Abstract
Among oysters, species of Crassostrea (Sacco, 1897) are the most attractive to aquaculture. In Brazil, the genus is represented by C. rhizophorae (Guilding, 1828) and C. brasiliana (Lamarck, 1819). Because the maturation and breeding technology is not well developed for these species, aquaculturists need a reliable method to decide the correct time to place spat collectors in the field, and to identify both species, which are morphologically similar. In this study a specific Multiplex PCR protocol was developed, using one pair of universal primers from 18S rDNA as a positive control and a pair of specific primers for each target species. The sensitivity and specificity of the protocol was evaluated. It detected C. rhizophorae DNA in low concentrations, and C. brasiliana DNA in even lower concentrations. Further, the Multiplex PCR proved efficient in detecting DNA in concentrations equivalent to that of a single larva of each species, either separated or combined, when mixed with total DNA extract of a plankton sample representing 1000 L of filtered water. Field tests confirmed the applicability of the protocol, which holds the promise to become an important tool for aquaculture or conservation programs, allowing for the continuous monitoring of the life cycle of C. brasiliana and C. rhizophorae, by detecting the right periods of larval release and settlement.
Multiplex PCR; oysters; plankton; specific primers
AQUACULTURE AND FISHERIES
A molecular method to detect and identify the native species of southwestern Atlantic Crassostrea (Mollusca: Ostreidae)
Sandra LudwigI,1 1 Corresponding author. E-mail: sand.ludwig@gmail.com ; Raquel PatellaI; Sérgio StoievI; Gisela Castilho-WestphalII; Marcus V. F. GirottoII; Antonio OstrenskyII
ILaboratório de Ecologia Molecular e Parasitologia Evolutiva, Departamento de Zoologia, Universidade Federal do Paraná. Caixa Postal 19073, 81531-980 Curitiba, PR, Brazil
IIGrupo Integrado de Aquicultura e Estudos Ambientais, Universidade Federal do Paraná. Curitiba, Paraná, Brazil
ABSTRACT
Among oysters, species of Crassostrea (Sacco, 1897) are the most attractive to aquaculture. In Brazil, the genus is represented by C. rhizophorae (Guilding, 1828) and C. brasiliana (Lamarck, 1819). Because the maturation and breeding technology is not well developed for these species, aquaculturists need a reliable method to decide the correct time to place spat collectors in the field, and to identify both species, which are morphologically similar. In this study a specific Multiplex PCR protocol was developed, using one pair of universal primers from 18S rDNA as a positive control and a pair of specific primers for each target species. The sensitivity and specificity of the protocol was evaluated. It detected C. rhizophorae DNA in low concentrations, and C. brasiliana DNA in even lower concentrations. Further, the Multiplex PCR proved efficient in detecting DNA in concentrations equivalent to that of a single larva of each species, either separated or combined, when mixed with total DNA extract of a plankton sample representing 1000 L of filtered water. Field tests confirmed the applicability of the protocol, which holds the promise to become an important tool for aquaculture or conservation programs, allowing for the continuous monitoring of the life cycle of C. brasiliana and C. rhizophorae, by detecting the right periods of larval release and settlement.
Key words: Multiplex PCR; oysters; plankton; specific primers.
Oyster harvesting is probably one of the oldest forms of marine extractivism, possibly dating from the dawn of civilization. The development of modern oyster cultivation techniques has boosted production, which supplies the demands of an ever growing market (SPENCER 1990). Oyster cultivation has important socio-economic impacts for economically disadvantaged coastal populations. It provides additional income for traditional communities, which are losing their long-established sources of income due to the collapse of fisheries throughout the world (ERSE & BERNARDES 2008).
