
Intestinal mRNA expression profile and bioinformatics analysis in a methamphetamine-induced mouse model of inflammatory bowel disease
Introduction
Methamphetamine abuse has become a serious global public health concern. In 2018, over 1.9 million people aged 12 years and above used methamphetamine in the United States, which equates to about 0.7% of the country’s population (1,2). The China: 2019 Drug Situation Report reported methamphetamine to be the most widely used drug in China, with 1.186 million users.
The production and use of methamphetamine create the perfect conditions for adverse medical and social complications (3). Methamphetamine use results in significant damage to almost all systems in the human body, including the neurologic, cardiac, pulmonary, and dental systems (3). This damage triggers a series of hyper-acute, acute, and chronic complications, including the release of the neurotransmitters dopamine, norepinephrine, and serotonin, as well as activation of the cardiovascular and central nervous systems, vasoconstriction, pulmonary hypertension, atherosclerotic plaque formation, cardiac arrhythmias, and cardiomyopathy (4-6). As a result, the user’s immune system is significantly impacted, rendering the body susceptible to infections and exacerbating the severity of disease (7). Methamphetamine use is also associated with adverse psychological effects, such as euphoria, paranoia, agitation, mood disturbances, anxiety, psychosis, depression, delusions, and violent behavior (3,8).
The release of dopamine and other neurotransmitters induced by methamphetamine can also have multiple harmful effects on the gastrointestinal system. However, abdominal complications due to methamphetamine use are uncommon and often ignored by clinicians (9); consequently, the resulting gastrointestinal pathology has rarely been reported. Although some cases of life-threatening abdominal complications caused by methamphetamine-induced mesenteric ischemia have been described previously (3,9,10), systematically organized domestic studies are lacking. A previous study (11) reported that gut dysbiosis impaired the intestinal barrier and impaired innate immune function in methamphetamine-dependent rats, leading to inflammation and infection. Another cellular study found that methamphetamine can cause NLRP3 inflammasome overexpression and severe intestinal inflammatory injury (12). Furthermore, inhibiting the expression of microRNA (miRNA) 181c, which directly targets and regulates tumor necrosis factor-α, has been shown to induce intestinal epithelial apoptosis, causing damage to the intestinal barrier (13). However, the mechanism of methamphetamine use impairs the immune function and the intestinal barrier remains unclear.
To better understand the integrated mechanism of immune function and intestinal barrier, we established a mouse model to evaluate the effects of methamphetamine use on pathological alterations and transcriptomic responses in the gastrointestinal system, and demonstrated that methamphetamine causes a specific type of inflammatory bowel disease (IBD). The results of this study provide a greater understanding of the molecular mechanisms in response to methamphetamine use in mice.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7741).
Methods
Animals and drugs
Experiments were performed under a project license (2018NS0083) granted by Animal Care and Use Committee of Kunming Medical University, in compliance with the National guidelines for the care and use of animals. Ten male specific-pathogen-free C57BL/6 mice (8-week, weight, 23.56–25.49 g) were supplied by the Jingda Laboratory Animal Co., Ltd. (cat. no. 1107271911003159). The mice were housed in ventilated cages with 5 animals per cage and given ad libitum access to rodent chow and water. All experiments were conducted during the light period (between 07:00 a.m. and 07:00 p.m.)
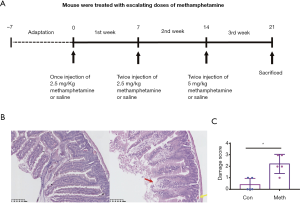
The mice were divided into two groups and were randomly allocated to receive either methamphetamine (treatment group, n=5) or saline vehicle solution (0.9% sodium chloride; control group, n=5). Treatment commenced when the mice reached 6 months old. The treatments were administered in the mouse housing room via intraperitoneal injection following an escalating dosing regimen to mimic patterns of drug abuse in humans (Figure 1A). During the first week of treatment, mice in the treatment group were injected once daily with 2.5 mg/kg of methamphetamine in the morning (between 8 and 10 a.m.). During the second week of treatment, the mice were injected twice daily with 5 mg/kg methamphetamine in the morning (between 8 to 10 a.m) and afternoon. In the third week of treatment, the mice were injected twice daily (Monday through Friday) with 10 mg/kg methamphetamine. For all 3 weeks, the control group was administered saline vehicle injections at the same frequency as the methamphetamine treatments. After the experiment, the mice were anesthetized and euthanized.
