Abstract
Cloning of the T-cell receptor genes is a critical step when generating T-cell receptor transgenic mice. Because T-cell receptor molecules are clonotypical, isolation of their genes requires reverse transcriptase-assisted PCR using primers specific for each different Valpha or Vß genes or by the screening of cDNA libraries generated from RNA obtained from each individual T-cell clone. Although feasible, these approaches are laborious and costly. The aim of the present study was to test the application of the non-palindromic adaptor-PCR method as an alternative to isolate the genes encoding the T-cell receptor of an antigen-specific T-cell hybridoma. For this purpose, we established hybridomas specific for trans-sialidase, an immunodominant Trypanosoma cruzi antigen. These T-cell hybridomas were characterized with regard to their ability to secrete interferon-gamma, IL-4, and IL-10 after stimulation with the antigen. A CD3+, CD4+, CD8- interferon-gamma-producing hybridoma was selected for the identification of the variable regions of the T-cell receptor by the non-palindromic adaptor-PCR method. Using this methodology, we were able to rapidly and efficiently determine the variable regions of both T-cell receptor chains. The results obtained by the non-palindromic adaptor-PCR method were confirmed by the isolation and sequencing of the complete cDNA genes and by the recognition with a specific antibody against the T-cell receptor variable ß chain. We conclude that the non-palindromic adaptor-PCR method can be a valuable tool for the identification of the T-cell receptor transcripts of T-cell hybridomas and may facilitate the generation of T-cell receptor transgenic mice.
NPA-PCR method; T-cell receptor; Trypanosoma cruzi; Trans-sialidase
Braz J Med Biol Res, March 2006, Volume 39(3) 345-354
The non-palindromic adaptor-PCR method for the identification of the T-cell receptor genes of an interferon-g-secreting T-cell hybridomaspecific for trans-sialidase, an immunodominant Trypanosoma cruzi antigen
M.I. Hiyane1,2, S.B. Boscardin2 and  Correspondence and Footnotes
Correspondence and Footnotes
 M.M. Rodrigues1,2
M.M. Rodrigues1,2
1Centro Interdisciplinar de Terapia Gênica, 2Departamento de Microbiologia, Imunologia e Parasitologia, Escola Paulista de Medicina, Universidade Federal de São Paulo, São Paulo, SP, Brasil
 References
References
 Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Correspondence and Footnotes
Abstract
Cloning of the T-cell receptor genes is a critical step when generating T-cell receptor transgenic mice. Because T-cell receptor molecules are clonotypical, isolation of their genes requires reverse transcriptase-assisted PCR using primers specific for each different Va or Vß genes or by the screening of cDNA libraries generated from RNA obtained from each individual T-cell clone. Although feasible, these approaches are laborious and costly. The aim of the present study was to test the application of the non-palindromic adaptor-PCR method as an alternative to isolate the genes encoding the T-cell receptor of an antigen-specific T-cell hybridoma. For this purpose, we established hybridomas specific for trans-sialidase, an immunodominant Trypanosoma cruzi antigen. These T-cell hybridomas were characterized with regard to their ability to secrete interferon-g, IL-4, and IL-10 after stimulation with the antigen. A CD3+, CD4+, CD8- interferon-g-producing hybridoma was selected for the identification of the variable regions of the T-cell receptor by the non-palindromic adaptor-PCR method. Using this methodology, we were able to rapidly and efficiently determine the variable regions of both T-cell receptor chains. The results obtained by the non-palindromic adaptor-PCR method were confirmed by the isolation and sequencing of the complete cDNA genes and by the recognition with a specific antibody against the T-cell receptor variable ß chain. We conclude that the non-palindromic adaptor-PCR method can be a valuable tool for the identification of the T-cell receptor transcripts of T-cell hybridomas and may facilitate the generation of T-cell receptor transgenic mice.