Among oysters, species of Crassostrea (Sacco, 1897) are the most attractive to aquaculture because they have large meatto-shell ratio and rapid growth rate. Within Crassostrea, the most widely reared species is Crassostrea gigas (Thunberg, 1793), the Pacific oyster. It was intentionally introduced in various continents for commercial purposes (YU & LI 2007). According to DIJKEMA (1997), C. gigas would not be able to reproduce in many regions where it had been introduced (a prediction proved incorrect) for the Atlantic waters of South America, since naturalized banks of the Pacific oysters are now known from Argentina (CASTAÑOS et al. 2009) to the state of Santa Catarina, Brazil (MELO et al. 2010).
As an invasive species, C. gigas causes serious environmental problems, such as displacement of native species (PATIL et al. 2005), alteration of the structure of natural communities, and introduction of parasites and epibionts (GALLO-GARCIA et al. 2004, FAO 2006). In order to avoid new introductions of C. gigas in Brazil, state and federal laws (IBAMA 1998) presently control the translocation of juveniles and adults. Such control has led to an increased interest in the cultivation of native oyster species.
In the southwestern Atlantic coast, two native species of Crassostrea are known, Crassostrea rhizophorae (Guilding, 1828) and Crassostrea brasiliana (Lamarck, 1819) (PIE et al. 2006b, IGNACIO et al. 2000). While C. rhizophorae grows slowly, achieving a relatively small final shell size (at least in southern Brazil), C. brasiliana grows faster and achieves a larger size, holding some economic promise (CHRISTO & ABSHER 2006). Consequently, where C. gigas cultivation is forbidden, C. brasiliana cultivation is likely the best alternative for aquaculture.
However, spat production of C. brasiliana appears to be one of the major bottlenecks for the culture of the species in Brazil. The maturation and breeding technology is not well developed for the species, and specialized laboratories are still scarce (POLI & LITLLEPAGE 1998). Aquaculturists capture juveniles directly from nature, resulting in significant environmental impacts to natural beds. An alternative to this practice is to use spat collectors, as presently done in Brazil. Nevertheless, studies on the reproductive cycles and settlement patterns of native oyster species are lacking in the country, hindering collecting efforts.
Larval and seedling morphologies of Crassostrea species are highly conservative, hindering species determination. Consequently, ecological studies or efforts to determine the adequate time for placing collectors in the environment for aquaculture purposes often suffer. Differentiation of larval forms of Crassostrea species requires the use of Scanning Electron Microscopy (CHRISTO et al. 2010), a method that does not provide rapid and precise identification of species in continuous studies. Previous life cycle studies on local Crassostrea spp. likely reflect a mix of congeners or even species of other bivalve genera (e.g. CHRISTO 2006) and thus are of little practical interest.
Fortunately, molecular protocols are being developed to allow simultaneous detection and identification of one or more species of interest in different types of samples, including plankton samples (MORGAN & ROGERS 2001, ANIL et al. 2002, KOTTA et al. 2006, PIE et al. 2006a). Indeed, identification of adult species of Crassostrea by molecular markers is becoming a common procedure (WANG & GUO 2007, PIE et al. 2006b, PATIL et al. 2005). However, the basic limitations that exist today to implementing molecular methods are the financial costs of using them in monitoring studies. Molecular methodologies were developed by PIE et al. (2006a) for screening larvae of the golden mussel, Limnoperna fortunei (Dunker, 1857) in zooplankton samples. That method has been shown to be efficient for continuous monitoring in rivers and lakes of the state of Parana, Brazil (BOEGER et al. 2007, DARRIGRAN et al. 2009, TSCHÁ et al. 2009).
In this paper we describe a Multiplex PCR protocol based on a set of primers that can be used to simultaneously detect/ identify, in plankton samples, larvae, seedlings and adults of C. brasiliana and C. rhizophorae. Our protocol may become an important tool for the continuous monitoring of the life cycle of C. brasiliana and C. rhizophorae, allowing researchers to detect temporal and spatial variations in larval availability in plankton and settlement. With this information in hand, aquaculturists may be able to select the best cultivation sites, and decide when to initiate spat collection, maximizing success in obtaining larvae and seedlings of each species of oysters. This protocol was successfully applied to plankton samples in the Southeast coast of Brazil to help coordinate the collection of naturally occurring spats for aquaculture.
