Manuscript accepted on : 03 September 2016
Published online on: --
Plagiarism Check: Yes
Mohd. Imran
Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Northern Border University, Rafha - 91911, P.O. BOX 840, Kingdom of Saudi Arabia.
Corresponding Author E-mail: imran_inderlok@yahoo.co.in.
DOI : http://dx.doi.org/10.13005/bbra/2280
ABSTRACT: In continuation of our work related to the identification of pyridazinone derivatives as Angiotensin Converting Enzyme (ACE) inhibitors, we report herein the molecular modelling studies and ACE inhibitory activity of some 4-benzylidene-2-substitutedbenzoyl-6-phenyl-4,5-dihydropyridazin-3(2H)-ones (3a-3l). These compounds were prepared by the reaction of 2-(2-substitutedbenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-ones (2a-2l) with benzaldehyde. Molecular docking studies were carried out to determine the most active inhibitor of the Angiotensin Converting Enzyme (ACE). The compound (3a) was identified as the most active inhibitor of the ACE. The in vitro enzymatic ACE inhibitory activity of the compound (3a) using Dojindo ACE Kit-WST test kit revealed that the compound (3a) had IC50 value of 4.40 μg/mL, wherein the standard drug Lisinopril had IC50 value of 0.99 μg/mL. The comparison of the binding pattern of Lisinopril and the compound (3a) with the active sites of the ACE enzyme revealed that the incorporation of free amino groups and//or free carboxylic acid groups in the chemical structure of the compounds (3a-3l) may provide more active and potent ACE inhibitors.
KEYWORDS: Molecular modelling, ACE inhibitor, Antihypertensive, Pyridazinones.
Download this article as:| Copy the following to cite this article: Imran M. Molecular Modelling Studies and Angiotensin Converting Enzyme Inhibitory Activity of Some 4-Benzylidene-2-Substitutedbenzoyl-6-Phenyl-4,5-Dihydropyridazin-3(2H)-Ones. Biotech Res Asia 2016;13(3). |
| Copy the following to cite this URL: Imran M. Molecular Modelling Studies and Angiotensin Converting Enzyme Inhibitory Activity of Some 4-Benzylidene-2-Substitutedbenzoyl-6-Phenyl-4,5-Dihydropyridazin-3(2H)-Ones. Biotech Res Asia 2016;13(3). Available from: https://www.biotech-asia.org/?p=15960 |
Introduction
Hypertension has become a major public health problem worldwide, including the Kingdom of Saudi Arabia.1-4 The life expectancy of the public is also increasing because of the development of new technologies and drugs.2-4 Accordingly, the burden of uncontrolled hypertension will also pose major problems in the health care system. Therefore, there is a need for more research in the field of hypertension.5,6 The 6-phenyl-4,5-dihydropyridain-3(2H)-one template has been reported to possess antihypertensive activity and many of its derivatives are either in clinical use or have been tested in clinical trials.7 Some of these pyridazinone derivatives were not found suitable for further development because of their side effects and low potency.7 Angiotensin Converting Enzyme (ACE) is an enzyme that is implicated in the pathogenesis of hypertension and many inhibitors of this enzyme are also in the market for the treatment of hypertension.8 Keeping this in mind, and in continuation of our work related to the identification of pyridazinone derivatives as Angiotensin Converting Enzyme (ACE) inhibitors7, we report herein the molecular modelling studies and ACE inhibitory activity of some 4-benzylidene-2-substitutedbenzoyl-6-phenyl-4,5-dihydropyridazin-3(2H)-ones (3a-3l).
Material and Methods
General
All computations were performed on an Intel(R) Core(TM) i5-2400 CPU @ 3.10 GHz capacity processor with memory of 8 GB RAM running with the Window 7 operating system. Glide 5.9 implemented in the Maestro 9.4 software package (Schrodinger, LLC, New York, NY, 2013) was used for molecular modeling studies. Melting points were measured in open capillary tubes and are uncorrected. The Rf value of the compounds was determined by using a mixture of benzene and acetone (9:1). IR (KBr) spectra were recorded on a JASCO, FTIR-4100 spectrophotometer. The 1H-NMR and 13C-NMR spectra were recorded on Bruker Ultrashield 500 Plus MHz spectrophotometer. All reagents used in the present work were of analytical grade. The standard drug Lisinopril used for the assessment of in vitro ACE inhibitory activity was procured from Sigma Aldrich, USA.
Preparation of the compounds
The compounds (2a-2l) and (3a-3l) were prepared according to the method provided in Figure 1.
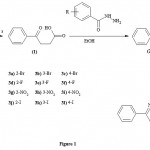 |
Figure 1
|
Preparation of 3-Benzoylpropionic acid (1)
It was prepared by the method provided in the literature.7
Preparation of 2-(2-bromobenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-one (2a)
3-Benzoylpropionic acid (1) (0.01 mole), 2-bromobenzohydrazide (0.01 mole), and ethanol (50 mL) were taken in a 100 mL round bottom flask. To this mixture sodium acetate (0.01 mole) was added and the mixture was refluxed for about 8 hours. The resulting mixture was reduced to half of its volume and then poured into the crushed ice. The solid separated was washed with water, and recrystallized from ethanol.
The compounds (2b-2l) were also prepared by similar method by reacting appropriate substituted benzohydrazides with 3-benzoylpropionic acid (1).
Preparation of (4Z)-4-benzylidene-2-(2-bromobenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-one (3a)
A mixture of 2-(2-bromobenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-one (2a) (0.01 mole) and benzaldehyde (0.01 mole) was taken in a round bottom flask containing 25 mL of absolute ethanol. Piperidine (0.5 mL) was added to the mixture and it was refluxed for about 8 hours. The mixture was reduced to half of its volume and then poured into the crushed ice. The solid separated was washed with water, and recrystallized from ethanol.
The compounds (3b-3l) were also prepared by similar method by reacting appropriate 2-(substitutedbenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-ones with benzaldehyde.
Molecular Modelling Studies
The compounds (3a-3l) were subjected to their molecular modelling studies. Angiotensin Converting Enzyme (ACE) is an enzyme that is involved in the pathogenesis of hypertension.8 The crystal structure of human Angiotensin Converting Enzyme (ACE) protein (PDB code: 1O86, Resolution – 2.00 Å) was retrieved from Protein Data Bank (PDB) and was utilized for molecular modelling studies.9 Protein was prepared with the Protein Preparation Wizard in Maestro using options: bond orders were assigned, hydrogen atoms were added, formal charges were treated and water molecules were deleted. The hydrogen bonding network was then optimized using the exhaustive sampling option and the protein was minimized to an RMSD limit from the starting structure of 0.3 Å using the Impref module of Impact with the OPLS_2005 force field. The prepared protein structure was used to generate Glide scoring grids for the subsequent docking calculations. Docking grids were generated with the default settings in Glide using the co-crystallized ligand to define the center of the grid box (20×20×20 Å). Default parameters were used and no constraints were included during grid generation. The three dimensional coordinates of the most active synthesized compound (3a) and lisinopril were generated using a Maestro module of Schrodinger. Ligands were prepared using LigPrep 2.6 with Epik 2.4 to expand protonation and tautomeric states at 7.0 ± 2.0 pH units and energy was minimized using the OPLS 2005 force field. The energy minimized ligands were docked to the protein structure using Glide XP docking since it incorporates structural and energetic information using the scoring function.10
Angiotensin Converting Enzyme Inhibitory Assay
The compound (3a) that showed highest ACE inhibitory activity according to the molecular modelling studies was subjected to ACE inhibitory assay using Dojindo ACE Kit-WST test kit, Dojindo Laboratories, Kumamoto, Japan.11,12 Briefly, The enzymatic reaction was initiated by the ACE and aminoacylase in the mixture containing 3HB-GGG (3-hydroxybutyrate glycylglycylglycine) and the ACE-inhibitor. The mixture was then incubated at 37oC for 60 minutes. During this incubation, the substrate, 3HB-GGG, was enzymatically cut into 3HB-G and G-G and then 3HB and G. The yield of 3HB was monitored indirectly through formazan concentration, which was measured at 450 nm after 10 minute reaction at 25oC. Testing procedures were run according to the manufacturer’s instructions using a 96-well plate without modification, and the inhibition rate was calculated based on a comparison of the optical absorbance of samples-treated wells (𝐴𝑠), control wells (𝐴𝑐), and blank wells (𝐴𝑏). Absorbance was measured at 450 nm using the microplate reader Biotek-ELX800 (BioTek, Vermont, USA). Inhibition rates were calculated using the following equation.
Inhibition rate (%) = [𝐴𝑐 – 𝐴𝑠 / 𝐴𝑐 − 𝐴𝑏] × 100
Samples were suspected to inhibit the ACE activity, and therefore inhibit the formation of formazan. The more strongly inhibitory the activity of the samples, the less color appeared in the final solution.
Statistical analysis
All ACE inhibitory activity data are presented as mean ± standard deviation (SD, n = 3). The data were analyzed by one-way analysis of variance (ANOVA) with Dunnett’s Multiple Comparison Test with respect to control group and standard groups using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego California USA). The results were considered significantly different at p < 0.05. The IC50 values were determined by linear regression calculator of GraphPad software.
Results and Discussion
Chemistry
The physical constants of the synthesized compounds (2a-2l) and (3a-3l) are summarized in Table 1.
Table 1: The physical constants of the compounds (2a-2l) and (3a-3l)
| Compd.
No. |
R | Molecular Formula | M.P. (±2oC) | Yield
(%) |
Rf Value |
| 2a | 2-Br | C17H13BrN2O2 | 112 | 60 | 0.72 |
| 2b | 3-Br | C17H13BrN2O2 | 133 | 60 | 0.69 |
| 2c | 4-Br | C17H13BrN2O2 | 145 | 75 | 0.80 |
| 2d | 2-F | C17H13FN2O2 | 156 | 55 | 0.66 |
| 2e | 3-F | C17H13FN2O2 | 125 | 70 | 0.75 |
| 2f | 4-F | C17H13FN2O2 | 155 | 44 | 0.70 |
| 2g | 2-NO2 | C17H13N3O4 | 170 | 50 | 0.67 |
| 2h | 3-NO2 | C17H13N3O4 | 141 | 45 | 0.69 |
| 2i | 4-NO2 | C17H13N3O4 | 158 | 40 | 0.79 |
| 2j | 2-I | C17H13IN2O2 | 177 | 55 | 0.68 |
| 2k | 3-I | C17H13IN2O2 | 151 | 65 | 0.72 |
| 2l | 4-I | C17H13IN2O2 | 132 | 50 | 0.81 |
| 3a | 2-Br | C24H17BrN2O2 | 133 | 50 | 0.90 |
| 3b | 3-Br | C24H17BrN2O2 | 142 | 45 | 0.82 |
| 3c | 4-Br | C24H17BrN2O2 | 177 | 60 | 0.77 |
| 3d | 2-F | C24H17FN2O2 | 144 | 60 | 0.72 |
| 3e | 3-F | C24H17FN2O2 | 119 | 50 | 0.76 |
| 3f | 4-F | C24H17FN2O2 | 166 | 55 | 0.69 |
| 3g | 2-NO2 | C24H17N3O4 | 143 | 70 | 0.71 |
| 3h | 3-NO2 | C24H17N3O4 | 138 | 65 | 0.83 |
| 3i | 4-NO2 | C24H17N3O4 | 146 | 60 | 0.75 |
| 3j | 2-I | C24H17IN2O2 | 190 | 65 | 0.68 |
| 3k | 3-I | C24H17IN2O2 | 179 | 70 | 0.70 |
| 3l | 4-I | C24H17IN2O2 | 194 | 45 | 0.83 |
The structures of the representative compounds 2a and 3a were confirmed on the basis of their IR, 1H-NMR, and 13C-NMR data. The spectral data of these compounds are given below.
2-(2-Bromobenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-one (2a)
IR (KBr) cm-1: 1675 and 1645 (C=O), 1615 (C=N), 1510 (C=C); 1H-NMR (DMSO-d6, 500 MHz) δ ppm: 2.45 (t, J = 8 Hz, 2H, -CO-CH2-), 2.96 (t, J = 8 Hz, 2H, -C(=N)-CH2-), 7.35-7.92 (m, 9H, Ar-H); 13C-NMR (DMSO-d6, 100 MHz) δ ppm: 23.1 (C5 of pyridazinone), 31.2 (C4 of pyridazinone), 117.7 (Ar-C), 126.7 (Ar-C), 127.1 (2C, Ar-C), 127.7 (2C, Ar-C), 130.0 (Ar-C), 130.1 (Ar-C), 130.6 (Ar-C), 131.7 (Ar-C), 135.3 (Ar-C), 135.6 (Ar-C), 146.8 (C6 of pyridazinone), 169.1 (C=O, C3 of pyridazinone)), 169.4 (C=O).
(4Z)-4-benzylidene-2-(2-bromobenzoyl)-6-phenyl-4,5-dihydropyridazin-3(2H)-one (3a)
IR (KBr) cm-1: 1680 and 1640 (C=O), 1610 (C=N), 1510 (C=C); 1H-NMR (DMSO-d6, 500 MHz) δ ppm: 2.85 (s, 2H, -C(=N)-CH2-), 6.81 (s, 1H, benzylidene-H of Ph-CH=C), 7.33-7.94 (m, 14H, Ar-H); 13C-NMR (DMSO-d6, 100 MHz) δ ppm: 33.3 (C4 of pyridazinone), 117.7 (Ar-C), 126.7 (2C, Ar-C), 127.1 (2C, Ar-C), 127.4 (4C, Ar-C), 127.7 (2C, Ar-C), 128.1 (C4 of pyridazinone), 130.0 (Ar-C), 130.1 (Ar-C), 130.6 (Ar-C), 131.7 (Ar-C), 133.0 (Ar-C), 134.1 (Ar-C), 135.6 (Ar-C), 137.1 (benzylidene-C of Ph-CH=C), 140.1 (C6 of pyridazinone), 153.8 (C=O, C3 of pyridazinone), 169.2 (C=O).
Molecular Modelling Studies
The molecular modelling studies revealed that almost all the functional groups of Lisinopril (docking score: -11.36; Figure 2) were occupied with either hydrogen bonding or other non-covalent lipophilic interactions and were important in exerting greater affinity towards ACE enzyme and ultimately ACE inhibitory activity. On the other hand, the compound (3a) (docking score: -6.54; Figure 3) was identified as the most active ACE inhibitor among the synthesized compounds. The compound (3b) (docking score: -5.29), compound (3c) (docking score: -4.68), compound (3d) (docking score: -5.06), compound (3e) (docking score: -5.44), compound (3f) (docking score: -4.44), compound (3g) (docking score: -5.87), compound (3h) (docking score: -5.93), compound (3i) (docking score: -5.07), compound (3j) (docking score: -4.60), compound (3k) (docking score: -3.41), and the compound (3l) (docking score: -4.14) also showed some ACE inhibitory activity. The ACE inhibitory activity of the compounds (3a-3l) by molecular docking score was not found significant when compared to the standard drug Lisinopril. In general terms, larger is the docking score in negative value, more is the active binding with the target receptor .e.g. active site of ACE.
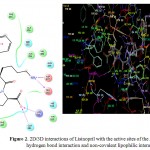 |
Figure 2: 2D/3D interactions of Lisinopril with the active sites of the ACE showing hydrogen bond interaction and non-covalent lipophilic interactions.
|
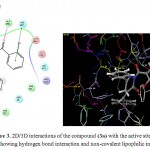 |
Figure 3: 2D/3D interactions of the compound (3a) with the active sites of the ACE showing hydrogen bond interaction and non-covalent lipophilic interactions.
|
Angiotensin Converting Enzyme Inhibitory Assay
Based on the molecular modelling results, the compound (3a) was selected for further in vitro ACE inhibitory assay using Dojindo ACE Kit-WST test kit, Dojindo Laboratories, