Abstract
Background:
Congenital heart diseases (CHD) are the most common of all birth defects, affecting nearly 0.9% of all live births. Nkx2-5 mutations were reported to cause CHD but data in Kurdish populations of Iran are limited.Objectives:
In this experimental study, we performed high resolution melt (HRM) mutation scanning of Nkx2-5 exons of non-syndrome patients.Patients and Methods:
Thirty nine patients with atrial septal defect and 57 patients with ventricular septal defect, 4 patients possessing both defects as case groups and 50 healthy controls. Then we grouped samples according to HRM graph and sequenced several samples from each group.Results:
HRM analysis showed 2 deviated curves for exon 1 and one group for exon 2A and exon 2B. Then, 2 samples of exon 1 that showed different HRM curves, 3 samples of another group from this exon and 5 samples of exon 2A, 2B and healthy controls were randomly sequenced. The results of sequencing confirmed the HRM analysis, and one polymorphism (A65G) was identified in 2 atrial septal defects with deviated curves.Conclusions:
The environmental and effective factors on the heart development within embryonic evolution as well as the possibility of the existence of the mutation in coding genes of the other cardiac transcription factors such as GATA4 and TBX5 can be the reasons for the lack of the pathogenic mutation in this study. It is suggested in further related studies to investigate normal and abnormal cardiac tissue samples of these studied patients and coding genes of the other cardiac transcription factors.Keywords
High Resolution Melt Nkx2-5 Atrial Septal Defect Ventricular Septal Defect
1. Background
Congenital heart diseases (CHD) are the most common of all birth defects which contribute substantially to infant mortality, affecting nearly 0.9% of all live births [1, 2]. Recent genetics studies have shown that many of congenital heart diseases are caused by mutations mainly from a combination of one or more gene interactions with and responses to environment. Complexity in heart development reflects the expression of various genes [3, 4]. Chromosomal and monogenetic abnormalities (about 8%), multifactorial ones (9%) and environmental teratogens (2%) are the main genetic factors for CHD. Moreover, CHD is correlated with structural and numeral chromosomal abnormalities (12% - 14%), like 21, 13 and 18 trisomy, turner, etc. [3, 5-7]. Congenital heart diseases also may be caused in result of defect in different parts of heart [3]. The role of several transcription factors like Nkx2-5, which is the most important one in the progress of mesoderm to the heart tissue, in the processes of cardiac morphogenesis was confirmed. Nkx2-5 also activates synthesis of the other transcription factors such as the members of GATA and MEF2 family. Mutation in this gene causes atrial and ventricular wall defects, Fallot and tricuspid valve abnormalities [3, 8]. Nkx2-5, which is a member of Nk home box gene family, locates on chromosome 5q34 and has been preserved during evolution. This gene has 10,209 bp lengths and contains two exons which encode a protein of 324 amino acids [3, 9]. Ventricular septal defect (VSD) and atrial septal defect (ASD) are the most common types of CHD and account for 50% of all cases of CHD [10]. Monogenic etiologies for ASD and VSD have principally been discovered by studying large families with autosomal dominant forms of septal defects using traditional linkage approaches [11, 12]. The first genetic etiology for ASD was the discovery that mutations in the transcription factor, TBX5, are a reason of septation defects in the Holt-Oram syndrome, which is characterized by cardiac and upper limb malformations [13]. Mutations in the cardiac transcription factor, Nkx2-5, were recognized in families who primarily showed non-syndromic ASD and atrioventricular conduction abnormalities [14-18]. High resolution melting (HRM) is a mutation scanning technique that detects the progressive change in fluorescence caused by the release of an intercalating DNA dye from a DNA duplex as it is denatured by peripheral increases in temperature [19]. It is an in-tube method requiring the inclusion of a saturating intercalating dye in the PCR mix and the addition of a high resolution melt step after PCR. The technique has already been employed to scan for somatic mutations in the KIT, BRAF, EGFR, ERBB2, TP53 and KRAS genes [20-24]. The main purpose of this study determines the role of gene mutations in CHD and genotype-phenotype correlation in ASD and VSD them by HRM, that is a highly effective scanning technique. The cause of CHD in the most cases is unknown, but the molecular evolution and gene analysis of the heart have led to identifying some mutations linked to the CHD. Therefore, the role of Nkx2-5 gene in patients with atrial and ventricular wall defects referred to Sanandaj and Kermanshah hospitals in Kurdish population has been investigated.