MATERIAL AND METHODS
Adult oysters, spats and plankton samples were obtained from Guaratuba Bay, Paraná, Brazil (25º51.80'S48º38.20'W). Plankton samples (n = 34) were obtained bimonthly from January to June 2010, from three regions within the Guaratuba Bay (Fig. 1) by using a water pump to filter 1000 L of water through a plankton net (mesh size = 65 µm). To determine the volume of plankton samples, the water was pumped up into a container. The time was counted until the container volume was filled. This time is the equivalent to the total volume sampled in 1000 L of pumped water.
Spats (n = 246) were obtained from January to April, 2010, using collectors consisting of two sets of three-PVC plates. Collectors were placed in the Cabaraquara region of the Guaratuba Bay (Fig. 1), positioned at two depths, 30 cm and 1 m, and for thirty days. All samples were fixed in 92% ethanol in the field and then transported to the laboratory. DNA extraction and application of the multiplex PCR protocol proceeded as described above.
Pooled DNA from plankton samples was obtained in the following manner: a manual vacuum pump was used to filter the samples through paper filter. Organisms accumulated on the filter were then used for DNA extraction. The total DNA was extracted using the EZ-DNA kit (Biosystems, Brazil), following the manufacturer's instructions. Adult oysters were identified using the protocol developed by PIE et al. (2006a).
A fragment of the mitochondrial 16S rDNA from tissues of adult oyster specimens was amplified with universal primers (16SAr and 16SBr, Tab. I) using the following PCR protocol: 4 min at 94ºC/32 cycles of 94ºC for 20s, 56ºC for 40s and 72ºC for 1min/72ºC for 1min with 25 mL reactions with, 2.5U of AmpliTaq DNA Polymerase (Invitrogen®), 1X of PCR Buffer, 3mM of MgCl2, 0.4mM of dNTPs, 0.4 µM of each primer, 2 ng/µL of extract of DNA and double-distilled water to complete total volume. PCR products were electrophoresed on 1.5% agarose gels and stained with ethidium bromide for band visualization and photo-documentation. Positive reactions were purified using the Minelute® (Qiagen, Germany) kit and cycle sequencing was carried out using the following final concentrations: 0.16µM of each primer, 0.25X reaction Buffer, 0.5 µL of BigDye v.3 (Applied Biosystems®), and 0.2-0.3 ng/µL of template DNA. Thermocycling conditions included an initial denaturation of 1 min at 96ºC, followed by 35 cycles of 10s at 96ºC, 5s at 50ºC, and 4min at 60ºC. Products were purified using SephadexTM G-50 medium (GE Healthcare Bio-Sciences AB) and both strands were sequenced on an ABI 3130 Automatic Sequencer. The sequences obtained were edited using the Staden package (STADEN et al. 1998). Representative sequences of each target species were deposited in the GenBank database under accession numbers HQ652329-HQ652331.
In order identify unique regions for designing specific primers, we compared the mitochondrial 16S rDNA sequences of C. brasiliana and C. rhizophorae with GenBank sequences of closely related mollusk species. Sequences from the following species, deposited on GenBank, were used: C. gigas (S66183.1), Crassostrea sp. (AY007426), Crassostrea nippona (Seki, 1934, AY510450.1), Crassostrea rivularis (Gould, 1861, EF122383), Crassostrea belcheri (Sowerby II, 1871, EU815968.1), Saccostrea cuccullata (Born, 1778, DQ280032.1), Ostrea edulis (Linnaeus, 1758, AF458911.1) and Saccostrea glomerata (Gould, 1850, AY160756.1). All sequences were aligned using BioEdit (HALL 1999) and ClustaX l.2 (THOMPSON et al. 1997), and then adjusted visually. The identification of unique regions was based on the frequency of mismatches between the target species sequences and the sequences of the remaining species in the dataset, giving higher weight to transversions and gaps. We designed our primers using the software Primer Premier® (SINGH et al. 1998, Fig. 1), and conducted a preliminary test of primer specificity using BLAST (GenBank: www.ncbi.nlm.nih.gov/blast). If a given BLAST search resulted in close matching with sequences of other species, the primer pair in question was considered inadequate and new primer combinations were sought.