Tissue collection, histopathological analysis, and total RNA isolation
All intestinal tissues were harvested from the mice at 9:00 AM. The tissue samples were collected according to a standard protocol, and histopathological analysis was performed as described previously (14,15). The ileum was fixed in 10% buffered formalin for 24 hours, embedded in paraffin, and stored at 4 °C. The tissue blocks were then serially cut into 4-µm sections and mounted on glass slides. Hematoxylin and eosin (HE) staining was performed to examine the ileum for histopathological abnormalities, and the results were evaluated by a histopathologist (15). Immunohistochemical staining was performed using the standard method. Anti-ZO1 (cat.no.ab96587; 1:500) and anti-claudin 1 (cat.no.ab15098; 1:500) antibodies were purchased from Abcam.
Total RNA was isolated from the tissues using RNA-Bee reagent following the manufacturer’s protocol. The RNA purity was examined using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). Then, the total RNA concentration in each sample was measured with the Qubit® RNA assay for the Qubit®2.0 Fluorometer (Life Technologies, CA, USA). After that, the integrity of the RNA was assessed using the RNA Nano 6000 Assay Kit with the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Samples with RNA integrity numbers <7.0 were excluded from subsequent experiments.
cDNA library preparation and sequencing
Sequencing libraries were generated using the NEB Next® UltraTM RNA Library Prep Kit for Illumina® (NEB#E7530, USA), following the manufacturer’s recommendations. Messenger RNA (mRNA) was purified using poly-T oligo-attached magnetic beads and then fragmented with divalent cations in reaction systems [NEB Next First Strand Synthesis Reaction Buffer (5X)] under elevated temperatures. Synthesis of first-strand cDNA was performed using the random hexamer primers with M-MuLV Reverse Transcriptase (RNase H), followed by second-strand cDNA synthesis (DNA Polymerase I and RNase H). The remaining overhangs were converted into blunt ends (exonuclease/polymerase). The NEB Next Adaptor, which has a hairpin loop structure, was ligated to the 3' ends of the DNA fragments. The library fragments were purified using the AMPure XP system (Beckman Coulter, Beverly, USA), and cDNA fragments of 250–300-bp in length were selected. Next, 3 µL of USER Enzyme (NEB) was incubated with size-selected, adaptor-ligated cDNA [37 °C for 15 minutes followed by 5 minutes at 95 °C before polymerase chain reaction (PCR)] with Phusion High-Fidelity DNA polymerase, universal PCR primers, and Index (X) Primer. At last, the products were purified and assessed using the AMPure XP and Agilent Bioanalyzer 2100 systems. The index-coded samples were clustered with the cBot Cluster Generation System using a TruSeq PE Cluster Kit v3-cBot-HS (Illumina), and then sequenced on an Illumina HiSeq platform, with 125-bp/150-bp paired-end reads.
Validation of the RNA sequencing profiles by quantitative PCR (qPCR)
To validate the DEGs identified through RNA sequencing (RNA-seq), 7 genes with differential expression patterns were selected for qPCR using the same RNA samples used for transcriptome profiling. The mouse CDC73 gene was used as an internal control. Gene-specific qPCR primers were designed using Primer Premier 5.0 software (Table S1), and the efficiency was evaluated with an amplification plot and a melting curve. Reverse transcription of total RNA into cDNA (1 µg) was performed using a PrimeScript™ RT reagent kit, following the manufacturer’s protocol. Finally, real-time qPCR was conducted using an SYBR® Premix Ex Taq™ II kit and a LightCycler 480 system.
Statistical analysis
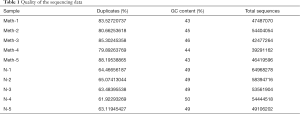
Raw data (raw reads, “fastq” format) were processed through in-house Perl scripts. The percentages of duplicates, total sequences, and GC content of the clean data (removing reads containing adapters, reads containing ploy-N, and low-quality reads from the raw data) were calculated (Table 1). All the following analyses were performed on high-quality, clean data.
Full table
Hisat 2v2.0.5 was used to align paired-end clean reads to the reference genome. The read numbers mapped to each gene were counted using FeatureCounts v1.5.0-p3. The number of fragments per kilobase of transcript sequence per million (FPKM) of base pairs sequenced was calculated based on the gene length and the read counts mapped to this gene; currently, this is the most popular method for estimating gene expression levels.
Differentially expressed genes (DEGs) between the control and methamphetamine-treated groups were identified. Differential expression analysis of the groups was performed using the DESeq2 R package (1.16.1). The resulting P values were adjusted using the Benjamini-Hochberg method. Genes with an adjusted P value <0.01 and |log2 fold-change| >2 were considered to be DEGs. The DEGs were visualized with TBtools (16). The web tool Metascape (http://metascape.org) (17-23) was used to perform functional enrichment analysis of the DEGs.
