
Mortality prediction for patients with acute respiratory distress syndrome based on machine learning: a population-based study
Introduction
Acute respiratory distress syndrome (ARDS) is a non-hydrostatic pulmonary oedema and hypoxemia associated with a variety of aetiologies. The morbidity, mortality (about 40%), and financial cost of ARDS are high (1,2). Predicting the outcome of patients with ARDS remains challenging (3), and no scoring system has been validated till date (4). Although using the PaO2/FiO2 ratio is the most common method of describing the severity of pulmonary dysfunction, it does not provide an accurate assessment of ARDS severity and outcome (5). Other scoring systems, such as Acute Physiology and Chronic Health Evaluation (APACHE-II), Oxygenation Saturation Index (OSI), Oxygenation Index (OI), and the Lung Injury Score can be used to predict the survival of patients in the intensive care units (ICUs). However, controversies exist since these scores offer limited prognostic information and poor predicting power (3,6,7). A more comprehensive and powerful prediction system is urgently needed.
Recently, machine learning (ML) (8) has been widely used in detecting adverse health events in hospital settings. Studies (9-12) have applied ML to predict the mortality of patients in ICU settings and have shown improved predictive ability. However, few studies have evaluated the role of ML in predicting mortality in ARDS. Ding et al. (13) and Zhang (14) developed a random forest (RF) model and a neural network model, respectively; however, their power was compromised due to either small sample size or lack of external validation.
This study aimed to develop an effective and robust mortality prediction ML model specific to ARDS. Our hypotheses included the following: firstly, the ML-based model can be used in mortality prediction of ARDS with better performance than the existing scoring systems; secondly, the ML-based model can be robust in a multi-source dataset. We present the following article in accordance with the STROBE reporting checklist (15) (available at http://dx.doi.org/10.21037/atm-20-6624).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data source
We extracted patient data from the Medical Information Mart for Intensive Care database (MIMIC-III) (16) and Telehealth Intensive Care Unit Collaborative Research Database (eICU-CRD) (17). MIMIC-III is a publicly available database that includes 53,423 distinct hospital admissions for adult patients (aged ≥18 years) in critical care units at a tertiary non-profit medical centre from 2001 to 2012. eICU-CRD is a multi-source ICU database with high granularity data for over 200,000 admissions to ICUs monitored by eICU programmes across the United States. The release of eICU-CRD is intended to build upon the success of MIMIC-III and potentially expand the scope of studies by ensuring data availability from multiple centres.
We used the same inclusion and exclusion criteria as the MIMIC-III and eICU-CRD databases. The inclusion diagram and study design are presented in Figure 1. Patients ≥18 years old with ARDS according to the International Classification of Diseases (ICD)-9 code (518.82 or 518.5) were selected. The diagnostic criteria for ARDS were as described in previous studies (18) with adjustment according to the Berlin definition (19). For patients receiving high flow oxygen or non-invasive ventilation, the PaO2/FiO2 values were not always available, and the values were calculated as the ratio of PaO2 to the nearest FiO2 available before the corresponding blood gas measurement. The bilateral infiltrates were confirmed by text matching with the keywords ‘edema’ OR (‘bilateral’ AND ‘infiltrate’) running against all radiographic reports (20,21). Patients with congestive heart failure were excluded based on the ICD-9 code 428 (instead of pulmonary wedge pressure), since the information of pulmonary wedge pressure was severely missing in these patients (22,23). A random sample of 25% of patients labelled to have ARDS was manually reviewed, and the extraction and pre-processing proposal were evaluated and tilted accordingly. Discrepancies were settled by a joint evaluation of the overall data available, but blinded to the exposure variables. Patients with the following criteria were excluded: ICU stay less than 24 h, on non-invasive ventilation, and post-surgical patients in the cardiovascular ICU.
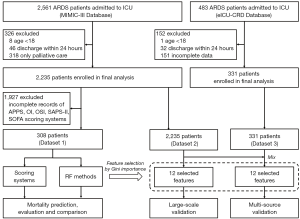
To compare the performance of our proposed ML methods with the existing scoring systems, from MIMIC-III, we selected 308 patients who met the above-mentioned criteria and had complete records of all the scores (see below), and named it Dataset 1. The scores calculated included the Simplified Acute Physiology Score II (SAPS-II), Sepsis-related Organ Failure Assessment (SOFA), OSI, OI, and APPS (3). A total of 2,235 patients in MIMIC-III (Dataset 2) and 331 patients in eICU-CRD (Dataset 3) who met the above-mentioned inclusion criteria were selected as the study population. Note that Dataset 1 is a subset of Dataset 2.