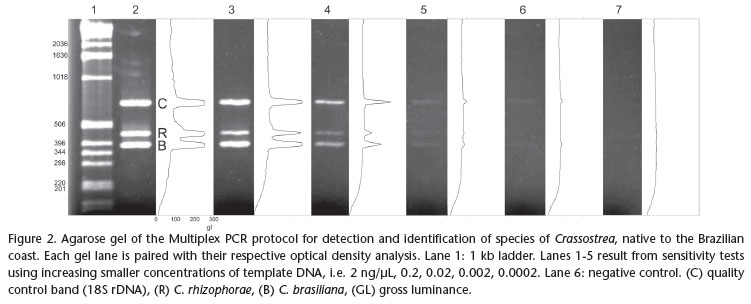
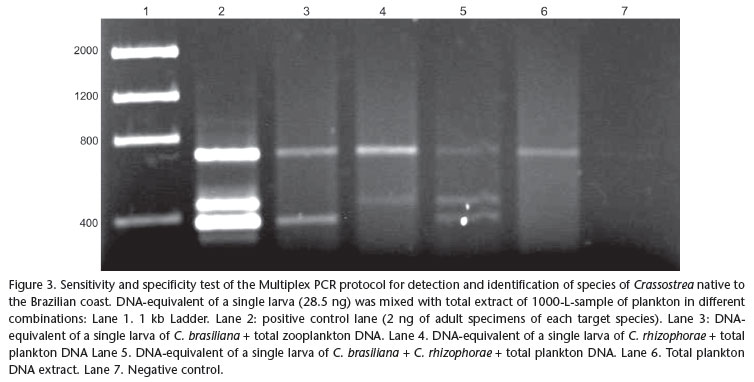
A Multiplex PCR was designed using one universal 18S rDNA primer pair (7F and 1100R, Tab. I) as positive control for the quality and presence of DNA in the sample, and primer pairs specific for the native species of Crassostrea. Thus, electrophoresis of a sample that does not contain either one of the target Crassostrea spp. sequences should result in a 18S rDNA stretch amplified by the universal primers. The absence of all fragments in a gel is interpreted as false negative which may have resulted from degraded DNA in the sample. The Multiplex reaction was optimized in an Eppendorf Mastercycler® Gradient in order to maximize sensitivity and specificity of the protocol, while attempting to attain electrophoresis bands of similar density for the same template concentration. The resulting electrophoretic gels (2% agarose) were subjected to densitometry with the software Gel-Pro Analyzer v4.0® (Media Cybernetics Inc., Silver Spring, MD), to improve the visualization of amplified fragments and separate the species identified by peaks corresponding to fragment size and position. A positive control, representing a sample of tissue of adult individuals of known identity and a negative control (lacking template DNA) were added to the protocol to confirm that the amplification was successful, and to control for possible contamination. The sensitivity of the optimized Multiplex PCR protocol was tested using extracts from adult tissue with 10-10.000 fold dilutions of an initial concentration of 2 ng/µL (0.2, 0.02, 0.002, 0.0002 ng/µL, respectively). A combined test for sensitivity and specificity was performed using the equivalent of the DNA content of a single bivalve larva (28.5 ng according to PIE et al. 2006a) added to the full extract of a zooplankton sample considered negative for larvae of both species of Crassostrea (1000 L of water filtered with 100 µm mesh size). Treatments were designed to simulate the absence, the presence of a single larva, and the simultaneous presence of one larva of each target species, submitted to the optimized multiplex PCR protocol.
Further, tests of the molecular protocol were conducted with plankton samples and spats described above. These samples were processed to evaluate the effectiveness of the proposed protocol in simulated field studies.