Results
Methamphetamine-treated intestinal damage mice model
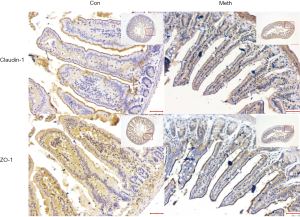
Ten ileum samples, one from each of the mice (Figure 1A), were collected for HE and immunohistochemical staining. Intestinal epithelial damage (14), intestinal barrier permeability (24), and the inflammation scores were assessed. The methamphetamine-treated mice displayed epithelial ulceration, and the epithelial damage was more pronounced than that of the control mice (Figure 1B,C). The intestinal barrier permeability of the methamphetamine-treated mice was also significantly decreased. The expression of the tight junction proteins ZO-1 and claudin-1 was significantly decreased in the methamphetamine-treated mice (Figure 2).
The identification and characterization of DEGs in the methamphetamine-treated mouse model
Ten ileum samples from the control and methamphetamine-treated mice were sequenced as described. After the removal of low-quality reads and reads containing adapters or ploy-N, 510554764 clean reads were obtained (Table 1).
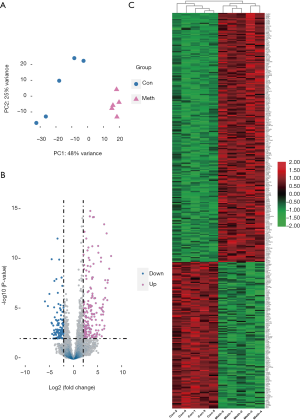
The principal component analysis (PCA) showed that the global transcriptome between the two groups were significantly different (Figure 3A). In the methamphetamine group, 322 differentiated expressed genes (|log2 fold-change| >2 and adjusted P value <0.01) were identified, including 119 upregulated genes and 203 downregulated genes (Figure 3B). The gene expression patterns of both the methamphetamine-treated and control mice were mainly clustered and showed distinct patterns (Figure 3C).
Next, Gene Ontology (GO) enrichment analysis of the DEGs was conducted; 418 GO terms were identified, including 292 biological processes, 74 cellular components, and 53 molecular functions. Of these terms, “cellular protein catabolic process” (GO:0044257), “IRE1-mediated unfolded protein response” (GO:0036498), and “response to incorrect protein” (GO:0035966) were the most enriched terms for biological processes. The most enriched terms for cellular components were “lysosome” (GO:0005764), “lytic vacuole” (GO:0000323), and “vacuolar membrane” (GO:0005774), while the most enriched terms for molecular functions included “ubiquitin-like protein transferase activity” (GO:0019787), “ubiquitin-protein ligase activity” (GO:0061630), and “ubiquitin-protein transferase activity” (GO:0004842) (Figure 4A).
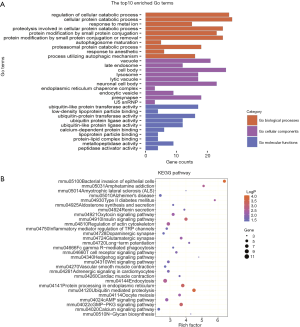
DEGs can be used to study genetic fingerprints and to select treatments; therefore, we further conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. Using a cutoff of P<0.05, the DEGs in the methamphetamine-treated mice were found to be significantly correlated with 29 KEGG pathways. According to the KEGG pathway annotation analysis (Table S2, Figure 4B), the DEGs from the comparison between methamphetamine-treated mice and controls were significantly correlated with immune processes, vascular smooth muscle, glutamatergic and dopaminergic synapses, protein processing, and type II diabetes mellitus, indicating that these processes show differential activity in response to methamphetamine use. These results suggested that the ileum tissue of the methamphetamine-treated mice displayed substantial more DEGs with dramatic mRNA expression changes compared with that of the control group.
The enrichment analysis of the DEGs
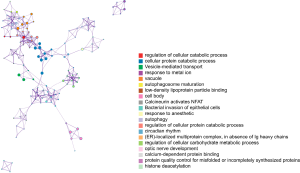
All statistically enriched terms in the GO and KEGG analyses were filtered with accumulative hypergeometric P values and enrichment factors, and the remaining significant terms with similarities >0.3 (Kappa-test score) were clustered into a tree (17). The network was visualized with Cytoscape (v3.1.2) with a “force-directed” layout and with the edge bundled for clarity. The term with the lowest P value in each cluster was selected, and its term description is shown as a label (Figure 5). The following 20 clusters were selected: regulation of cellular catabolic process (GO:0031329), cellular protein catabolic process (GO:0044257), vesicle-mediated transport (R-MMU-5653656), response to metal ion (GO:0010038), vacuole (GO:0005773), autophagosome maturation (GO:0097352), low-density lipoprotein particle binding (GO:0030169), cell body (GO:0044297), calcineurin activates nuclear factor of activated T cells (NFAT, R-MMU-2025928), bacterial invasion of epithelial cells (mmu05100), response to anesthetic (GO:0072347), autophagy (GO:0006914), regulation of cellular protein catabolic process (GO:1903362), circadian rhythm (GO:0007623), endoplasmic reticulum (ER)-localized multiprotein complex, in absence of Ig heavy chains (CORUM:414), regulation of cellular carbohydrate metabolic process (GO:0010675), optic nerve development (GO:0021554), calcium-dependent protein binding (GO:0048306), protein quality control for misfolded or incompletely synthesized proteins (GO:0006515), and histone deacetylation (GO:0016575).