Key words: NPA-PCR method, T-cell receptor, Trypanosoma cruzi, Trans-sialidase
Introduction
Transgenic mice expressing specific T-cell receptors (TCR) are a valuable tool for the study of the immune responses to auto-antigens, infectious agents and neoplastic cells (for reviews, see Refs. 1-6). A critical step when generating these transgenic mice is the isolation of the TCR genes. Because TCR molecules are clonotypical, isolation of these genes requires the identification of the individual TCR expressed by a T-cell clone or hybridoma. This can be accomplished using a panel of TCR-specific monoclonal antibodies. Although monoclonal antibodies are available for the identification of the different Vß chains of the mouse TCR, antibodies specific for many of the murine Va chains do not exist. Alternatively, cloning can be accomplished by reverse transcriptase-assisted PCR using primers specific for each different Va or Vß genes or by the screening of cDNA libraries generated from RNA obtained from each individual T-cell clone. Although feasible, these approaches are laborious and costly.
In 1992, Chen and Platsoucas (7) developed the non-palindromic PCR method for the amplification of transcripts with variable or unknown 5' ends, such as TCR and immunoglobulins. Among the advantages of this methodology is the fact that only one 5' end extension primer is required. Since it was first described, this methodology has been successfully used to clone the genes encoding TCR transcripts of T cells isolated from humans and mice (8-12).
Chagas' disease, caused by the protozoan parasite Trypanosoma cruzi, afflicts 16 to 18 million people in Latin America (13). The cell-mediated immune response (CMI) participates in multiple aspects of Chagas' disease. During the acute phase of infection both CD4 and CD8 T cells have been described as important mediators of protective immunity. Type 1 cells (Th1 and Tc1) are thought to mediate protective immunity in part by their ability to secrete interferon-g (IFN-g) (14). During the chronic phase of the disease, most individuals have a strong CMI to parasite antigens. These T cells may help to maintain the low levels of parasitemia preventing the reactivation of acute infection (15). On the other hand, type 1 CMI may be the cause of heart damage during the chronic phase of infection (16,17).
T. cruzi trypomastigotes express an enzyme denominated trans-sialidase (TS) on their surface. This enzyme is required for the parasite to obtain sialic acid from host glycoproteins (18). Serological studies have shown that TS is highly immunogenic during natural human infection with T. cruzi. Almost all individuals develop antibodies that recognize an epitope composed of amino acid repetitions located at the carboxy-terminal region of TS (19). Chagasic patients also generate antibodies specific for the N-terminal region of TS that strongly inhibit its enzymatic activity (20-22).
In addition to antibodies, the catalytic domain of TS induces type 1 CMI in chagasic patients (23). The role of CMI specific for the catalytic domain of TS was studied during acute infection in mice. Immunization with DNA vaccines elicited strong type 1 CMI and protective immunity against acute infection in BALB/c or C57BL/6 mice (24-26). These experiments provided strong evidence that CMI to TS plays a protective role during acute T. cruzi infection. Nevertheless, the participation of CMI to TS during the chronic phase of infection is still unclear. To study this problem, we will need more powerful experimental models using transgenic CD4 and/or CD8 T cells specific for TS. The first step toward the generation of these transgenic animals is the isolation of TCR genes specific for the antigen. To this end, in the present study we established and characterized T-cell hybridomas specific for TS. Using these T-cell hybridomas, we determined whether the non-palindromic adaptor (NPA)-PCR method would be able to easily identify the TCR transcripts.
Material and Methods
Animals
Female 5- to 8-week-old BALB/c mice were provided by CEDEME-UNIFESP.
Trans-sialidase recombinant protein production
Recombinant TS was produced in E. coli transformed with the plasmid pTScat7 (27). The purity of recombinant TS was determined by sodium dodecyl sulfate 10% polyacrylamide gel electrophoresis. A single band of 70 kDa was visualized in the gel. Protein concentration was estimated by the method of Bradford (BioRad, Hercules, CA, USA).
Immunization of mice and culture of lymph node cells
BALB/c mice were immunized with 25 µg of purified recombinant TS emulsified in complete Freund's adjuvant (Sigma, St. Louis, MO, USA). The emulsion was injected subcutaneously into the two hind footpads. Two weeks later, inguinal and popliteal lymph nodes (LN) were collected. LN cells (3 x 106/mL) were then cultured in RPMI medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10 mM HEPES, 2 g/L sodium bicarbonate, 2 mM L-glutamine, 50 µM 2-mercaptoethanol, 1 mM sodium pyruvate, 1% (v/v) of the nonessential amino acid solution, 1% (v/v) MEM vitamin solution (all purchased from Invitrogen), 100 U penicillin and streptomycin per mL (Sigma), 10 µg/mL recombinant TS, 20 units/mL recombinant human IL-2, and 2% (v/v) inactivated normal human serum. The cultures were maintained for 5 days at 37ºC in an atmosphere containing 5% CO2 and then harvested and used for fusion.
