
Microbial marauders: pancreatic microbiota and its impact on carcinogenesis
Over the past several decades the importance of the human microbiota, the collection of bacteria, viruses, fungi, protozoa, and archea that establish a commensal, symbiotic, and pathobiont relationship within every individual, in disease development and progression has been established (1-3). While the intestinal microbiota is the most logical organ system to begin such investigations, a myriad of data now exist that establish not only organ-based microbiota that impact diseases but that the intestinal microbiota is influential on non-intestinal pathophysiology as well (4-7). Most studies have focused on the role of bacteria, given the relative ease to investigate and manipulate these organisms and greater abundance compared to other components of the microbiota. The mechanisms whereby bacteria influence disease progression is variable and include microbial-derived metabolism, production of genotoxins, and activation of proinflammatory pathways (8). Given that alterations of the microbiota have been associated with autoimmune diseases and can impact immune processes (9,10), it is natural to surmise that the microbiota may impact carcinogenic processes through immune pathways (11-13). This has been established in colorectal cancer and melanoma but only recently has the role of the microbiota on pancreatic carcinogenesis through immune system regulation been proposed (14,15). In a recent study by Pushalkar and colleagues (16), they demonstrate that the microbiota facilitates an immune tolerant phenotype that allows the progression of pancreatic ductal adenocarcinoma (PDAC) in a murine model. The study thus has implications for our understanding and treatment of PDAC, a cancer that is now the third leading cause of cancer death in the United States and 12th leading cause of cancer death worldwide (17,18).
Pancreatic ductal carcinoma is the most common malignancy of the pancreas and has an annual incidence of approximately 55,000 cases in the United States and 460,000 cases worldwide (17,18). The tumor microenvironment is such that an inflammatory milieu is established that promotes carcinogenesis in the context of immune tolerance (19). The mechanisms whereby this environment is setup is poorly understood and contributes to the lethality of the disease. Expanding on their prior published work, Pushalkar and colleagues investigated the ability of the intestinal microbiota to alter PDAC progression in relation to immune cell subtypes. The authors argue that few data exist in support of the gut microbiota influencing carcinogenesis in an organ “remote” from the gastrointestinal lumen or its drainage. To address this in regard to the pancreas and PDAC, the authors first sought to establish the ability of endoluminal intestinal bacteria to migrate to the pancreas. In separate experiments using either fluorescently labeled Enterococcus faecalis or Escherichia coli, the investigators orally gavaged C57BL/6 wild-type (WT) mice with the bacteria and followed their presence in the pancreas over time. They report an initial increase in E. faecalis fluorescent detection at 30 minutes by flow cytometry with subsequent decrease by 3 hours. Likewise, green fluorescent labeled E. coli was detected in the pancreata of WT gavaged mice compared to control at 6 hours by immune fluorescent microscopy. These data demonstrate the ability of the pancreas to harbor bacteria but the clinical applicability is questionable given the high concentration oral gavage required to detect the bacteria and subsequent decline in abundance over time. In order to expand this to human samples, fluorescent in situ hybridization (FISH) with a universal 16S rRNA bacterial probe was performed to assess the presence of bacteria within the pancreata of either human PDAC specimens or the Pdx1Cre; LSL-KrasG12D; Trp53R172H (KPC) transgenic mouse model of pancreatic cancer. Compared to non-cancer controls, both the malignant human and mouse pancreas had a greater amount of bacteria which was confirmed with quantitative polymerase chain reaction (qPCR). Interestingly, repopulation of the WT mice microbiota in which their intestinal microbiota was ablated with an antibiotic cocktail demonstrated increased intrapancreatic bacteria by qPCR when KPC feces was gavaged