2. Objectives
In this experimental study, we performed high resolution melt (HRM) mutation scanning of Nkx2-5 exons of non-syndrome patients.
3. Patients and Methods
3.1. Study Subjects
In this experimental study, we studied Kurdish population consisted of 50 healthy with rang 10 - 34 years as control group and 100 (62 females and 38 males) non-syndrome patients, 57 patients (26 males and 31 females) with VSD, 39 patients (12 males and 27 females) with ASD and 4 patients (2 males and 2 females) possessing both defects as case groups with rang 2 - 45 years from Kermanshah Imam Ali cardiovascular hospital and Sanandaj Tohid hospital. The medical college of Kurdistan Medical Sciences University and the Molecular and cellular biology research center of Kurdistan Medical University, have approved the present study. All of patients devoted their blood voluntarily.
3.2. DNA Extraction
Genomic DNA was extracted from whole blood by AccuPrep® genomic DNA extraction kit from Bioneer corporation, Korea.
3.3. Real Time PCR and HRM Assay
The primers were designed by CLC main workbench 5.5 for the coding exons of Nkx2-5 gene (Table 1) and were synthesized by German Metabion Company. Final primer sequences are listed in Table 1. PCR cycling and HRM analysis was performed on the Rotor-Gene 6000 (Corbett research) and 2x QuantiFast SYBR® Green kit (QIAGEN company, Germany). The amplification was initialed at 95°C for 5 minutes then followed by 45 cycles of two steps consisting of 95°C for 10 seconds, and 60°C for 30 seconds, one cycle of 95°C for 1 second, 60°C for 90 seconds and a HRM step from 72 - 95°C rising at 0.1°C per second.
3.4. Sequencing
The PCR products of patient samples were sequenced by research center for gastroenterology and liver disease of Shahid Beheshti University of Medical Sciences. The results were analyzed using CLC main workbench 5.5.
3.5. Statistical Analysis
χ2 test was conducted using SPSS-16 for gender prevalence.
4. Results
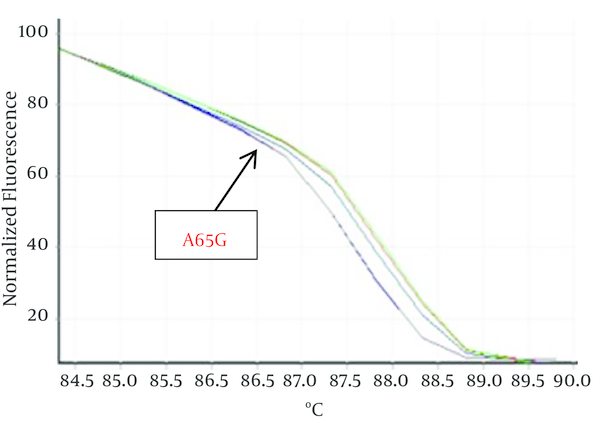
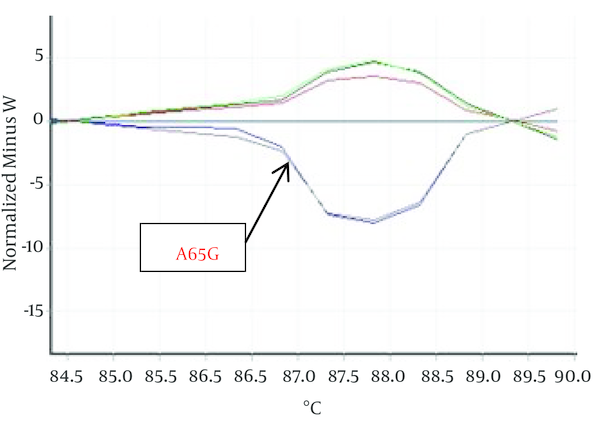
Hundred Kurdish patients with ASD and VSD were investigated in order to identify mutation in Nkx2-5 gene. Statistical results showed that VSD prevalence is not related to gender but ASD outbreak is relate to gender (P < 0.05). HRM curves of these samples were obtained by real time PCR and 2x QuantiFast SYBR® Green kit in order to perform mutation scanning using the specific Nkx2-5 gene primers. Figure 1 shows normalized plots and Figure 2 shows difference plots of each sample compared to wild-type controls for amplicons for exon 1. HRM curves analysis detected deviation in curves of 2 samples in exon 1, (Figures 1, 2) but we could not detect any deviation in exon 2 and healthy controls. This deviation could represent the amplified amplicons structural difference; therefore, to confirm the structural differences between samples, or in other words, mutations in these two samples sequencing was used. For this purpose, 2 samples of exon 1 with deviation curve, 3 samples of the other group of this exon were sequenced. Likewise, 5 samples of 2A and 2B exon and healthy controls were randomly sequenced. The sequencing results also confirmed the HRM curve analysis and one polymorphism A65G in 2 ASDs which has different melting curve was identified while any mutation was detected in 2A and 2B exons.