Fusion and screening of the T-cell hybridomas
Specific LN cells and BWa-ß-thymoma cells (kindly provided by Dr. P. Marrack, University of Colorado Health Science Center, CO, USA) were mixed at a 5:1 ratio. One milliliter FCS-free MEM (Invitrogen) containing 50% polyethylene glycol 1450 solution (w/v, Sigma) was added to the cell pellet under continuous gentle shaking at 37ºC. After 1 min at 37oC, 50 mL MEM was slowly added. Cells were centrifuged at 250 g for 6 min. The cells were resuspended in MEM supplemented with 2.86 g/L sodium bicarbonate, 0.6 g/L D-dextrose, 2 mM L-glutamine, 30 µM 2-mercaptoethanol, 1 mM sodium pyruvate, 1.1% (v/v) of the nonessential amino acid solution, 0.3% (v/v) essential amino acid solution, 80 U/mL penicillin G, 60 U/mL streptomycin sulfate (10% S-MEM), and 10% FCS (HyClone, Logan, UT, USA). Serial dilutions were made with this cell suspension. Cloning was achieved by limiting dilution. One hundred microliters of the suspension was poured onto a 96-well microplate and the remaining cell suspension was diluted 1:4, 1:16, and 1:64. Each dilution was poured onto a 96-well microplate. The cultures were maintained at 37ºC in an atmosphere containing 5% CO2. After 48 h, 50 µL 10% S-MEM supplemented with 3X HAT (Invitrogen) was added to each well. Every 5 days, 100 µL 10% S-MEM supplemented with 1X HAT/well was exchanged. If cell proliferation was observed, cells were transferred to 24-well plates and further expanded. Two weeks after fusion, the medium was changed to 10% S-MEM supplemented with 1X HAT (Invitrogen) and subsequently changed to 10% S-MEM. Established clones were tested for reactivity in the presence of the gene.
Cytokine production by cell hybridomas
For screening, 1 x 105 T-cell hybridoma cells and 1 x 106 syngeneic BALB/c spleen cells were co-cultured in the presence or absence of 10 µg/mL recombinant TS in 10% S-MEM. After 3 days, the supernatants were collected and the cytokine concentration was estimated by capture ELISA using antibodies purchased from Pharmingen (San Diego, CA, USA) exactly as described by Rodrigues et al. (28).
Fluorescence-activated cell sorting analysis of the T-cell hybridomas
Phenotypic analyses of hybridomas were carried out by fluorescence-activated cell sorting (FACS) using a Facscalibur Cytometer (Becton Dickinson, Mountain View, CA, USA). Hybridomas (106 cells) were incubated on ice for 45 min with hybridoma supernatants. Monoclonal antibodies anti-CD4 (GK1.5, rat IgG2), anti-CD8 (2.43, rat IgG2), anti-CD44 (KM-102, rat IgG), and anti-CD3 (145-2C11, hamster IgG) were used. After two washes, cells were incubated with fluorescein-isothiocyanate (FITC)- labeled goat anti-rat immunoglobulin G (KPL) or FITC-labeled goat anti-hamster IgG (KPL) for an additional 45 min on ice, washed, and fixed in PBS containing 1% (w/v) paraformaldehyde. FITC-labeled anti-TCR ß chain (H57-597, Pharmingen) or anti-Vß6 TCR (RR4-7, Pharmingen, kindly provided by Dr. Milena B.P. Soares, FIOCRUZ, Brazil) was used as described above without secondary antibody incubation.