versus WT feces. These data demonstrate the potential ability of bacteria to translocate from the gut to the pancreas and intestinal bacteria from animals with PDAC have a greater propensity to do so compared to non-cancer controls. Subsequently, using 16S rRNA sequencing, the authors noted the increased abundance of the phyla Proteobacteria (45%), Bacteroidetes (31%), and Firmicutes (22%) in human PDAC samples compared to normal controls. Furthermore, the bacterial composition in PDAC samples was distinct from normal controls, suggesting a disease and organ-specific microbiota. Given these findings, the authors demonstrated that the presence of a microbiota accelerates pancreatic carcinogenesis given that germ-free (GF) KPC mice as well as WT KPC littermates with antibiotic-depleted intestinal microbiota and orthotopically implanted xenografts had slower progression of their PDAC compared to respective microbiota-intact controls. In order to ascertain if a specific intestinal microbiota could promote pancreatic carcinogenesis, the researchers next repopulated the gut microbiota of Ptf1aCre; LSL-KrasG12D (KC) mice, which had been depleted with an antibiotic cocktail, using feces from either WT or KPC. They demonstrate increased tumor formation in microbiota-depleted KC mice when repopulated with feces from KPC mice versus WT controls, suggesting that specific bacterial species are able to accelerate pancreatic carcinogenesis in a genetically susceptible host. A similar phenotype was seen in GF KC mice that were repopulated with KPC stool. Finally, using longitudinal bacterial sequencing of KC versus WT mouse stool over time, there was an enrichment of Bifidobacterium in the KC mice with Bifidobacterium pseudolongum being the most abundant species. Hypothesizing that this species may be at least partially responsible for the acceleration of PDAC, B. pseudolongum was then repopulated into GF KC mice and also resulted in increased PDAC formation compared to sham-gavaged control GF KC mice.
Given the presented data and publications on the role of intratumoral immune suppression in other malignancies (20,21), the authors hypothesized that the microbiota induces an immune suppressive environment within PDAC that results in disease progression. To test this hypothesis, immune profiling was performed by flow cytometry on orthotopically implanted KPC-derived tumor cells xenografts in control and antibiotic-treated WT mice. Microbial depletion resulted in increased T-cells, reduced myeloid-derived suppressor cells (MDSC), and reduction in immunosuppressive CD206+ M2-like tumor associated macrophages (TAM). Concomitantly, an increase in M1-like TAM with higher expression of major histocompatibility complex II (MHC-II) was noted. A similar phenotype was observed in vitro when cell free extract from the stool of PDAC-bearing mice resulted in a reduction of MHC-II expression and upregulation of IL-10 in splenic macrophages compared to cell free extract from control mice not bearing a PDAC xenograft. Given this evidence of microbial mediated macrophage polarization and their prior published work that such polarization directs T-cell immunogenicity (22), the authors hypothesized that bacterial ablation would activate tumor infiltrating T-cells. Indeed, by assessing immune populations in WT mice orthotopically implanted with KPC-derived tumor cells, the investigators demonstrated that antibiotic-mediated microbiota depletion resulted in the TH1 polarization of CD4 T-cells with increased infiltration of cytotoxic CD8 T-cells. These CD4 and CD8 subsets of T-cells were subsequently noted to have increased expression of PD-1 and CD44, known cofactors in antitumor immunogenicity and memory function. In corollary to these experiments, the investigators then repopulated in intestinal microbiota of these antibiotic treated mice with that from KPC mice which reversed the intratumoral immune phenotype. Finally, macrophages exposed to cell free stool extract from PDAC-bearing KC mice mitigated the activation of CD4 and CD8 T-cells and prevented TH1 CD4 differentiation compared to macrophages exposed to cell free stool extract from WT mice. Together, these data support the hypothesis that the host microbiota promotes immunosuppression in PDAC via macrophage-mediated pathways that may be responsible for its progression.
