu (data not shown).
Discussion
Circulating DNA was first reported in 1948 in healthy humans (9) which was followed by detection of ctDNA in cancer patients in 1977 (10). Some of the earlier work was on patients with myelodysplastic syndrome and pancreatic cancer, whereby aberrations in the RAS-pathway were detected in the blood (11). These initial observations paved way to the development of ctDNA as a biomarker. ctDNA offers a unique non-invasive approach for studying tumor genomics, potential targets for treatment, and disease prognosis without the hazards and difficulties usually encountered in obtaining a surgical specimen.
Mechanisms regarding the release of ctDNA involve various physiological events that accompany tumor growth such as apoptosis, micro metastasis and necroptosis (12,13). ctDNA allows for quantification of somatic mutation burden along with detection of various cancer related genetic and epigenetic alterations including tumor mutations (14), promoter methylation (15), microsatellite instability, deletion, amplification, translocation of chromosome and loss of heterozygosity (16,17).
Patients with advanced cancers have been shown to have high rate of detection of genetic aberrations via next generation sequencing of ctDNA (6). A comparison has also been done with patients at early stages of cancers where higher level of ctDNA is detected in advanced stage versus early stage (14% in stage 1 versus 50% in stage III-IV) (3). Few studies have shown the concordance of ctDNA results with tissue based genomic sequencing. One study reported high concordance rates of 96%, 94%, 95% and 91% respectively between ctDNA and tissue biopsy in KRAS, MYC, KRAS G12V and EGFR amplification among patients with gastrointestinal malignancies (18). HER2/Neu amplification is seen in up to a quarter to a third of patients with gastric or gastroesophageal junction cancers (GE-junction) adenocarcinomas (19). This is a marker of more aggressive disease. However, in an era of anti-HER2 directed therapy, finding this aberration could be of great value. The overall survival benefit of such an approach has been already demonstrated in the ToGA trial, that lead to the approval of trastuzumab in patients with HER2 positive advanced gastric/GE junction adenocarcinomas (20).
HER2 positivity needs to be demonstrated, however, based on currently approved consensus criteria (IHC score of 3+ OR 2+ with a positive fluorescent in situ hybridization result). Two issues have been well documented with regards to HER2 measurement which include intratumoral heterogeneity which is more common in gastric cancer than breast cancer and therapeutic resistance (21). Overall rate of response to HER2 directed therapy in HER2 positive gastric cancer is 47% (20). Therefore due to potential therapeutic implications accurate measurement of HER2 status is becoming increasingly important. Various studies have shown the potential of liquid biopsy to detect HER2/Neu amplification along with showing concordance with routine IHC and FISH on tissue samples (22,23). Our own data shows that HER2/Neu amplification was detected via ctDNA testing in about 3.7% of all genetic alterations with concordance observed in 3 out of 5 available tissue samples.
Most studies have documented ctDNA levels in cancer patients against healthy individuals; however, a longitudinal approach is of more clinical relevance in terms of assessing disease response to various treatment modalities and evolving tumor dynamics. The non-invasive nature of ctDNA testing allows for repeated testing and monitoring evolution of tumors which is a clear advantage of the technique over traditional tissue biopsies that are usually only done at diagnosis (17). In our own study sample radiographic disease progression correlated with increase in somatic mutation burden on ctDNA testing. Further, new alterations are detected on disease progression which can be indicative of disease response.
The evolution of tumor genome captured by ctDNA testing has been demonstrated by two recent reports on patients with colorectal cancer where liquid biopsy was used to demonstrate acquisition of RAS-mutations while on anti-EGFR therapies as mechanisms of resistance (24,25). Levels have shown not to correlate with traditional tumor markers like CEA and CA 19-9 confirming the lack of prognostic significance of these traditional tumor markers.
With the recent approval of immunotherapy in mismatch repair deficient (dMMR), or microsatellite instability high (MSI-H) solid malignancies knowledge of MMR/MSI-H status is becoming increasingly important. Although IHC for MMR proteins can not be assessed with liquid biopsies, the technology can provide MSI status along with determination of the burden of somatic mutations, also known as tumor mutational burden (TMB). The TMB appears to correlate with response to immunotherapy and further illustrates the clinical utility of a liquid biopsy in predicting response to immunotherapy (26,27). We have been able to identify atleast one patient with gastric cancer via ctDNA testing who was MSI-H and is currently being treated with front line immunotherapy as he was chemotherapy ineligible (data not shown here).