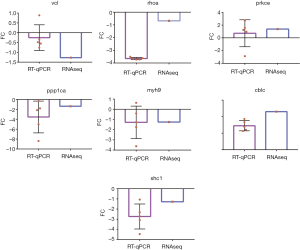
Validation of the DEGs
To verify the expression profiles of DEGs identified through RNA-seq, 7 genes were randomly selected for qPCR analysis. The results showed that the abundance patterns of the selected DEGs mRNA were generally similar (Figure 6).
Discussion
This study is the first to use RNA sequencing to document high-resolution gene expression in intestinal tissues from methamphetamine-treated mice. Previous studies of methamphetamine-treated intestinal tissues concentrated primarily on the mechanisms involved in amphetamine transport and efflux (25). A handful of recent studies reported that the administration of methamphetamine causes gut dysbiosis and activates proinflammatory factors in the epithelium (11-13). Alternatively, release of dopamine and other neurotransmitters induced by methamphetamine can also have multiple harmful effects on the gastrointestinal system. Case reports have also indicated that methamphetamine induces ischemic colitis through exerting a vasoconstrictive effect (26). However, no transcriptomic study had previously been conducted, and the gene expression patterns in methamphetamine-treated intestinal tissues was unclear. Therefore, we investigated the transcriptomic changes in the intestinal tissue of mice treated with methamphetamine.
Methamphetamine is considered to be a neurotoxin drug, and as it is mostly taken orally (27), the gastrointestinal tissue is the first tissue to be affected. Case reports have confirmed that methamphetamine has detrimental effects on the digestive system; however, no systematic experiments have verified whether methamphetamine induces intestinal dysfunction. In this study, a mouse model was used to comprehensively and quantitatively assess the small intestinal tissue of mice after treatment with methamphetamine. Histopathology revealed that methamphetamine treatment resulted in inflammatory infiltration and epithelial damage. Further, DEGs related to the immune system were identified from the tissue samples, and function enrichment analysis showed these genes to be involved in many biological processes. A previous animal model (7) also revealed increased permeability and endotoxins, and indicated epithelial ulceration after inducing inflammation in mice. Both the permeability of and endotoxins in the intestinal tissue of methamphetamine-treated mice were preceded by histological evidence of inflammation in the ileum colon junction. Another study reported that methamphetamine caused intestinal barrier lesions in a monolayer cell model and in methamphetamine-dependent people with inflammatory cytokine tumor necrosis factor-α release (13). A previous study (12) reported that methamphetamine administration can lead to severe intestinal inflammatory injury via NLRP3 inflammasome overexpression. A rhesus macaque model of chronic methamphetamine abuse (28) showed that methamphetamine enhanced the expression of genes associated with chemokines and chemokine receptors, promoting the apoptotic and proinflammatory pathways in immune cells in the brain. Conversely, another study (29) reported that methamphetamine inhibits mast cell activation and lipopolysaccharide-induced cytokine/chemokine production in C57BL/6J mice, and clinical studies (8,30) have also revealed that methamphetamine has a negative impact on the immune response. These conclusions suggest that methamphetamine significantly affects the immune system via a complex regulatory network.