Given the effect of macrophages on CD4 and CD8 cells and their subsequent anti-PDAC role with microbial depletion/ablation, the investigators sought to solidify this observation with adoptive transfer of T-cells isolated from orthotopic xenografts of KPC-derived tumor cells grown in control or antibiotic-treated WT mice to WT mice challenged with KPC subcutaneous implanted xenografts. Indeed, adoptive transfer of T-cells from PDAC orthotopic xenografts derived from microbiota-depleted mice resulted in a 50% reduction in PDAC subcutaneous xenograft size compared to adoptive transfer of T-cells from microbiota-intact mice. As anticipated, CD4/8 depletion resulted in a concomitant abrogation of the anti-tumor effect seen with antibiotic-mediated microbial depletion but no effect on control animals. Interestingly, antibiotic depletion of the intestinal microbiota also upregulated PD-1 expression in intratumoral CD4 and CD8 cells. Given clinical data on the role of PD-1 inhibition in the treatment of various malignancies (20,21), the authors further hypothesized that depletion of the microbiota would have a synergistic effect with anti-PD-1 blockade. As such, combined antibiotic microbial depletion with anti-PD-1 therapy resulted in decreased orthotopic xenograft size compared to anti-PD-1 treatment alone which correlated with increased CD4 and CD8 T-cell activation (Figure 1).
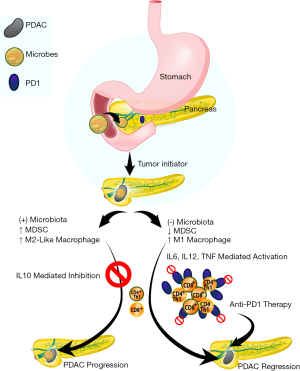
Finally, in order to demonstrate a mechanism for microbial-mediated progression of pancreatic carcinogenesis, the investigators interrogated pattern recognition receptors (PRRs). They had previously demonstrated that upregulation of these receptors, responsible for recognition of pathogens and initiating requisite pathways to eliminate it, resulted in PDAC progression by adaptive and innate immune suppression and subsequently hypothesized that the mechanism is mediated by the microbiota (23,24). Indeed, cell-free extract from the stool of KC mice induced PRR expression (TLR2, TLR4, and TLR5) in the HEK293 reporter cell line compared to cell-free extract from WT stool. Decreased PRR expression was conversely seen in PDAC xenografts derived from antibiotic treated mice compared to control. Finally, the tumoral immune suppression was evident by upregulation of TLR2 and TLR5 in macrophages isolated from orthotopic KPC xenografts and corresponded to larger tumors when mice were treated with the ligands for TLR2 and TLR5. These data suggest that an intestinal microbiota present in PDAC is able to activate immune suppressive macrophages via a TLR2 and TLR5 mechanisms to potentiate pancreatic carcinogenesis.
Until recently, investigations involving the role of the microbiome on pancreatic carcinogenesis had been limited to associative studies (25,26). Several groups have since provided insight into this evolving area of research with strong evidence supporting the role of the host microbiota in the acceleration of PDAC (27,28). Pushalkar et al. eloquently demonstrate this concept utilizing a variety of mouse models and further implicate the microbiota in innate and adaptive immune suppression as a potential mechanism for this phenotype. Several points are worthy of discussion and additional investigation. While the investigators allude to the observation of bacterial translocation into the pancreas as the basis for the microbial-mediated phenotypic changes, more recent data suggest that the microbiota can act in a long-distance manner without the need for bacteria within the PDAC microenvironment (27). Additionally, the high bacterial load gavage required for their observations does not mimic a clinical scenario and gavaged bacteria are part of the lower gastrointestinal flora, not upper. This observation also does not take into account studies demonstrating fecal microbial differences between control and PDAC patients (26). Furthermore, while a microbiota exists within the pancreas, such a microbiota may not necessarily be able to distinguish normal from diseased states (27). Regardless, it appears that the microbiota impacts pancreatic carcinogenesis through an immune-mediated mechanism which would be similar to other malignancies. The impact of the adaptive and innate immune systems is an active area of debate given that similar observations are seen in animal models that possess only innate immunity (27). Finally, investigations have focused on progression of PDAC, not initiation, given that all models used are already malignant cells (cell line-based work) or have the propensity to develop cancer (genetically engineered mouse models). Although studies have associated differences in either the oral or fecal microbiota with pancreatic cancer (25,26), further work is needed to determine if such differences in the intestinal microbial composition is able to initiate PDAC. This is clinically important point as identification of microbial initiators of PDAC would enable clinicians to potentially identify high risk patients and intervene to prevent pancreatic cancer development.
