Abstract
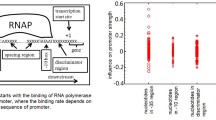
The energy of interaction between complementary nucleotides in promoter sequences of E. coli was calculated and visualized. The graphic method for presentation of energy properties of promoter sequences was elaborated on. Data obtained indicated that energy distribution through the length of promoter sequence results in picture with minima at −35, −8 and +7 regions corresponding to areas with elevated AT (adenine-thymine) content. The most important difference from the random sequences area is related to −8. Four promoter groups and their energy properties were revealed. The promoters with minimal and maximal energy of interaction between complementary nucleotides have low strengths, the strongest promoters correspond to promoter clusters characterized by intermediate energy values.
Similar content being viewed by others
References
Babb, K., McAlister, J.D., Miller, J.C., Stevenson, B., 2004. Molecular characterization of borrelia burgdorferi promoter/operator elements.Journal of Bacteriology,186(9):2745–2756.
Burr, T., Mitchell, J., Minchin, S., Busby, S., 2000. DNA sequence elements located immediately upstream of the −10 hexamer inEscherichia coli promoters: a systematic study.Nucleic. Acids. Res.,28:1864–1870.
Campbell, E.A., Muzzin, O., Chlenov, M., Sun, J.L., Olson, C.A., Weinman, O., Trester-Zedlitz, M.L., Darst, S.A., 2002. Structure of the bacterial RNA polymerase promoter specificity σ subunit.Mol. Cell,9:527–539.
Gourse, R.L., Ross, W., Gaal, T., 2000. UPs and downs in bacterial transcription initiation: role of the α subunit of RNA polymerase in promoter recognition.Mol. Microbiol.,37:687–695.
Gralla, J.D., 1996. Activation and repression ofE. coli promoters.Curr. Opin. Genet.,6(5):526–530.
Guo, Y., Gralla, J.D., 1998. Promoter opening via a DNA fork junction binding activity.Proc. Natl. Acad. Sci. USA,95(20):11655–11660.
Jones, C.H., Tatti, K.M., Moran, C.P.Jr., 1992. Effects of amino acid substitutions in the −10 binding region of sigma E fromBacillus subtilis.J. Bacteriol.,174(2):6815–6821.
Juang, Y.L., Helmann, J.D., 1994. A promoter melting region in the primary sigma factor ofBacillus subtilis. Identification of functionally important aromatic amino acids.J. Mol. Biol.,235(5):1470–1488.
Kainz, M., Roberts, J., 1992. Structure of transcription elongation complexes in vivo.Science,255:838–841.
Kajitani, M., Ishihama, A., 1983. Determination of the promoter strength in the mixed transcription system. II. Promoters of ribosomal RNA, ribosomal protein S1 and recA protein operons fromEscherichia coli.Nucleic. Acids. Research,11(12):3873–3888.
Kanehisa, M., Goto, S., Kawashima, S., Kuno, Y., Hattori, M., 2004. The KEGG resources for deciphering the genome.Nucleic. Acids. Res.,32:D277-D280.
Kudritskaya, Z.G., Danilov, V.I., 1976. Quantum mechanical study of bases interactions in various associates in atomic dipole approximation.J. Theor. Biol.,59:301–318.
Meier, T., Schickor, P., Wedel, A., Cellai, L., Heumann, H., 1995. In vitro transcription close to the melting point of DNA: analysis of Thermotoga maritima RNA polymerase-promoter complexes at 75 degree C using chemical probes.Nucleic. Acids. Research,23(6):988–994.
Mori, H., Isono, K., Horiuchi, T., Miki, T., 2000. Functional genomics ofEscherichia coli in Japan.Res. Microbiol.,151:121–128.
Murakami, K.S., Masuda, S., Campbell, E.A., Muzzin, O., Darst, S.A., 2002. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.Science,296:1285–1290.
Roberts, C.W., Roberts, J.W., 1996. Base-specific recognition of the nontemplate strand of promoter DNA byE. coli RNA polymerase.Cell,86(3):495–501.
Ross, W., Ernst, A., Gourse, R.L., 2001. Fine structure ofE. coli RNA polymerase-promoter interactions: α subunit binding to the UP element minor groove.Genes and Development,15(5):491–506.
Spassky, A., Kirkegaard, K., Buc, H., 1985. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex withEscherichia coli RNA polymerase.Biochemistry,24(11):2723–2731.
Vogel, S.K., Schulz, A., Rippe, K., 2002. Binding affinity ofEscherichia coli RNA polymerase σ54, holoenzyme for the glnAp2, nifH and nifL promoters.Nucleic. Acids. Research,30(18):4094–4101.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Berezhnoy, A.Y., Shckorbatov, Y.G. Dependence of the E. coli promoter strength and physical parameters upon the nucleotide sequence. J Zheijang Univ Sci B 6, 1063–1068 (2005). https://doi.org/10.1631/jzus.2005.B1063
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.2005.B1063