Methamphetamine may cause a special type of IBD
In this study, RNA-seq analysis yielded 322 DEGs, including 119 that were downregulated and 203 that were upregulated, which shows that methamphetamine significantly affected the intestinal transcriptome of the mice. To further characterize the gene expression data from the RNA-seq, we found that the DEGs were known to be associated with IBD. To date, approximately 240 single nucleotide polymorphisms (SNPs) have been found to be associated with IBD through genome-wide association study (GWAS) (31-35). Among the DEGs in this study, G protein-coupled receptor 137 (Gpr137), MOB family member 4, protein (Mob4), MUS81 structure-specific endonuclease subunit (Mus81), ash1 (absent, small, or homeotic)-like (Ash1l), chemokine (C-C motif) receptor 5 (Ccr5), decapping exoribonuclease (Dxo), glutathione peroxidase 4 (Gpx4), guanine nucleotide-binding protein (G protein), beta 2 (Gnb2), leucyl/cysteinyl aminopeptidase (Lnpep), mutS homolog 5 (Msh5), peptidase (mitochondrial processing) alpha (Pmpca), pyruvate kinase liver and red blood cell (Pklr), ras homolog family member A (Rhoa), ring finger protein 5 (Rnf5), stromal cell-derived factor 2-like 1 (Sdf2l1), Ski2 like RNA helicase (Skiv2l), and ubiquitin-specific peptidase 4 (Usp4) were located near SNPs that are strongly associated with IBD. In the tissues of the methamphetamine-treated mice, the gene expressions of Gpx4, Msh5, Mus81, Dxo, Gpr137, Usp4, Gnb2, Mob4, and Skiv2l were upregulated, and Rnf5, Rhoa, Ash1l, Ccr5, Pmpca, Pklr, Sdf2l1, and Lnpep were downregulated (Figure 7).
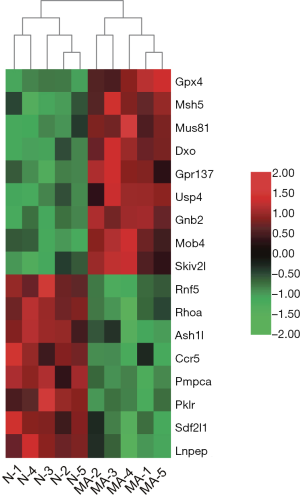
In this study, the pathology and transcriptome results suggest that methamphetamine may result in a special type of IBD. However, in this study, no further experiments were conducted and additional studies are needed to investigate this possibility. Furthermore, the human intestine is complex and comprises many different cell types, including intestinal and inflammatory cells. The “canonical” RNA-seq analyses performed in this study have notable limitations, and single-cell RNA-seq experiments are needed to confirm and expand our findings.
Methamphetamine downregulates the tight junction proteins by regulating autophagy and cellular catabolic processes
Increased intestinal permeability and intestinal mucosal barrier damage are major characteristics of IBD (36). The mucosal barrier is an important part of innate immunity, and the intestinal barrier plays a significant role in protecting the intestines from pathogenic infection (37,38). The intestinal mechanical barrier is formed by intestinal epithelial cells through a complex interaction with tight junction proteins, which defend the lumen from microbes, toxins, and molecules (39-41). Claudin 1, which is mainly localized in intestinal epithelial cells, regulates paracellular diffusion, and seals the epithelial monolayer along with adapter and scaffolding protein (42). The reduction of Zonula occludes 1 (ZO1) expression disintegrates tight junctions causing the breakdown of the barrier, which is composed of scaffolding proteins that bind and anchor claudin proteins to form a seal to regulate paracellular diffusion (43).
In this study, pathways such as “bacterial invasion epithelial cells (mmu05100)” were clustered, which suggests that gut microbiota dysbiosis is associated with methamphetamine-induced intestinal dysfunction. Notably, the related genes RhoA, Vcl, Arpc1a, Arpc4, Myh9, Pik3ca, and Ssh2 are shown to be associated with focal adhesion and actin stabilization. A previous study (7) also reported that methamphetamine reduced claudin-1 and ZO-1 immunofluorescence in rats. Actin cytoskeleton rearrangement and membrane remodeling is a significant process of reduction or morphological redistribution in sealing tight junction proteins (44). It leads to intestinal hyperpermeability, in which contents normally restricted to the lumen may translocate into the lamina propria and circulatory system to promote systemic inflammation (45,46).
Actin cytoskeleton remodeling is also crucial for the maturation of nascent phagosomes, which involves lysosome fusion and the digestion of engulfed materials in phagosomes. In this study, GO biological processes, such as “autophagy (GO:0006914)” and “autophagosome maturation (GO:0097352)”, GO cellular components, such as “vacuole (GO:0005773)”, and the Reactome Pathways gene set “Vesicle-mediated transport (R-MMU-5653656)” were clustered. The cellular processes “regulation of cellular catabolic process (GO:0031329)”, “cellular protein catabolic process (GO:0044257)”, “regulation of cellular protein catabolic process (GO:1903362)”, and “protein quality control for misfolded or incompletely synthesized proteins (GO:0006515)” were also clustered. These findings suggest that methamphetamine might participate in the regulation of autophagy and protein catabolic processes. However further experiments are needed for verification.