In summary, the field of research investigating the role of the microbiome in carcinogenesis continues to expand and has offered novel insight into pancreatic carcinogenesis. With PDAC death rates showing no signs of slowing, paradigm shifting research such as this is desperately needed to gain insight into this deadly disease.
Acknowledgements
Figure illustration courtesy of Lisa Thomas.
Funding: American Cancer Society - Norma and Rich DiMarco Mentored Research Scholar Grant (MRSG-17- 228-01-TBG; RM Thomas) and Gatorade Trust through funds distributed by the University of Florida, Department of Medicine (M Zajac-Kaye).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Thomas RM, Jobin C. The Microbiome and Cancer: Is the 'Oncobiome' Mirage Real? Trends Cancer 2015;1:24-35. [Crossref] [PubMed]
- Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014;146:1489-99. [Crossref] [PubMed]
- Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967-70. [Crossref] [PubMed]
- Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity 2014;40:843-54. [Crossref] [PubMed]
- Paun A, Yau C, Danska JS. The Influence of the Microbiome on Type 1 Diabetes. J Immunol 2017;198:590-5. [Crossref] [PubMed]
- Ussar S, Griffin NW, Bezy O, et al. Interactions between Gut Microbiota, Host Genetics and Diet Modulate the Predisposition to Obesity and Metabolic Syndrome. Cell Metab 2015;22:516-30. [Crossref] [PubMed]
- Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest 2014;124:4204-11. [Crossref] [PubMed]
- Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800-12. [Crossref] [PubMed]
- Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol 2011;7:569-78. [Crossref] [PubMed]
- Rothhammer V, Borucki DM, Tjon EC, et al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018;557:724-8. [Crossref] [PubMed]
- Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004;4:478-85. [Crossref] [PubMed]
- Ivanov II, Frutos Rde L, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008;4:337-49. [Crossref] [PubMed]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012;336:1268-73. [Crossref] [PubMed]
- Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97-103. [Crossref] [PubMed]
- Grivennikov SI, Wang K, Mucida D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012;491:254-8. [Crossref] [PubMed]
- Pushalkar S, Hundeyin M, Daley D, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018;8:403-16. [Crossref] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2017. [cited 2018 Sep 21]. Available online: https://www-cancer-org.lp.hscl.ufl.edu/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and- figures/2017/cancer-facts-and-figures-2017.pdf
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Hiraoka N, Onozato K, Kosuge T, et al. Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clin Cancer Res 2006;12:5423-34. [Crossref] [PubMed]
- Postow MA, Chesney J, Pavlick AC, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 2015;372:2006-17. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Seifert L, Werba G, Tiwari S, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532:245-9. [Crossref] [PubMed]
- Daley D, Mani VR, Mohan N, et al. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J Exp Med 2017;214:1711-24. [Crossref] [PubMed]
- Zambirinis CP, Levie E, Nguy S, et al. TLR9 ligation in pancreatic stellate cells promotes tumorigenesis. J Exp Med 2015;212:2077-94. [Crossref] [PubMed]
- Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 2018;67:120-7. [Crossref] [PubMed]
- Ren Z, Jiang J, Xie H, et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget 2017;8:95176-91. [Crossref] [PubMed]
- Thomas RM, Gharaibeh RZ, Gauthier J, et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Sethi V, Kurtom S, Tarique M, et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018;155:33-7.e6. [Crossref] [PubMed]