RESULTS
We designed and choose two sets of specific primer pairs to amplify solely a fragment of the mitochondrial 16S rDNA from C. brasiliana and C. rhizophorae, respectively (Tab. I). The Multiplex reaction was optimized with the following program: 4 min at 94ºC/32 cycles of 94ºC for 20s, 59ºC for 44s and 72ºC for 15s/72ºC for 1min. in 10 µL reactions with 1U of TaqPlatinum (Invitrogen®), 1X of PCR Buffer, 3 mM of MgCl2, 0.4 mM of dNTPs, 2 µM of each specific primer of C. rhizophorae and 1.2 µM of each specific primer of C. brasiliana, 0.1 µM of universal primer 7F, 0.1 µM of universal primer 1100R and, 2 ng/µL of extract of DNA and double-distilled water to complete total volume. After optimization of the reaction, the samples are processed more quickly. The initial reaction took two hours and ten minutes to complete. After optimization, the reaction time was reduced to one hour and thirty minutes. The sizes of the resulting fragments are as follows: 18S rDNA control: app. 700 bp long; C. rhizophorae: app. 450 bp long; C. brasiliana: app. 370 bp long.
The results obtained from the dilutions of DNA extracts from tissue samples of adult oyster demonstrate that the protocol detects low concentrations of oyster DNA (Fig. 2). The protocol detected the presence of the DNA of C. rhizophorae in concentrations as low as 0.002 ng/¼L, and as low as 0.0002 ng/¼L, in the case of C. brasiliana.
Sensitivity and specificity of the primers proposed for the target species were also demonstrated by the test using extracts of zooplankton samples mixed with a DNA equivalent to that of a single larva (Fig. 3). Independent of the combination of each treatment, the method did successfully detect the target DNA of a single larva despite eventual competition with DNA of non-target organisms present in the samples. No nonspecific bands were detected in any treatment.
A total of 34 plankton samples were tested using the method proposed in this contribution. Larvae of C. brasiliana and C. rhizophorae were successfully detected every month of this experiment in Cabaraquara. In Parati, larvae of both species were detected only in January, whereas larvae of C. brasiliana represented the only species detected from April to June; no larvae of the target species were detected in the samples from February and March, 2010. In Vicente (sampled only after February, 2010), larvae of C. brasiliana and C. rhizophorae were detected only in April (Tab. II), both being absent in February, March, and June of 2010.
Finally, of the 246 spats collected, 38 (app. 16%) were identified as C. brasiliana; 58 oyster spats (app. 24%) were identified as C. rhizophorae; 82 were identified as a possible Crassostrea sp. spat but could not be identified as either target species (specimens for which the control band was amplified but no specific band was visible); and 68 (28%) were not adequately preserved and did not yield accurate results (see discussion for more details). Both species were detected in collectors removed every month; unidentified spats were also present every month (Fig. 4). Crassostrea rhizophorae was relatively more frequent in January and February whereas C. brasiliana was more prevalent in February and March, and less prevalent in April. Unidentified spats were detected in greater numbers in April (these results were obtained after sequencing the 18S fragment). Samples with degraded DNA (detected by the absence of amplification of the universal/control 18S band) were more abundant in January and February but were significantly reduced in the last two months of the sampling period.
DISCUSSION
Laboratory and field tests support the efficiency of our Multiplex PCR protocol in detecting the target species even when DNA concentrations are low. Results from our simulations suggests that it is possible to detect at least one larva of each target species per 1000 L of filtered water in total plankton samples, without amplifying other species. If amplification of non-target species had happened, non-specific amplification bands would have been detected in our gels.