Methamphetamine use impairs mesenteric vasodilator function
Methamphetamine can have adverse and potentially fatal effects on arteries and blood vessels; these include elevated blood pressure, acute vasospasm, and atherosclerotic cardiovascular disease (4-6). Methamphetamine may lead to cardiovascular complications through catecholamine toxicity, due to its high-affinity interactions with binding sites on the dopamine, norepinephrine, and serotonin receptors (47). Several clinical and postmortem studies have reported an association between amphetamine use and cardiovascular disease (4-6,48,49), and several clinical case reports (9,10,26,50,51) have reported that methamphetamine use may be a cause of nonobstructive intestinal ischemia. One study reported that methamphetamine use may be associated with significant microvascular compromise, which increases the risk of mesenteric ischemia (50); however, the mechanisms of these pathological responses in the intestinal tissue of methamphetamine users are still largely unknown (4). In this study, enrichment analysis revealed that the DEGs were predominantly enriched in pathways associated with the hormone and cardiovascular systems, including adrenergic signaling in cardiomyocytes (mmu04261), renin secretion (mmu04924), cardiac muscle contraction (mmu04260), aldosterone synthesis and secretion (mmu04925), and vascular smooth muscle contraction (mmu04270). Furthermore, the “calcineurin activates nuclear factor of activated T cells (NFAT)” pathway was clustered, which plays a crucial role in cardiovascular and skeletal muscle development in vertebrates (52). To some extent, these findings verify the previous conjecture that methamphetamine causes intestinal vascular vasospasm, vasoconstriction, ischemia, with chronic methamphetamine resulting in impaired mesenteric vasodilator function.
The other DEG-enriched pathways were associated with amphetamine addiction (mmu05031), glutamatergic synapses (mmu04724), long-term potentiation (mmu04720), dopaminergic synapses (mmu04728), nervous system diseases such as Alzheimer’s disease (mmu05010) and amyotrophic lateral sclerosis (mmu05014), type II diabetes mellitus (mmu04930), the insulin signaling pathway (mmu04910), protein processing in the endoplasmic reticulum (mmu04141), ubiquitin-mediated proteolysis (mmu04120), oocyte meiosis (mmu04114), the hedgehog signaling pathway (mmu04340), N-glycan biosynthesis (mmu00510), and inflammatory mediator regulation of transient receptor potential (TRP) channels (mmu04750). A previous study reported that amphetamine-regulated transcription in the central nervous system modulates behavioral motor function via a central corticotropin-releasing factor receptor-dependent mechanism and peripheral cholinergic pathways (53). These results suggest that methamphetamine may induce sensitivity in both the central nervous system (54,55) and the autonomic nervous system, as well as sensitivity to stimulation, rendering responses more dependent on enteric nerves.
Conclusions
In conclusion, this study has uncovered new molecules that are potentially important in the pathogenesis of a special type of methamphetamine-associated IBD. It is also the first study to subject intestinal tissue samples from methamphetamine-treated mice to RNA-seq technology. These discoveries may offer new insights for future studies on methamphetamine abuse and IBD, and emphasize the association between intestinal complications and methamphetamine abuse.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (Grant No. 3171101074, 81860100, 31860306, and 81870458), Science and Technology Department of Yunnan Province (Grant No. 2018DH006, 2018NS0086, 202001AV070010), Yunnan Province Clinical Research Center (2019ZF012), and Yunling Scholar (Grant No. YLXL20170002).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7741
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-7741
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7741). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (2018NS0083) granted by Animal Care and Use Committee of Kunming Medical University, in compliance with the National guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- SAMHSA: 2019 National Survey of Drug Use and Health (NSDUH) Releases [Internet]. Rockville; Substance Abuse and Mental Health Services Administration. September, 2020. Available online: https://www.samhsa.gov/data/release/2019-national-survey-drug-use-and-health-nsduh-releases
- SAMHSA: National Survey Of Substance Abuse Treatment Services (N-SSATS): 2018, Data On Substance Abuse Treatment Facilities [Internet]. Rockville; Substance Abuse and Mental Health Services Administration. September 16, 2019. Available online: https://www.samhsa.gov/data/report/national-survey-substance-abuse-treatment-services-n-ssats-2018-data-substance-abuse
- Lineberry TW, Bostwick JM. Methamphetamine abuse: a perfect storm of complications. Mayo Clin Proc 2006;81:77-84. [Crossref] [PubMed]
- Kevil CG, Goeders NE, Woolard MD, et al. Methamphetamine Use and Cardiovascular Disease. Arterioscler Thromb Vasc Biol 2019;39:1739-46. [Crossref] [PubMed]
- Ben-Yehuda O, Siecke N. Crystal Methamphetamine: A Drug and Cardiovascular Epidemic. JACC Heart Fail 2018;6:219-21. [Crossref] [PubMed]
- Won S, Hong RA, Shohet RV, et al. Methamphetamine-associated cardiomyopathy. Clin Cardiol 2013;36:737-42. [Crossref] [PubMed]
- Persons AL, Bradaric BD, Dodiya HB, et al. Colon dysregulation in methamphetamine self-administering HIV-1 transgenic rats. PLoS One 2018;13:e0190078. [Crossref] [PubMed]
- Prakash MD, Tangalakis K, Antonipillai J, et al. Methamphetamine: Effects on the brain, gut and immune system. Pharmacol Res 2017;120:60-7. [Crossref] [PubMed]
- Zou X, Huang H, Yang L, et al. Methamphetamine consumption and life-threatening abdominal complications: A case report. Medicine (Baltimore) 2018;97:e0647. [Crossref] [PubMed]
- Choi K, Imrie A, Lourie R, et al. Intestinal ischemia due to methamphetamine use: A case report. Int J Surg Case Rep 2019;58:11-3. [Crossref] [PubMed]
- Ning T, Gong X, Xie L, et al. Gut Microbiota Analysis in Rats with Methamphetamine-Induced Conditioned Place Preference. Front Microbiol 2017;8:1620. [Crossref] [PubMed]
- Zhao J, Shen S, Dai Y, et al. Methamphetamine Induces Intestinal Inflammatory Injury via Nod-Like Receptor 3 Protein (NLRP3) Inflammasome Overexpression In Vitro and In Vivo. Med Sci Monit 2019;25:8515-26. [Crossref] [PubMed]
- Shen S, Zhao J, Dai Y, et al. Methamphetamine-induced alterations in intestinal mucosal barrier function occur via the microRNA-181c/ TNF-alpha/tight junction axis. Toxicol Lett 2020;321:73-82. [Crossref] [PubMed]
- Jilling T, Lu J, Jackson M, et al. Intestinal epithelial apoptosis initiates gross bowel necrosis in an experimental rat model of neonatal necrotizing enterocolitis. Pediatr Res 2004;55:622-9. [Crossref] [PubMed]
- Meriwether D, Sulaiman D, Volpe C, et al. Apolipoprotein A-I mimetics mitigate intestinal inflammation in COX2-dependent inflammatory bowel disease model. J Clin Invest 2019;129:3670-85. [Crossref] [PubMed]
- Chen C, Chen H, Zhang Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020;13:1194-202. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med 1990;9:811-8. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Oughtred R, Stark C, Breitkreutz B, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 2019;47:D529-41. [Crossref] [PubMed]
- Li T, Wernersson R, Hansen R, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 2017;14:61-4. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Piñero J, Bravo À, Queralt-Rosinach N, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45:D833-9. [Crossref] [PubMed]
- Meddings JB, Gibbons I. Discrimination of site-specific alterations in gastrointestinal permeability in the rat. Gastroenterology 1998;114:83-92. [Crossref] [PubMed]
- Robertson SD, Matthies H, Galli A. A closer look at amphetamine-induced reverse transport and trafficking of the dopamine and norepinephrine transporters. Mol Neurobiol 2009;39:73-80. [Crossref] [PubMed]
- Prendergast C, Hassanein AH, Bansal V, et al. Shock with intestinal ischemia: a rare complication of methamphetamine use. Am Surg 2014;80:101-2. [Crossref] [PubMed]
- Chomchai C, Chomchai S. Global patterns of methamphetamine use. Curr Opin Psychiatry 2015;28:269-74. [Crossref] [PubMed]
- Najera JA, Bustamante EA, Bortell N, et al. Methamphetamine abuse affects gene expression in brain-derived microglia of SIV-infected macaques to enhance inflammation and promote virus targets. BMC Immunol 2016;17:7. [Crossref] [PubMed]
- Xue L, Geng Y, Li M, et al. Inhibitory effects of methamphetamine on mast cell activation and cytokine/chemokine production stimulated by lipopolysaccharide in C57BL/6J mice. Exp Ther Med 2018;15:3544-50. [Crossref] [PubMed]
- Mravčík V, Florián Z, Nečas V, et al. Infectious and other somatic comorbidity in problem drug users - results of a cross-sectional study with medical examination. Epidemiol Mikrobiol Imunol 2016;65:56-62. [PubMed]
- Wawrzyniak M, Scharl M. Genetics and epigenetics of inflammatory bowel disease. Swiss Med Wkly 2018;148:w14671. [PubMed]
- Thiruvengadam SS, O'Malley M, LaGuardia L, et al. Gene Expression Changes Accompanying the Duodenal Adenoma-Carcinoma Sequence in Familial Adenomatous Polyposis. Clin Transl Gastroenterol 2019;10:e00053. [Crossref] [PubMed]
- Frenkel S, Bernstein CN, Sargent M, et al. Genome-wide analysis identifies rare copy number variations associated with inflammatory bowel disease. PLoS One 2019;14:e0217846. [Crossref] [PubMed]
- de Lange KM, Moutsianas L, Lee JC, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256-61. [Crossref] [PubMed]
- Coenen MJH. NUDT15 genotyping in Caucasian patients can help to optimise thiopurine treatment in patients with inflammatory bowel disease. Transl Gastroenterol Hepatol 2019;4:81. [Crossref] [PubMed]
- Wang F, Graham WV, Wang Y, et al. Interferon-γ and Tumor Necrosis Factor-α Synergize to Induce Intestinal Epithelial Barrier Dysfunction by Up-Regulating Myosin Light Chain Kinase Expression. Am J Pathol 2005;166:409-19. [Crossref] [PubMed]
- Schnoor M, Garcia Ponce A, Vadillo E, et al. Actin dynamics in the regulation of endothelial barrier functions and neutrophil recruitment during endotoxemia and sepsis. Cell Mol Life Sci 2017;74:1985-97. [Crossref] [PubMed]
- Shen L, Black ED, Witkowski ED, et al. Myosin light chain phosphorylation regulates barrier function by remodeling tight junction structure. J Cell Sci 2006;119:2095-106. [Crossref] [PubMed]
- Anderson JM, Van Itallie CM. Physiology and function of the tight junction. Cold Spring Harb Perspect Biol 2009;1:a002584. [Crossref] [PubMed]
- Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 2009;9:799-809. [Crossref] [PubMed]
- Dörfel MJ, Huber O. Modulation of tight junction structure and function by kinases and phosphatases targeting occludin. J Biomed Biotechnol 2012;2012:807356. [Crossref] [PubMed]
- Lu Z, Ding L, Lu Q, et al. Claudins in intestines: Distribution and functional significance in health and diseases. Tissue Barriers 2013;1:e24978. [Crossref] [PubMed]
- Reinhold AK, Rittner HL. Barrier function in the peripheral and central nervous system-a review. Pflugers Arch 2017;469:123-34. [Crossref] [PubMed]
- Cornu R, Chrétien C, Pellequer Y, et al. Small silica nanoparticles transiently modulate the intestinal permeability by actin cytoskeleton disruption in both Caco-2 and Caco-2/HT29-MTX models. Arch Toxicol 2020;94:1191-202. [Crossref] [PubMed]
- Estes JD, Harris L, Klatt N, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathogens 2010;6:e1001052. [Crossref] [PubMed]
- Wesemann DR, Nagler CR. The Microbiome, Timing, and Barrier Function in the Context of Allergic Disease. Immunity 2016;44:728-38. [Crossref] [PubMed]
- Han DD, Gu H. Comparison of the monoamine transporters from human and mouse in their sensitivities to psychostimulant drugs. BMC Pharmacol 2006;6:6. [Crossref] [PubMed]
- McKetin R, Lubman DI, Baker AL, et al. Dose-related psychotic symptoms in chronic methamphetamine users: evidence from a prospective longitudinal study. JAMA Psychiatry 2013;70:319-24. [Crossref] [PubMed]
- Darke S, Duflou J, Kaye S. Prevalence and nature of cardiovascular disease in methamphetamine-related death: A national study. Drug Alcohol Depend 2017;179:174-9. [Crossref] [PubMed]
- Anderson JE, Brown IE, Olson KA, et al. Nonocclusive mesenteric ischemia in patients with methamphetamine use. J Trauma Acute Care Surg 2018;84:885-92. [Crossref] [PubMed]
- Carlson TL, Plackett TP, Gagliano RA Jr, et al. Methamphetamine-induced paralytic ileus. Hawaii J Med Public Health 2012;71:44-5. [PubMed]
- Schulz RA, Yutzey KE. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev Biol 2004;266:1-16. [Crossref] [PubMed]
- Tebbe JJ, Ortmann E, Schumacher K, et al. Cocaine- and amphetamine-regulated transcript stimulates colonic motility via central CRF receptor activation and peripheral cholinergic pathways in fed, conscious rats. Neurogastroenterol Motil 2004;16:489-96. [Crossref] [PubMed]
- Lecomte T, Dumais A, Dugre JR, et al. The prevalence of substance-induced psychotic disorder in methamphetamine misusers: A meta-analysis. Psychiatry Res 2018;268:189-92. [Crossref] [PubMed]
- Fleckenstein AE, Volz TJ, Riddle EL, et al. New insights into the mechanism of action of amphetamines. Annu Rev Pharmacol Toxicol 2007;47:681-98. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