Amplification, cloning and sequencing of the a and ß variable segment genes of the TCR
Hybridoma 09 RNA was purified using TRIzol (Invitrogen) according to manufacturer instructions. RNA was precipitated with ethanol, washed, and resuspended in nuclease-free water. The NPA-PCR method was used for the amplification of TCR transcripts. Briefly, 5 µg of total RNA from hybridoma 09 was used as template for double-stranded cDNA synthesis using either the Ca3-NotI or Cß3-NotI primer (Table 1) and Superscript (Invitrogen). NPA (Table 1) were ligated onto the ends of the double-stranded cDNA, resulting in the addition of the adaptors at both the 5' and 3' ends. After the ligation was completed, the adaptor was removed from the 3' end by NotI restriction nuclease digestion, but was retained at the 5' end. A single strand of the adaptor, called adaptor primer (Table 1), was used as a sense 5' end amplification primer for both rounds of amplification performed. Two Ca primers, designated mCa2 and mCa1, and two Cß primers, designated mCß2 and mCß1 (Table 1), were used as 3' anti-sense amplification primers for the first and second amplification reactions, respectively. mCa1 and mCß1 were located 5' to the primers mCa2 and mCß2 used for the first amplification. The products resulting from the second amplification were sized ~500 bp to the TCR a chain, and ~600 bp to the TCR ß chain. Each DNA band was purified and ligated into the pMOSblue cloning vector. The transformation resulted in 24 colonies of the TCR a chain and 26 of the TCR ß chain. Twenty colonies of each chain were screened by colony hybridization with either the 32P-labeled mCaS or mCßS probe (Table 1). Plasmids containing inserts representing each TCR chain were sequenced. Automatic sequencing of double-stranded DNA was performed using the BigDye Terminator Cycle Sequencing kit (Perkin-Elmer, Foster City, CA, USA) and an ABI PRISM 377 sequencer (Perkin-Elmer). U19 and T7 oligonucleotides (Invitrogen) present in the flanking regions of the pMOSblue vector were used. The comparison of the nucleic acid and the deduced amino-acid sequences obtained with the GenBank data base and ncbi/BLAST software allowed the identification of both TCR chains.
Amplification, cloning and sequencing of the genes encoding complete cDNA of the TCR a and ß chains
The cDNA reaction was performed with 5 µg of total RNA from hybridoma 09 using the ThermoScript RT-PCR system (Invitrogen). The oligo (dT) primer supplied in the kit was used for the first strand cDNA reaction. Primers specific for the variable regions and constant regions of each TCR chain (Table 1) were used to amplify the genes encoding the complete cDNA of a and ß chains. The products of amplification were sized ~800 or ~900 bp in the case of a or ß chain cDNA fragments, respectively. These PCR products were cloned into pMOSblue and sequenced.
Results
Characterization of T-cell hybridomas specific for Trypanosoma cruzi trans-sialidase
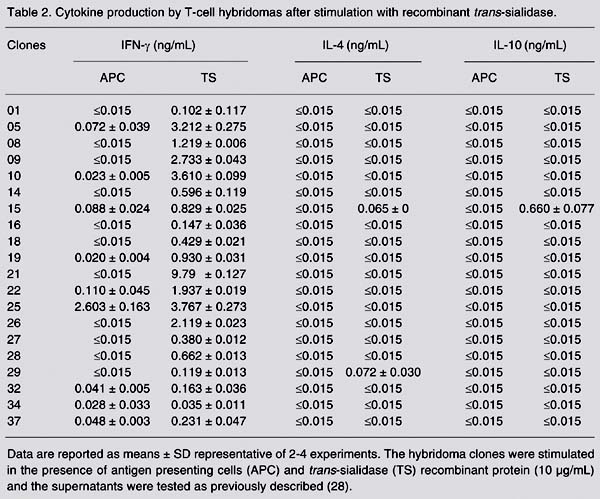
Twenty clones of T-cell hybridomas were established after fusion of LN cells and BWa-ß-thymoma cells. These hybridomas were stimulated in vitro with recombinant TS in the presence of syngeneic spleen cells. The culture supernatants were tested for the presence of cytokines. Eight hybridomas produced more than 1.2 ng/mL IFN-g. In contrast, only one hybridoma produced 0.065 or 0.660 ng IL-4 or IL-10, respectively (Table 2). By FACS analysis, all hybridomas expressed CD4 and CD3 but none expressed the CD8 surface marker. As shown in Figure 1, T-cell hybridomas H09, H10, H21, and H26 expressed CD3 and CD4 but not CD8. All four hybridomas expressed the TCR ß chain and CD44. Control cells (BWa-ß-) did not express CD3, CD4, CD8, or TCR ß (Figure 1).