The results of our tests using environmental samples have demonstrated that our protocol permits the detection of larvae in plankton samples and the rapid identification of spats from collectors. Our results also suggest that the major obstacle to spat identification using PCR is poor preservation of fixed specimens, which results in loss of DNA integrity. Indeed, field personnel failed to use enough ethanol when fixing spat samples in the months of January and February, resulting in significant loss of samples (Fig. 4). The fixation protocol was applied correctly on the spats removed from collectors in March and April, which resulted in a reduction in the percentage of degraded DNA (Fig. 4). Fixation was not a problem for plankton samples, as no false negative was detected.
Thus, the proposed protocol represents an important tool for continuous monitoring of the life cycle of C. brasiliana and C. rhizophorae, detecting temporal and spatial variations of larval availability in the plankton and settlement for each species. Although the sampling period of the field test was limited, it has provided evidence that there are differences in the spatial and temporal occurrence of larvae of the two targeted species of Crassostrea. Further, the identification and quantification of spats indicates distinct patterns of fluctuation in the settlement of C. rhizophorae and C. brasiliana in the same location (Cabaraquara, Fig. 1), which could represent important information on the differences in the life cycle of both species.
A rapid analysis of figure 4 strongly suggests that, in the region sampled, the peak of settlement of C. rhizophorae occurs between January and February, and that of C. brasiliana, between February and March. During this preliminary study we have also detected fluctuation in the availability of spats of other oyster species. These could represent either native species of other genera (not Crassostrea), or invasive species, as reported from other regions of the Southwestern Atlantic Coast (see MELO et al. 2010, VARELA et al. 2007). Regardless of whether these organisms are native or not, their presence in the samples provides further evidence that the primers we designed are very specific, as they did not amplify the DNA of morphologicallysimilar spats (no cross-reaction).
If the life cycle and settlement of the two native Crassostrea species are asynchronous, as suggested by the results of the field test, we can predict the time individuals of each species settle by monitoring their larvae in the plankton. Based on such data, aquaculturists could ensure the identity of the majority of the seeds they collect by exposing the collectors in the environment during the right period and on the right site. The choice sites for installation of collectors appears to be another parameter to be considered to ensure the collection of oysters for aquaculture. From April to June, only larvae of C. brasiliana, the most adequate species for culture (at least in certain regions of the Southwestern Atlantic coast), were detected in plankton samples. The method developed in this study may also be used to complement morphological identification of spats and adult oysters, thus facilitating and maximizing success in selecting spats of the desired species by growers.
Even though we do not expect that false positive reactions will occur when using the proposed method, we recommend that researchers using it regularly sequence specific bands of the Multiplex PCR in order to confirm its precision.
Unfortunately, the present method is not capable to quantify larval density in plankton samples. However, our protocol could be easily adapted to allow an inference of the number of specimens of each target species though calibration with Real Time PCR, as developed by ENDO et al. (2009) for L. fortunei. Subsampling of the plankton samples may also provide an alternative method to quantify the relative density of larvae of each species.
ACKNOWLEDGMENTS
This study is part of the MSc Thesis of SL, Graduate student in Zoology, Universidade Federal do Paraná. The Conselho de Desenvolvimento Científico e Tecnológico (CNPq) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) have provided support for this study and scholarship assistance, respectively. The authors also thank the important contribution of Walter A. Boeger (UFPR); without his help this work would not have been possible.
LITERATURE CITED
Submitted: 14.III.2011; Accepted: 30.V.2011.
Editorial responsibility: Paulo da C. Lana
- ANIL, A.C.; T.K VENKA; S.S. SAWANT; M. DILEEPKUMAR; V.K. DHARGALKAR; N. RAMAIAH; S.N. HARKANTRA; Z.A. ANSARI. 2002. Marine Bioinvasion: Concern for ecology and shipping. Current Science 83: 214-218.
- BOEGER, W.A; M.R. PIE; R.M. FALLEIROS; G. DARRIGRAN; M.C.D. MANSUR; C.E. BELZ. 2007. Testing a molecular protocol to monitor the presence of golden mussel larvae (Limnoperna fortunei) in plankton samples. Journal of Plankton Research 29: 1015-1019.