Show Normalized Plots
Show Difference Plots
Primers for the Amplification of Coding Exons of Nkx2-5 Genes
| Exon | Primer sequence |
|---|---|
| 1F | 5' CCGCTTTCTGCCGCCCAC 3' |
| 1R | 5' TCCTCACCTTTCTTTTCGGC 3' |
| 2AF | 5' CCCTTACCATTACTGTGC 3' |
| 2AR | 5' TAACCGTAGGGATTGAGG 3' |
| 2BF | 5' AGTGCTGGTGCGCGATGG 3' |
| 2BR | 5' TGTTGAGGTGGGATCGGTC 3' |
5. Discussion
In this work, following the results of sequencing and comparing them with the reference gene, only one polymorphism was detected, A65G located in exon 1, that was considered as common polymorphisms. Possibility of the existence of the mutation in coding genes of the other cardiac transcription factors such as GATA4 and TBX5 can be the reasons for the lack of the pathogenic mutation in this study. Since the first report implicating Nkx2-5 mutations in human CHD, the coding region of Nkx2-5 has been consistently searched for additional disease-associated sequence alterations. There are many studies where germ line Nkx2-5 mutations have been analyzed or identified in CHD Nkx2-5 mutations were identified in different countries. Different Nkx2-5 germ line mutations, most of which lead to amino acid change (nonsynonymic mutations) have been reported. These mutations are single nucleotide substitutions, deletions and insertions. The Nkx2-5 mutations are spread along the gene, and except for one at a splice site, all are located in the coding region [25].
Benson and Schott et al. introduce germ line mutation as one of the pathologic factor but having observed several missense mutations in one single patient they consider it as a somatic mutation [4, 14]. Draus et al. argue that the reason for occurring several missense mutations in one patient could be the use of formalin for sample fixation and also the long period of fixation [26]. Reamon-Buettner and Borlak conduct a similar study on fresh tissue and report that germ line mutation is the cause of disease [25].
In this work, following the results of sequencing and comparing them with the reference gene, only one polymorphism was observed, A65G located in exon 1, that was considered as common polymorphisms. The result obtained in the present study, in line with Benson and Schott et al. [4, 14] confirms possibility somatic mutation as the cause of congenital heart disease in the examined patient. Also statistical results showed that VSD prevalence is not related to gender but ASD outbreak is relate to gender. ASD shows a female preponderance, with a male/female ratio of 1 to 2. In general, heart malformation related to Nkx2-5 gene mutation with autosomal dominant pattern included ASD, VSD and AV block, but similar structural disorders resulting from mutation in the other cardiac transcription factor genes such as GATA4 and TBX5 have also been reported [27, 28]. Thus, congenital heart diseases are considered as multifactorial diseases, for which genes and environment interactions determine the incidence and severity of the disease. Among the environmental risk factors contributing to the disease, chemicals, drugs, infectious agents and maternal diseases could be mentioned [29]. The result of this research indicates that the ASD and VSD heart diseases are of multifactorial nature, i.e. , the incidence of these diseases is caused by mutation in other cardiac transcription factors or by environmental factors. Considering the findings of the previous studies, causes of failure to observe mutations in the examined samples may be attributed to environmental factors as well as the factors influencing the heart genesis during fetal development. The most well-recognized nongenetic causes of CHD are environmental teratogens (dioxins, polychlorinated biphenyls, pesticides), maternal exposures (alcohol, isotretinoin, thalidomide, antiepileptic medications), and infectious agents (e.g., rubella) [30-32]. Familial CHD mutations also occur as autosomal dominant, autosomal recessive or X-linked traits that are expressed with high penetrance and with variable clinical manifestations. Congenital heart diseases are genetically heterogeneous. In other words, mutations in different genes cause an identical malformation. This finding emphasizes the highly interdependent roles of molecules involved in heart development.