Identification of TCR transcripts by NPA-PCR
The NPA-PCR method was used for the amplification of TCR transcripts from hybridoma 09. The alignment of three clones identified the a chain as the TCRAV18S2 variable segment and TCRAJ17 junctional segment. The alignment of two clones of the ß chain identified TCRBV6S19 as the variable region and TCRBJ1S1 as the junctional region. The sequences obtained by NPA-PCR allowed us to design specific primers for the amplification of the a and ß variable regions of hybridoma 09.
cDNA from hybridoma 09 was utilized as template for RT-PCR using specific primers for the a chain gene or ß chain gene. The nucleic acid and deduced amino acid sequences revealed that the clone representing the TCR a chain transcript contained the TCRAV18S2 variable region and the TCRAJ17 junctional region (Figure 2A). The clone representing the TCR ß chain transcript contained the TCRBV6S19 variable region and TCRBJ1S1 junctional region (Figure 2B). These results confirmed the data obtained by the NPA-PCR method described above.
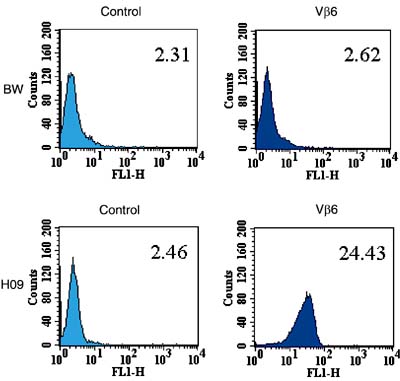
To confirm the presence of the TCRBV06 gene product on the surface of hybridoma 09, we stained these cells with an antibody specific for murine Vß6 (RR4-7). As shown in Figure 3, the fluorescent geometric mean of hybridoma 09 was ten times higher than in non-labeled cells or control cells (BWa-ß-). The detection of the receptor on the surface of hybridoma 09 confirmed that the ß chain gene of the TCR is in fact being functionally expressed as a protein on the surface of the hybridoma cells.
Expression of surface molecules in hybridomas (H) 09, 10, 21 and 26. T-cell hybridomas indicated at the top of the figure were stained with mAbs specific for the surface markers indicated, followed by FITC-conjugated goat anti-hamster (secondary) or FITC-conjugated goat anti-rat (secondary 2) antibody, and analyzed by FACS. The relative geometric mean fluorescence obtained with each mAb is indicated in the right upper corner of each histogram. TCR = T-cell receptor.
Deduced amino acid sequence of TCR a and ß chains from hybridoma 09. Representation of sequences obtained from TCR-H09 of a chain containing the TCRAV18S2 variable segment, TCRAJ17 junctional segment and constant region 1 (A) and ß chain containing the TCRBV6S19 variable region, TCRD diversity region, TCRBJ1S1 junctional region, and constant region (B).
Expression of Vß6 on the surface of hybridoma (H) 09. H09 and BWa-ß-cells were stained with an FITC-labeled mAb specific for murine Vß6. The relative geometric mean fluorescences are indicated in the right upper corner of each histogram.
Cytokine production by T-cell hybridomas after stimulation with recombinant trans-sialidase.
Discussion
In the present study, we established and characterized T-cell hybridomas specific for TS, an immunodominant T. cruzi antigen. We selected one CD3+ and CD4+ IFN-g-secreting T-cell hybridoma to test whether we could use the NPA-PCR method to easily identify its TCR transcripts. We demonstrated that the NPA-PCR method can be useful to rapidly and efficiently determine the variable regions of both TCR transcripts of this T-cell hybridoma. These observations are important because a number of advantages can be credited to this methodology. First, the NPA-PCR method uses the same reagents (primers) for the identification of TCR transcripts of any T-cell clone or hybridoma. Second, these primers are much more reliable and less expensive than the specific monoclonal antibodies used for the identification of TCR. Third, because a number of antibodies specific for the TCR Va families are not available, the screening using specific antibodies may end up being useless. Finally, this method does not require the laborious generation and screening of cDNA libraries.