- CASTAÑOS, C.; M. PASCUAL; A.P. CAMACHO. 2009. Reproductive biology of the nonnative oyster, Crassostrea gigas (THUNBERG, 1793), as a key factor for its successful spread along the rocky shores of northern patagonia, Argentina. Journal of Shellfish Research 28: 837-847.
- CHRISTO, S.W. 2006. Biologia Reprodutiva e Ecologia de Ostras do gênero Crassostrea Sacco, 1897 na Baía de Guaratuba (Paraná-Brasil): Um Subsídio ao Cultivo. PhD Thesis, Departamento de Zoologia, Universidade Federal do Paraná. Available online at: http://dspace.c3sl.ufpr.br:8080/dspace/bitstream/1884/5198/1/Tese.pdf [Accessed: 05/XI/2009 ]
- CHRISTO, S.W. & T.M. ABSHER. 2006. Reproductive period of Crassostrea rhizophorae (Guilding, 1828) and Crassostrea brasiliana (Lamark, 1819) (Bivalvia: Ostreidae) in Guaratuba bay, Paraná, Brazil. Journal of Coastal Research 39: 1215-1218.
- CHRISTO, S.W.; T.M. ABSHER; G. BOEHS. 2010. Morphology of the larval shell of three oyster species of the genus Crassostrea SACCO, 1897 (Bivalvia: Ostreidae). Brazilian Journal of Biology 70: 645-650. doi: 10.1590/S1519-69842010000300023.
- Darrigran, G.; A.W. Boeger; C. Damborenea; M. Maroñas. 2009. Evaluation of sampling and analysis techniques for early detection of Limnoperna fortunei (Mytilidae) in limit areas of its distribution. Brazilian Journal of Biology 69: 979-980.
- DIJKEMA, R. 1997. Molluscan fisheries and culture in the Netherlands, p. 115-135. In: C.L. MACKENZIE JR (Ed.). The history, present condition and future of the molluscan fisheries of North and Central America and Europe. Seattle, NOAA Technical Report NMFS, vol. 3.
- ERSE, E. B. & M.A. BERNARDES. 2008. Levantamento de estoques da ostra Crassostrea sp. em bancos naturais no litoral paranaense. Biotemas 21: 57-63.
- ENDO, N; K. SATO & Y. NOGATA. 2009. Molecular based method for the detection and quantification of larvae of the golden mussel Limnoperna fortunei using real-time PCR. Plankton Benthos and Research 4: 125-128
- FAO 2006. FAO Fisheries and Aquaculture Department [online ] Rome. Available online at: http://www.fao.org/fishery/culturedspecies/Crassostrea_gigas/en [Acessed: 29/XI/2010 ]
- GALLO-GARCIA, M.C; M.G. ULLOA-GÓMEZ & D.E. GODÍNEZ-SIORDIA. 2004. Evaluation of two treatments in polychaete worm intensity associated with Crassostrea gigas (THUNBERG, 1873) oyster valves. Ciencias Marinas 30: 455-464.
- HALL, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/ NT. Nucleic Acid Symposium Series 41: 95-98.
- IBAMA 1998. Portaria 145 de 29 de outubro de 1998. Available online at: www.cprh.pe.gov.br/downloads/Portaria-145-29-10-1998.doc [Accessed: 29/XI/2010 ]
- IGNACIO, B.L; T.M. ABSHER; C. LAZOSKI & A.M. SOLÉ-CAVA. 2000. Genetic evidence of the presence of two species of Crassostrea (Bivalvia: Ostreidae) on the coast of Brazil. Marine Biology 136: 987-991.
- KOTTA, J.; I. KOTTA; M. SIMM; A. LANKOV; V. LAURINGSON; A. POLLUMAE & H. OJAVEER. 2006. Ecological Consequences of biological invasions: three invertebrate case studies in the north-eastern Baltic Sea. Helgoland Marine Research 60: 106-112.
- MELO, C.M.R; F.C. SILVA; C. GOMES; A.M. SOLÉ-CAVA & C. LAZOSKI. 2010. Crassostrea gigas in natural oyster banks in Southern Brazil. Biological Invasions 12: 441-449.
- MORGAN, T.S & A.D. ROGERS. 2001. Specificity and sensitivity of microsatellite markers for identification of larvae. Marine Biology 139: 967-973. doi: 10.1007/s002270100589
- PATIL, J.G; R.M. GUNASEKERA; B.E. DEAGLE & N.J. BAX. 2005. Specific Detection of Pacific Oyster (Crassostrea gigas) Larvae in Plankton Samples Using Nested Polymerase Chain Reaction. Marine Biotechnology 7: 11-20.
- PIE, M.R.; W.A. BOEGER; L. PATELLA & R. FALLEIROS. 2006a. A Fast and accurate molecular method for the detection of larvae of the golden mussel Limnoperna fortunei (Mollusca: Mytilidae) in plankton samples. Journal of Molluscan Studies 72: 218-219.
- PIE, M.R,; R.O. RIBEIRO; W.A. BOEGER; A. OSTRENSKY; R.M. FALLEIROS & L. ANGELO. 2006b. A simple PCR-RFLP method for the discrimination of native and introduced oyster species (Crassostrea brasiliana, C. rhizophorae and C. gigas, Bivalvia: Ostreidae) cultured in Southern Brazil. Aquaculture Research 37: 1598-1600.
- POLI, C.R. & J. LITLLEPAGE. 1998. Desenvolvimento do Cultivo de Moluscos no Estado de Santa Catarina, p. 163-182. In: Anais Aquicultura Brasil 1998. Recife, I Congresso Sul-Americano de Aquicultura. Available online at: http://www.lmm.ufsc.br/data/files/cultivo_em_sc.pdf [Acessed: 30/XI/2010 ]
- SINGH, V.K; A.K. MANGALAM; S. DWIVEDI & S. NAIK. 1998. Primer Premier: program for design of degenerate primers from a protein sequence. BioTechnique 24: 318-319.
- SPENCER, B.E. 1990. Cultivation of Pacific Oysters. MAFF Directorate of Fisheries Research, Lowestoft 63: 1-47.
- STADEN, R; K.F. BEAL; J.K. BONFIELD. 1998. The Staden Package. Computer Methods in Molecular Biology 132: 115-130.
- THOMPSON, J.D; T.J. GIBSON; F. PLEWNIAK; F. JEANMOUGIN & D.G. HIGGINS. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876-4882.
- TSCHÁ, M.K; W.A. BOEGER; L. PATELLA; A. OSTRENSKY & M.R. PIE. 2009. Aumentando a sensibilidade e a agilidade do monitoramento de Limnoperna fortunei utilizando o método molecular, p. 223-228. In: G. DARRIGRAN & C. DAMBORENEA (Eds). Introdução a Biologia das Invasões. O Mexilhão Dourado na América do Sul: biologia, dispersão, impacto, prevenção e controle. São Carlos, Cubo Editora, XIII+246p.
- VARELA, E.S; C.R. BEASLEY; H. SCHENEIDER; I. SAMPAIO; N.S. MARQUES-SILVA & C.H. TAGLIARIO. 2007. Molecular Phylogeny of Mangrove Oysters (Crassostrea) from Brazil. Journal of Molluscan Studies 73: 229-234.
- WANG, Y & X. GUO. 2007. Development and Characterization of EST-SSR Markers in the Eastern Oyster Crassostrea virginica Marine Biotechnology 9: 500-511. doi: 10.1007/s10126-007-9011-7.
- YU, H & Q. LI. 2007. EST-SSR markers from the Pacific oyster, Crassostrea gigas. Molecular Ecology Notes 7: 860-862.
Publication Dates
-
Publication in this collection
16 Sept 2011 -
Date of issue
Aug 2011
History
-
Received
14 Mar 2011 -
Accepted
30 May 2011