The recent joint review by the American society of Clinical Oncology and College of American Pathologists (5), was a comprehensive analysis of the analytical and clinical value of liquid biopsy. Liquid biopsy though is a promising area for future cancer research it is currently limited by lack of prospective larger studies and heterogeneity that is inherent in different types of cancers. Ongoing studies like the NEXT-2 trial where treatment was directed based on the aberrations detected via ctDNA testing will provide further insight for integrating ctDNA testing into clinical practice (28). The main limitation of our study was the small population size and retrospective design. However, we were able to show that liquid biopsy is a feasible test for patients with advanced gastric cancers with a quick turnaround time. The feasibility and value of ctDNA testing would have to be individually studied for different tumor types for understanding its limitations and strengths in addition to evaluating the concordance between tissue based testing and liquid biopsy.
Acknowledgements
The authors are deeply indebted to the Center of Individualized Medicine (CIM), Mayo Clinic, for their support and guidance.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Wang J, Yu JC, Kang WM, et al. Treatment strategy for early gastric cancer. Surg Oncol 2012;21:119-23. [Crossref] [PubMed]
- Kim K, Shin DG, Park MK, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res 2014;86:136-42. [Crossref] [PubMed]
- Chen A, Li S, Jaboni J, et al. Mutation detection in circulating tumor DNA in stage I vs. later stage cancer samples. J Clin Oncol 2016;34:e23046. [Crossref]
- De Mattos-Arruda L, Olmos D, Tabernero J. Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol 2011;7:1385-97. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Catenacci DVT, Nagy RJ, Braiteh FS, et al. Cell free circulating tumor DNA (ctDNA) landscape in patients with advanced gastroesophageal adenocarcinoma (GEC). J Clin Oncol 2017;35:47. [Crossref]
- Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 2015;10:e0140712. [Crossref] [PubMed]
- Kim ST, Lee WS, Lanman RB, et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget 2015;6:40360. [Crossref] [PubMed]
- Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994;3:67-71. [PubMed]
- Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. [Crossref] [PubMed]
- Choi JJ, Reich CF 3rd, Pisetsky DS. The role of macrophages in the in vitro generation of extracellular DNA from apoptotic and necrotic cells. Immunology 2005;115:55-62. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414-23. [Crossref] [PubMed]
- Schwarzenbach H, Chun FK, Muller I, et al. Microsatellite analysis of allelic imbalance in tumour and blood from patients with prostate cancer. BJU Int 2008;102:253-8. [Crossref] [PubMed]
- Tsujiura M, Ichikawa D, Konishi H, et al. Liquid biopsy of gastric cancer patients: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol 2014;20:3265-86. [Crossref] [PubMed]
- Riviere P, Fanta PT, Ikeda S, et al. The mutational landscape of gastrointestinal malignancies as reflected by circulating tumor DNA. Mol Cancer Ther 2018;17:297-305. [Crossref] [PubMed]
- Gravalos C, Jimeno A. HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol 2008;19:1523-9. [Crossref] [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [Crossref] [PubMed]
- Hofmann M, Stoss O, Shi D, et al. Assessment of a HER2 scoring system for gastric cancer: results from a validation study. Histopathology 2008;52:797-805. [Crossref] [PubMed]
- Shoda K, Masuda K, Ichikawa D, et al. HER2 amplification detected in the circulating DNA of patients with gastric cancer: a retrospective pilot study. Gastric Cancer 2015;18:698-710. [Crossref] [PubMed]
- Shoda K, Ichikawa D, Fujita Y, et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer 2017;20:126-35. [Crossref] [PubMed]
- Diaz LA Jr, Williams RT, Wu J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012;486:537-40. [Crossref] [PubMed]
- Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532-6. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Kasi PM. Mutational burden on circulating cell-free tumor-DNA testing as a surrogate marker of mismatch repair deficiency or microsatellite instability in patients with colorectal cancers. J Gastrointest Oncol 2017;8:747-8. [Crossref] [PubMed]
- Lee J, Kim ST, Kim KM, et al. Cell-free DNA sequencing-guided therapy in a prospective clinical trial: NEXT-2 trial—A feasibility analysis. J Clin Oncol 2016;34:11534. [Crossref]