On the basis of these demonstrated advantages, we believe that this methodology can significantly contribute to the identification of TCR from T-cell hybridomas or clones, that can be used to generate TCR-transgenic mice. More specifically, in our case, the TCR genes isolated from hybridoma 09 may allow us to establish a TCR transgenic mouse specific for an immunodominant T. cruzi antigen.
 Address for correspondence: M.M. Rodrigues, CINTERGEN, EPM, UNIFESP, Rua Mirassol, 207, 04044-010 São Paulo, SP, Brasil. Fax: +55-11-5571-1095. E-mail: mrodrigues@ecb.epm.br
Address for correspondence: M.M. Rodrigues, CINTERGEN, EPM, UNIFESP, Rua Mirassol, 207, 04044-010 São Paulo, SP, Brasil. Fax: +55-11-5571-1095. E-mail: mrodrigues@ecb.epm.br
Research supported by FAPESP and CNPq. M.I. Hiyane and M.M. Rodrigues are recipients of fellowships from CNPq. S.B. Boscardin was the recipient of a fellowship from FAPESP. Received April 8, 2005. Accepted October 14, 2005.
- 1. Furtado GC, Olivares-Villagomez D, Curotto de Lafaille MA et al. (2001). Regulatory T cells in spontaneous autoimmune encephalomyelitis. Immunological Reviews, 182: 122-134.
- 2. Haskins K (2004). T-cell receptor transgenic (TCR-Tg) mice from two diabetogenic CD4+ islet-antigen-specific T-cell clones. Journal of Autoimmunity, 22: 107-109.
- 3. Chen J, Eisen HN & Kranz DM (2003). A model T-cell receptor system for studying memory T-cell development. Microbes and Infection, 5: 233-240.
- 4. Bumann D (2003). T cell receptor-transgenic mouse models for studying cellular immune responses to Salmonella in vivo FEMS Immunology and Medical Microbiology, 37: 105-109.
- 5. Morrot A & Zavala F (2004). Effector and memory CD8+ T cells as seen in immunity to malaria. Immunological Reviews, 201: 291-303.
- 6. Ganss R, Limmer A, Sacher T et al. (1999). Autoaggression and tumor rejection: it takes more than self-specific T-cell activation. Immunological Reviews, 169: 263-272.
- 7. Chen PF & Platsoucas CD (1992). Development of the non-palindromic adaptor polymerase chain reaction (NPA-PCR) for the amplification of alpha- and beta-chain T-cell receptor cDNAs. Scandinavian Journal of Immunology, 35:539-549.
- 8. Lin WL, Kuzmak J, Pappas J et al. (1998). Amplification of T-cell receptor alpha- and beta-chain transcripts from mouse spleen lymphocytes by the nonpalindromic adaptor-polymerase chain reaction. Hematopathology and Molecular Hematology, 11: 73-88.
- 9. Slachta CA, Jeevanandam V, Goldman B et al. (2000). Coronary arteries from human cardiac allografts with chronic rejection contain oligoclonal T cells: persistence of identical clonally expanded TCR transcripts from the early post-transplantation period (endomyocardial biopsies) to chronic rejection (coronary arteries). Journal of Immunology, 165: 3469-3483.
- 10. Oleszak EL, Lin WL, Legido A et al. (2001). Presence of oligoclonal T cells in cerebrospinal fluid of a child with multiphasic disseminated encephalomyelitis following hepatitis A virus infection. Clinical and Diagnostic Laboratorial Immunology, 8: 984-992.
- 11. Sakkas LI, Xu B, Artlett CM et al. (2002). Oligoclonal T-cell expansion in the skin of patients with systemic sclerosis. Journal of Immunology, 168: 3649-3659.
- 12. Xu B, Sakkas LI, Goldman BI et al. (2003). Identical alpha-chain T-cell receptor transcripts are present on T cells infiltrating coronary arteries of human cardiac allografts with chronic rejection. Cellular Immunology, 225: 75-90.