Finally, and as a way of conclusion, the range of heart malformations implicates genomic context, maternal-fetal environment, cardiac biomechanics, and other factors as important influences that impact the clinical consequences of CHD mutations. So we should further conduct investigation of Nkx2-5 mutation in sample tissue of such patient and also patient suffering from other CHD then compare result of sequencing blood and tissue samples to tell for confirming that somatic mutation or germ line mutation is the major cause behind this disease.
Acknowledgements
References
-
1.
Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112(4):707-20. [PubMed ID: 23410880]. https://doi.org/10.1161/CIRCRESAHA.112.300853.
-
2.
Moodie D. Adult congenital heart disease: past, present, and future. Tex Heart Inst J. 2011;38(6):705-6. [PubMed ID: 22199442].
-
3.
Ramegowda S, Ramachandra N. An understanding the genetic basis of congenital heart disease. Indian J Hum Genet. 2005;11(1):14-23. https://doi.org/10.4103/0971-6866.16289.
-
4.
Benson DW. Genetic origins of pediatric heart disease. Pediatr Cardiol. 2010;31(3):422-9. [PubMed ID: 20033147]. https://doi.org/10.1007/s00246-009-9607-y.
-
5.
Chaoui R, Korner H, Bommer C, Goldner B, Bierlich A, Bollmann R. [Prenatal diagnosis of heart defects and associated chromosomal aberrations]. Ultraschall Med. 1999;20(5):177-84. [PubMed ID: 10595385]. https://doi.org/10.1055/s-1999-8912.
-
6.
Rauch R, Hofbeck M, Zweier C, Koch A, Zink S, Trautmann U, et al. Comprehensive genotype-phenotype analysis in 230 patients with tetralogy of Fallot. J Med Genet. 2010;47(5):321-31. [PubMed ID: 19948535]. https://doi.org/10.1136/jmg.2009.070391.
-
7.
Nakashima Y, Yanez DA, Touma M, Nakano H, Jaroszewicz A, Jordan MC, et al. Nkx2-5 suppresses the proliferation of atrial myocytes and conduction system. Circ Res. 2014;114(7):1103-13. [PubMed ID: 24563458]. https://doi.org/10.1161/CIRCRESAHA.114.303219.
-
8.
Bartlett H, Veenstra GJ, Weeks DL. Examining the cardiac NK-2 genes in early heart development. Pediatr Cardiol. 2010;31(3):335-41. [PubMed ID: 19967350]. https://doi.org/10.1007/s00246-009-9605-0.
-
9.
Banerjee-Basu S, Baxevanis AD. Molecular evolution of the homeodomain family of transcription factors. Nucleic Acids Res. 2001;29(15):3258-69. [PubMed ID: 11470884].
-
10.
Pierpont ME, Basson CT, Benson DJ, Gelb BD, Giglia TM, Goldmuntz E, et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):3015-38. [PubMed ID: 17519398]. https://doi.org/10.1161/CIRCULATIONAHA.106.183056.
-
11.
Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126(6):1037-48. [PubMed ID: 16990131]. https://doi.org/10.1016/j.cell.2006.09.003.
-
12.
Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci. 2006;63(10):1141-8. [PubMed ID: 16568242]. https://doi.org/10.1007/s00018-005-5532-2.
-
13.
Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet. 1997;15(1):30-5. [PubMed ID: 8988165]. https://doi.org/10.1038/ng0197-30.
-
14.
Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281(5373):108-11. [PubMed ID: 9651244].
-
15.
Gioli-Pereira L, Pereira AC, Mesquita SM, Xavier-Neto J, Lopes AA, Krieger JE. NKX2.5 mutations in patients with non-syndromic congenital heart disease. Int J Cardiol. 2010;138(3):261-5. [PubMed ID: 19073351]. https://doi.org/10.1016/j.ijcard.2008.08.035.
-
16.
Ouyang P, Saarel E, Bai Y, Luo C, Lv Q, Xu Y, et al. A de novo mutation in NKX2.5 associated with atrial septal defects, ventricular noncompaction, syncope and sudden death. Clin Chim Acta. 2011;412(1-2):170-5. [PubMed ID: 20932824]. https://doi.org/10.1016/j.cca.2010.09.035.
-
17.
Wang J, Xin YF, Liu XY, Liu ZM, Wang XZ, Yang YQ. A novel NKX2-5 mutation in familial ventricular septal defect. Int J Mol Med. 2011;27(3):369-75. [PubMed ID: 21165553]. https://doi.org/10.3892/ijmm.2010.585.
-
18.
Costa MW, Guo G, Wolstein O, Vale M, Castro ML, Wang L, et al. Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ Cardiovasc Genet. 2013;6(3):238-47. [PubMed ID: 23661673]. https://doi.org/10.1161/CIRCGENETICS.113.000057.
-
19.
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49(6 Pt 1):853-60. [PubMed ID: 12765979].
-
20.
Willmore-Payne C, Holden JA, Tripp S, Layfield LJ. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005;36(5):486-93. [PubMed ID: 15948115]. https://doi.org/10.1016/j.humpath.2005.03.015.
-
21.
Willmore-Payne C, Holden JA, Layfield LJ. Detection of epidermal growth factor receptor and human epidermal growth factor receptor 2 activating mutations in lung adenocarcinoma by high-resolution melting amplicon analysis: correlation with gene copy number, protein expression, and hormone receptor expression. Hum Pathol. 2006;37(6):755-63. [PubMed ID: 16733218]. https://doi.org/10.1016/j.humpath.2006.02.004.
-
22.
Nomoto K, Tsuta K, Takano T, Fukui T, Fukui T, Yokozawa K, et al. Detection of EGFR mutations in archived cytologic specimens of non-small cell lung cancer using high-resolution melting analysis. Am J Clin Pathol. 2006;126(4):608-15. [PubMed ID: 16938658]. https://doi.org/10.1309/N5PQNGW2QKMX09X7.
-
23.
Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. [PubMed ID: 17184525]. https://doi.org/10.1186/1471-2407-6-295.
-
24.
Krypuy M, Ahmed AA, Etemadmoghadam D, Hyland SJ, Australian Ovarian Cancer Study G, DeFazio A, et al. High resolution melting for mutation scanning of TP53 exons 5-8. BMC Cancer. 2007;7:168. [PubMed ID: 17764544]. https://doi.org/10.1186/1471-2407-7-168.
-
25.
Reamon-Buettner SM, Borlak J. NKX2-5: an update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD). Hum Mutat. 2010;31(11):1185-94. [PubMed ID: 20725931]. https://doi.org/10.1002/humu.21345.
-
26.
Draus JJ, Hauck MA, Goetsch M, Austin E3, Tomita-Mitchell A, Mitchell ME. Investigation of somatic NKX2-5 mutations in congenital heart disease. J Med Genet. 2009;46(2):115-22. [PubMed ID: 19181906]. https://doi.org/10.1136/jmg.2008.060277.
-
27.
Nemer M. Genetic insights into normal and abnormal heart development. Cardiovasc Pathol. 2008;17(1):48-54. [PubMed ID: 18160060]. https://doi.org/10.1016/j.carpath.2007.06.005.
-
28.
Clark KL, Yutzey KE, Benson DW. Transcription factors and congenital heart defects. Annu Rev Physiol. 2006;68:97-121. [PubMed ID: 16460268]. https://doi.org/10.1146/annurev.physiol.68.040104.113828.
-
29.
Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115(23):2995-3014. [PubMed ID: 17519397]. https://doi.org/10.1161/CIRCULATIONAHA.106.183216.
-
30.
Kopf PG, Walker MK. Overview of developmental heart defects by dioxins, PCBs, and pesticides. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2009;27(4):276-85. [PubMed ID: 19953399]. https://doi.org/10.1080/10590500903310195.
-
31.
Zhu H, Kartiko S, Finnell RH. Importance of gene-environment interactions in the etiology of selected birth defects. Clin Genet. 2009;75(5):409-23. [PubMed ID: 19459879]. https://doi.org/10.1111/j.1399-0004.2009.01174.x.
-
32.
Dewan P, Gupta P. Burden of Congenital Rubella Syndrome (CRS) in India: a systematic review. Indian Pediatr. 2012;49(5):377-99. [PubMed ID: 22700664].