For the missing variables in some patients, we used the mean values (for the variables with normal distribution) or median values (for the variables that were not normally distributed) of non-missing data of the corresponding variables as alternatives in each dataset. The detailed frequency of the present/missing data is provided in the Appendix Section 1.
ML model development and feature selection with Dataset 1
An RF model was built in Dataset 1 using the scikit-learn library (24) in Python. Studies have shown that the RF method in most classification tasks is better than other classifiers (8,25). The outcome measures included in-hospital mortality, 30-day mortality, and 1-year mortality. As shown in Figure S1, the training and testing was based on an 8-fold cross-validation, which means that the dataset was divided into 8 folds, and in each run, 7 were used for training and the remaining 1 was used for testing. During the training procedure, we also used a nested 8-fold cross-validation grid-search scheme to find the optimal hyperparameters. The details of the hyperparameter tuning are provided in the Appendix Section 2.
For feature selection, a total of 90 variables were collected in Dataset 1, including demographic data, ventilator settings, laboratory hemodynamic variables, physiological information, and other clinical data that may be relevant to the mortality of patients with ARDS. To investigate the contribution of different variables, an importance measure, the Gini importance, was used (26), which was computed as the total reduction of the criterion brought by that feature. We selected the top 45 (50%) features with high importance. Among these, only 12 features that could also be obtained in Dataset 3 from eICU-CRD were used to establish the ML models. To assess the impact on the results, the predictive performances of RF models with 45 variables and 12 variables were evaluated and compared in Dataset 1 using 8-fold cross-validation. The details of the feature selection are provided in the Appendix Section 3.
Performance comparisons between the ML method and existing scoring systems with Dataset 1
We calculated the SAPS-II, SOFA, OSI, OI, and APPS scores for Dataset 1. OI and OSI were calculated for each patient using the following formulae:
We then tested the prediction performance using logistic regression. OSI, OI, APPS, SAPS-II, and SOFA scores were individually used as the inputs for the regression model (5-7,27).
The classification models’ performance was evaluated using the area under the receiver operating characteristic curve (AUROC). The accuracy, sensitivity, and specificity were calculated from the receiver operating characteristics (ROC) curve with the threshold with the highest accuracy. In addition, we performed calibration using the Hosmer-Lemeshow test (28) to compare the predicted and observed probabilities of death.
RF model evaluation with Dataset 2 alone and with multi-source data (Dataset 2 and Dataset 3)
An RF model was built with the 12 features for predicting the mortality of patients with ARDS in Dataset 2, and the classification models’ performance was evaluated using the above-mentioned scheme.
Training and testing were further performed using the combined data of Dataset 2 and Dataset 3. The whole procedure was the same as that described in “ML model development and feature selection with Dataset 1” section, with the same 12 features. We calculated AUROC, accuracy, sensitivity, and specificity, and performed the calibration using the Hosmer-Lemeshow test for Dataset 2 and Dataset 3, respectively, to independently evaluate the performance in these two datasets. Since only in-hospital mortality was recorded in Dataset 3, the predicting performance of the RF model was only evaluated for in-hospital mortality with this combined multi-source dataset. The classification performance of the SAPS-II and APACHE scoring systems were also calculated since other scoring systems were unavailable in the eICU-CRD database (Dataset 3).
Statistical analysis
Statistical analyses were performed using R (http://www.R-project.org) and EmpowerStats software (X&Y Solutions, Inc., Boston MA, USA). Continuous variables were presented as mean ± standard deviation, and categorical variables were presented as counts and percentages. Differences between groups were tested using the Pearson chi-square or Fisher exact test for categorical variables, and with t-test, Mann-Whitney, or Kruskal-Wallis test for numerical variables.
The AUROCs were compared using the Delong method (29). A P value <0.05 was considered statistically significant. Discrimination was also evaluated by plotting and comparing the predicted probabilities of death among survivors and non-survivors.
Results (Figures 2-6)

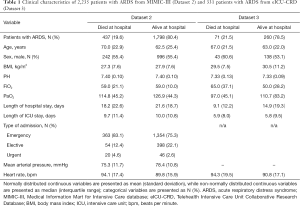
Patients’ characteristics
Demographics and clinical data of Dataset 2 and Dataset 3 are shown in Table 1. The in-hospital mortality rate was 19.6% in Dataset 2 and 21.5% in Dataset 3.
Full table
Feature selection
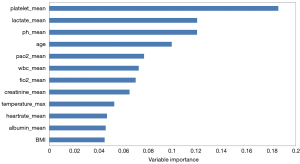
The AUROCs of RF models with 45 variables were 0.914, 0.909, and 0.910 for in-hospital, 30-day, and 1-year mortality prediction, respectively in Dataset 1 by using 8-fold cross-validation, and the AUROCs of the 12-variable based model were 0.891, 0.883, and 0.892, respectively, showing no significant difference. The 12 features finally used included age, white blood cell (WBC) count, serum creatinine, albumin, platelet count, pH, lactate level, FiO2, PaO2, heart rate, temperature, and body mass index (BMI). For the laboratory results and vital signs, mean values during the first 24 h of hospitalisation were calculated and used as the selected features, except temperature for which the maximum values were used.
Figure 2 shows the top 12 important features in the RF-based in-hospital mortality prediction model with Dataset 1, in which the platelet count and lactate level are the two strongest predictors.
Performance comparisons between the existing scoring systems and RF model with Dataset 1
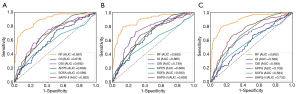
As shown in Table 2 and Figure 3, with Dataset 1, the AUROC for in-hospital mortality prediction was 0.891, which was significantly higher than the predictive values of the SAPS-II, SOFA, APPS, OSI, and OI scores (AUROC, 0.586–0.694; all P<0.001). The AUROC for the 30-day mortality prediction was 0.883, which was also significantly higher than the predictive values of these existing scores (AUROC 0.583–0.739; all P<0.001). The AUROC for the 1-year mortality prediction was 0.892, which was also significantly superior to the predictive values of these existing scores (AUROC 0.569–0.732; all P<0.001).
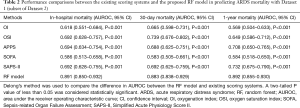
Full table
The calibration results are provided in Table S1 and Figure 4A. These statistics and plots suggest good fits for these scoring systems (all P>0.05), of which our RF model achieved better performance in predicting 30-day mortality (P=0.915) and 1-year mortality (P=0.823).
Discrimination plots of the RF-based mortality prediction model with Dataset 1 are shown in Figure S2, indicating the significant differences between the predicted probabilities of death among the survivors and non-survivors using each prediction algorithm.
Prediction performance of the RF model with Dataset 2 alone and with multi-source data (Dataset 2 and Dataset 3)
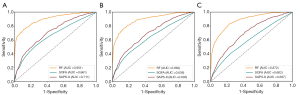
As shown in Table 3 and Figure 5 with Dataset 2, the AUROC was 0.901 for the in-hospital mortality prediction, 0.882 for the 30-day mortality prediction, and 0.872 for the 1-year mortality prediction.

Full table
The calibration results are provided in Table S2 and Figure 4B. These statistics and plots suggest good fits for these scoring systems and our RF model (all P>0.05).
Discrimination plots of the RF-based mortality prediction model with Dataset 2 are shown in Figure S3, indicating the significant differences between the predicted probabilities of death among the survivors and non-survivors using each prediction algorithm.
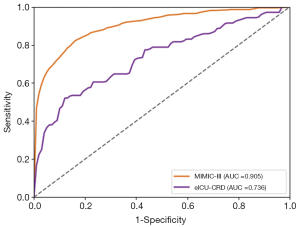
As shown in Table S3 and Figure 6, when refitting the RF model in the multi-source data consisting of Dataset 2 and Dataset 3, the AUROC for the in-hospital mortality prediction was 0.905 and 0.736 for Dataset 2 and Dataset 3, respectively. With SAPS-II and APACHE scoring systems, the AUROC for in-hospital mortality prediction with Dataset 3 was 0.511 and 0.528, respectively. We also studied the predictive value of our RF model with only Dataset 3 for training and testing, and the AUROC was 0.696 [95% confidence interval (CI), 0.621–0.770], lower than the performance trained with multi-source data consisting of Dataset 2 and Dataset 3.
The calibration results are provided in Table S3 and Figure 4C. These statistics and plots suggest good fits for these scoring systems (all P>0.05), except for our RF model for the in-hospital mortality prediction of MIMIC-III data (P=0.021). The predictive value of our RF model for MIMIC-III data has been justified in our previous results.
Discrimination plots of the RF-based mortality prediction model with multi-source data are shown in Figure S4, indicating the significant differences between the predicted probabilities of death among the survivors and non-survivors using the RF algorithm in different training sets.
Discussion
We found that ML improved the ARDS mortality prediction performance compared to the OI, OSI, SAPS-II, SOFA, and APPS scoring systems, and showed relatively stable performance with multi-source data.
The existing scoring systems combined with logistic regression have been used for mortality prediction in ARDS (3,6,7). Similar to the previous studies, our study achieved an AUROC of 0.618, 0.692, 0.692, 0.586, and 0.694 (Figure 3A), respectively, for in-hospital mortality, which was significantly poorer than the performance of the RF-based model in our study. Since the reasons for the death of patients with ARDS are complicated, it is difficult for these scoring systems to predict mortality accurately since they are generally a linear combination of explanatory variables. However, the generalizability of these scoring systems in different ARDS cohorts may be limited. For example, while the APPS score achieved an AUROC of 0.800 in predicting the in-hospital mortality (7), we achieved an AUROC of only 0.694. In another external validation study (3) using APPS, the AUROC was 0.62. This may be because APPS is a simple scoring system that incorporates limited information regarding the age, PaO2/FiO2 ratio, and plateau pressure.
Studies have demonstrated that ML is effective in predicting ICU mortality. Pirracchio et al. (10) adopted the Super ICU Learner Algorithm in mortality prediction for patients in the ICU achieving an AUROC of 0.88, which was better than previous scoring systems. Marafino et al. (30) used ML to predict in-hospital ICU mortality and yielded excellent predictive performance. Zhang et al. (11) applied a gradient boosting machine, and achieved an AUROC of 0.748. Ding et al. (13) built an RF model to predict ARDS events in a small sample of 296 patients, and achieved an AUROC of 0.82. Zhang (14) developed a neural network model with a genetic algorithm to predict 90-day mortality in a database of 745 patients, and in the testing cohort of 272 patients achieved an AUROC of 0.821. These two studies on ARDS indicated that ML was superior to the traditional APACHE score; however, the robustness requires further validation owing to the relatively small sample size or lack of multi-source validation. Similarly, our results demonstrated the ability of ML to predict the mortality of patients with ARDS, achieving comparable or even better performance than Ding et al. (13) and Zhang (14) in a larger and multi-source dataset. Compared with existing scoring systems, the RF-based algorithm has the following advantages. Firstly, the RF algorithm can automatically learn the interaction and non-linear effects between the predictors from the data, and thus may be more suitable for high-dimensional data. Secondly, with cross-validation, the optimal model could be selected. Lastly, the RF algorithm may be more powerful in dealing with the unbalanced datasets by applying an unbiased estimation for the generalisation error (31).
In the feature selection, 45 variables (50%, 90 in total) with higher Gini importance were selected based on Dataset 1 from MIMIC-III, but only 12 variables were available in Dataset 3 from eICU-CRD. The predictive performances of RF models with 45 variables and 12 variables were compared in Dataset 1. The AUROCs of the 45-variable based model were 0.914, 0.909, and 0.910 for in-hospital, 30-day, and 1-year mortality prediction, while the 12-variable based model achieved AUROCs of 0.891, 0.883, and 0.892, respectively, showing no significant difference and limiting impact on the results.
The 12 variables with the highest importance in predicting the in-hospital mortality are shown in Figure 2. Such variables have also been reported in previous studies (32-34). Advanced age, PaO2, FiO2, and creatinine are well-recognised independent risk factors for mortality in ARDS (35-37). Similarly, WBC count, temperature, and heart rate on the first day were screened as predictors of ARDS events by Ding et al. (13), while age, albumin, and FiO2 were identified as important variables associated with the 90-day mortality by Zhang (14). Acute kidney injury is associated with a high mortality, especially during ARDS development (37,38). Fever above 38.5–39.5 °C increases both the ventilatory (high respiratory drive: large tidal volume, high respiratory rate) and metabolic (increased O2 consumption) demands, further impairing the cardio-ventilatory reserve (39). Some studies have suggested that control of elevated body temperature resulting in normothermia (35.5–37 °C) could lower both the ventilatory and metabolic demands (40). However, no studies have been reported supporting the role of temperature control in preventing ARDS and improving survival. Low pH and high lactate levels are characteristic of metabolic acidosis, which is known to be predictive of mortality in extreme acidosis or critically ill patients (37,41,42). Hypoalbuminemia, a factor known to be predictive of poor prognosis (43), disrupts the oncotic balance between fluids in the pulmonary circulation and lung alveoli. In patients who are seriously ill, hypoalbuminemia may be a marker of leaky capillaries rather than a cause of hydrostatic oedema (18).
In our study, we tested and reported a very similar prediction performance of our model in two datasets from MIMIC-III with different sizes (Dataset 1 and Dataset 2) (Tables 2,3). This may indicate that sample size has a limited impact on the prediction performance of our ML-based method. We combined the eICU-CRD dataset (Dataset 3) and Dataset 2, and the results showed that the in-hospital mortality prediction performance in Dataset 3 decreased (AUROC, 0.736). There may be two explanations for this relatively lower performance in Dataset 3. Firstly, the feature selection was based on the feature importance with the MIMIC-III dataset. The distributions of these selected features in the eICU-CRD dataset can be different from those of the MIMIC-III dataset, and thus may affect the final prediction performance. Secondly, the data in MIMIC-III are from a single centre (Beth Israel Deaconess Medical Center), while the data in eICU-CRD are from multiple centres. This may cause a certain level of heterogeneity in Dataset 3 and thus lower the performance. In addition, we showed that the predictive value of the model trained with multi-source data was better than that with Dataset 3 only.
The model proposed in our study represents a momentous step towards building tools for the habitual identification of patients with ARDS who are at greatest risk of hospital death. The findings of several mortality-associated parameters in our study are unexpected but not new, possibly providing bases for future therapeutic innovation.
Our study has some limitations. Firstly, this was a retrospective study and some valuable features may have been missing. In MIMIC-III and eICU-CRD, some variables were not directly available, and the lack of such features may limit the selection of clinical variables for modelling. Although we have filled in the missing data with the median or mean values, these values were not the real values. Secondly, the performance of our model in a multi-source dataset is not as good as that in a single-source dataset. To validate our model, by incorporating additional predictive variables, a larger external cohort is warranted. Thirdly, the mortality in our patients was relatively low compared to that in other studies, varying from 4.6% to 52% (44-46). This could be because the patients with non-invasive ventilation or in the surgical ICU recruited in our study had a relatively better prognosis.
Conclusions
Using the MIMIC-III database, we successfully established an RF-based ML model for predicting the mortality of patients with ARDS. The RF model achieved significantly improved performance compared to the traditional scoring systems. Based on the multi-source dataset obtained from both the MIMIC-III and the eICU-CRD database, the generalisation ability of this prediction model was verified.
Acknowledgments
Funding: This work was supported by Seed Funding from Guangzhou Science and Technology Planning Project (No. 201903010073), Natural Science Foundation of Guangdong Province (No. 2020A1515010571), Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions (No. 2019SHIBS0003), Guangdong Key Basic Research Grant (No. 2018B030332001), Guangdong Pearl River Talents Plan (No. 2016ZT06S220), Shenzhen Science and Technology Project (JCYJ20200109114014533), SZU Top Ranking Project - Shenzhen University (860/000002100108) and Tencent “Rhinoceros Birds” Scientific Research Foundation for Young Teachers of Shenzhen University.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6624
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6624). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bernard GR, Artigas A, Brigham KL, et al. The American-European Consensus Conference on ARDS. Definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818-24. [Crossref] [PubMed]
- Association's TAL. Lung Disease Data: 2008. 2008. Available online: http://action.lung.org/site/DocServer/lung-disease-data-2008-report.pdf
- Bos LD, Schouten LR, Cremer OL, et al. External validation of the APPS, a new and simple outcome prediction score in patients with the acute respiratory distress syndrome. Ann Intensive Care 2016;6:89. [Crossref] [PubMed]
- Santos RS, Silva PL, Rocco JR, et al. A mortality score for acute respiratory distress syndrome: predicting the future without a crystal ball. J Thorac Dis 2016;8:1872-6. [Crossref] [PubMed]
- Villar J, Blanco J, del Campo R, et al. Assessment of PaO(2)/FiO(2) for stratification of patients with moderate and severe acute respiratory distress syndrome. BMJ Open 2015;5:e006812 [Crossref] [PubMed]
- DesPrez K, McNeil JB, Wang C, et al. Oxygenation Saturation Index Predicts Clinical Outcomes in ARDS. Chest 2017;152:1151-8. [Crossref] [PubMed]
- Villar J, Ambros A, Soler JA, et al. Age, PaO2/FIO2, and Plateau Pressure Score: A Proposal for a Simple Outcome Score in Patients With the Acute Respiratory Distress Syndrome. Crit Care Med 2016;44:1361-9. [Crossref] [PubMed]
- Fernández-Delgado M, Cernadas E, Barro S, et al. Do we need hundreds of classifiers to solve real world classification problems? J Mach Learn Res 2014;15:3133-81.
- Desautels T, Calvert J, Hoffman J, et al. Prediction of Sepsis in the Intensive Care Unit With Minimal Electronic Health Record Data: A Machine Learning Approach. JMIR Med Inform 2016;4:e28 [Crossref] [PubMed]
- Pirracchio R, Petersen ML, Carone M, et al. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): a population-based study. Lancet Respir Med 2015;3:42-52. [Crossref] [PubMed]
- Zhang Z, Zheng B, Liu N, et al. Mechanical power normalized to predicted body weight as a predictor of mortality in patients with acute respiratory distress syndrome. Intensive Care Med 2019;45:856-64. [Crossref] [PubMed]
- Nemati S, Holder A, Razmi F, et al. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Crit Care Med 2018;46:547-53. [Crossref] [PubMed]
- Ding XF, Li JB, Liang HY, et al. Predictive model for acute respiratory distress syndrome events in ICU patients in China using machine learning algorithms: a secondary analysis of a cohort study. J Transl Med 2019;17:326. [Crossref] [PubMed]
- Zhang Z. Prediction model for patients with acute respiratory distress syndrome: use of a genetic algorithm to develop a neural network model. PeerJ 2019;7:e7719 [Crossref] [PubMed]
- von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet 2007;370:1453-7. [Crossref] [PubMed]
- Johnson AEW, Pollard TJ, Lu S, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016;3:160035 [Crossref] [PubMed]
- Pollard TJ, Johnson AEW, Raffa JD, et al. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data 2018;5:180178 [Crossref] [PubMed]
- Jia X, Malhotra A, Saeed M, et al. Risk factors for ARDS in patients receiving mechanical ventilation for > 48 h. Chest 2008;133:853-61. [Crossref] [PubMed]
- Ferguson ND, Fan E, Camporota L, et al. The Berlin definition of ARDS: an expanded rationale, justification, and supplementary material. Intensive Care Med 2012;38:1573-82. [Crossref] [PubMed]
- Herasevich V, Yilmaz M, Khan H, et al. Validation of an electronic surveillance system for acute lung injury. Intensive Care Med 2009;35:1018-23. [Crossref] [PubMed]
- McKown AC, Brown RM, Ware LB, et al. External Validity of Electronic Sniffers for Automated Recognition of Acute Respiratory Distress Syndrome. J Intensive Care Med 2019;34:946-54. [Crossref] [PubMed]
- Fuchs L, Feng M, Novack V, et al. The Effect of ARDS on Survival: Do Patients Die From ARDS or With ARDS? J Intensive Care Med 2019;34:374-82. [Crossref] [PubMed]
- Jentzer JC, van Diepen S, Barsness GW, et al. Cardiogenic Shock Classification to Predict Mortality in the Cardiac Intensive Care Unit. J Am Coll Cardiol 2019;74:2117-28. [Crossref] [PubMed]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine learning in Python. J Mach Learn Res 2011;12:2825-30.
- Churpek MM, Yuen TC, Winslow C, et al. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med 2016;44:368. [Crossref] [PubMed]
- Kuhn M, Johnson K. Applied Predictive Modeling. New York, NY: Springer, 2013.
- Heffner JE, Brown LK, Barbieri CA, et al. Prospective validation of an acute respiratory distress syndrome predictive score. Am J Respir Crit Care Med 1995;152:1518-26. [Crossref] [PubMed]
- Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med 2007;35:2052-6. [Crossref] [PubMed]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. [Crossref] [PubMed]
- Marafino BJ, Park M, Davies JM, et al. Validation of Prediction Models for Critical Care Outcomes Using Natural Language Processing of Electronic Health Record Data. JAMA Netw Open 2018;1:e185097 [Crossref] [PubMed]
- Khoshgoftaar TM, Golawala M, Van Hulse J, et al. An empirical study of learning from imbalanced data using random forest. 19th IEEE International Conference on Tools with Artificial Intelligence (ICTAI 2007); 29-31 Oct. 2007; Patras, Greece. IEEE, 2007:310-7.
- de Prost N, Pham T, Carteaux G, et al. Etiologies, diagnostic work-up and outcomes of acute respiratory distress syndrome with no common risk factor: a prospective multicenter study. Ann Intensive Care 2017;7:69. [Crossref] [PubMed]
- Tignanelli CJ, Hemmila MR, Rogers MAM, et al. Nationwide cohort study of independent risk factors for acute respiratory distress syndrome after trauma. Trauma Surg Acute Care Open 2019;4:e000249 [Crossref] [PubMed]
- Dai Q, Wang S, Liu R, et al. Risk factors for outcomes of acute respiratory distress syndrome patients: a retrospective study. J Thorac Dis 2019;11:673-85. [Crossref] [PubMed]
- Luo L, Shaver CM, Zhao Z, et al. Clinical Predictors of Hospital Mortality Differ Between Direct and Indirect ARDS. Chest 2017;151:755-63. [Crossref] [PubMed]
- Seeley E, McAuley DF, Eisner M, et al. Predictors of mortality in acute lung injury during the era of lung protective ventilation. Thorax 2008;63:994-8. [Crossref] [PubMed]
- Panitchote A, Mehkri O, Hastings A, et al. Factors associated with acute kidney injury in acute respiratory distress syndrome. Ann Intensive Care 2019;9:74. Erratum in: Ann Intensive Care 2019 Jul 23;9(1):84. doi: 10.1186/s13613-019-0558-z. [Crossref] [PubMed]
- De Jong A, Verzilli D, Jaber S. ARDS in Obese Patients: Specificities and Management. Crit Care 2019;23:74. [Crossref] [PubMed]
- Petitjeans F, Leroy S, Pichot C, et al. Hypothesis: Fever control, a niche for alpha-2 agonists in the setting of septic shock and severe acute respiratory distress syndrome? Temperature (Austin) 2018;5:224-56. [Crossref] [PubMed]
- Manthous CA, Hall JB, Olson D, et al. Effect of cooling on oxygen consumption in febrile critically ill patients. Am J Respir Crit Care Med 1995;151:10-4. [Crossref] [PubMed]
- Allyn J, Vandroux D, Jabot J, et al. Prognosis of patients presenting extreme acidosis (pH <7) on admission to intensive care unit. J Crit Care 2016;31:243-8. [Crossref] [PubMed]
- Gunnerson KJ, Saul M, He S, et al. Lactate versus non-lactate metabolic acidosis: a retrospective outcome evaluation of critically ill patients. Crit Care 2006;10:R22. [Crossref] [PubMed]
- Jellinge ME, Henriksen DP, Hallas P, et al. Hypoalbuminemia is a strong predictor of 30-day all-cause mortality in acutely admitted medical patients: a prospective, observational, cohort study. PLoS One 2014;9:e105983 [Crossref] [PubMed]
- Zeiberg D, Prahlad T, Nallamothu BK, et al. Machine learning for patient risk stratification for acute respiratory distress syndrome. PLoS One 2019;14:e0214465 [Crossref] [PubMed]
- Esteban A, Anzueto A, Frutos F, et al. Characteristics and outcomes in adult patients receiving mechanical ventilation: a 28-day international study. JAMA 2002;287:345-55. [Crossref] [PubMed]
- Esteban A, Frutos-Vivar F, Muriel A, et al. Evolution of mortality over time in patients receiving mechanical ventilation. Am J Respir Crit Care Med 2013;188:220-30. [Crossref] [PubMed]