- 13. Morel CM & Lazdins J (2003). Chagas disease. Nature Reviews. Microbiology, 1: 14-15.
- 14. Tarleton RL (2003). Chagas disease: a role for autoimmunity? Trends in Parasitology, 19: 447-451.
- 15. Sartori AM, Shikanai-Yasuda MA, Amato Neto V et al. (1998). Follow-up of 18 patients with human immunodeficiency virus infection and chronic Chagas' disease, with reactivation of Chagas' disease in three patients. Clinical Infectious Diseases, 26: 177-179.
- 16. Soares MB, Silva-Mota KN, Lima RS et al. (2001). Modulation of chagasic cardiomyopathy by interleukin-4: dissociation between inflammation and tissue parasitism. American Journal of Pathology, 159: 703-709.
- 17. Pontes-de-Carvalho L, Santana CC, Soares MB et al. (2002). Experimental chronic Chagas' disease myocarditis is an autoimmune disease preventable by induction of immunological tolerance to myocardial antigens. Journal of Autoimmunity, 18: 131-138.
- 18. Schenkman S, Eichinger D, Pereira ME et al. (1994). Structural and functional properties of Trypanosoma trans-sialidase. Annual Review of Microbiology, 48: 499-523.
- 19. Cazzulo JJ & Frasch ACC (1992). SAPA/transsialidase and cruzipain: two antigens from Trypanosoma cruzi contain immunodominant but enzymatically inactive domains. FASEB Journal, 6: 32593264.
- 20. Leguizamon MS, Campetella O, Russomando G et al. (1994). Antibodies inhibiting Trypanosoma cruzi trans-sialidase activity in sera from human infections. Journal of Infectious Diseases, 170: 1570-1574.
- 21. PereiraChioccola VL, Schenkman S & Kloetzel JK (1994). Sera from chronic chagasic patients and rodents infected with Trypanosoma cruzi inhibit transsialidase by recognizing its aminoterminal and catalytic domain. Infection and Immunity, 62: 29732978.
- 22. Pereira-Chioccola VL, Fragata-Filho AA, Levy AM et al. (2003). Enzyme-linked immunoassay using recombinant trans-sialidase of Trypanosoma cruzi can be employed for monitoring of patients with Chagas' disease after drug treatment. Clinical and Diagnostic Laboratory Immunology, 10: 826-830.
- 23. Ribeirao M, Pereira-Chioccola VL, Renia L et al. (2000). Chagasic patients develop a type 1 immune response to Trypanosoma cruzi trans-sialidase. Parasite Immunology, 22: 49-53.
- 24. Costa F, Franchin G, Pereira-Chioccola VL et al. (1998). Immunization with a plasmid DNA containing the gene of trans-sialidase reduces Trypanosoma cruzi infection in mice. Vaccine, 16: 768-774.
- 25. Fujimura AE, Kinoshita SS, Pereira-Chioccola VL et al. (2001). DNA sequences encoding CD4+ and CD8+ T-cell epitopes are important for efficient protective immunity induced by DNA vaccination with a Trypanosoma cruzi gene. Infection and Immunity, 69: 5477-5486.
- 26. Fralish BH & Tarleton RL (2003). Genetic immunization with LYT1 or a pool of trans-sialidase genes protects mice from lethal Trypanosoma cruzi infection. Vaccine, 21: 3070-3080.
- 27. Ribeirão M, Pereira-Chioccola VL, Eichinger D et al. (1997). Temperature differences for trans-glycosylation and hydrolysis reaction reveal an acceptor binding site in the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Glycobiology, 7: 1237-1246.
- 28. Rodrigues MM, Ribeirão M, Pereira-Chioccola VL et al. (1999). Predominance of CD4 Th1 and CD8 Tc1 cells revealed by characterization of the cellular immune response generated by immunization with a DNA vaccine containing a Trypanosoma cruzi gene. Infection and Immunity, 67: 3855-3863.
Correspondence and Footnotes
Publication Dates
-
Publication in this collection
06 Mar 2006 -
Date of issue
Mar 2006
History
-
Accepted
14 Oct 2005 -
Received
08 Apr 2005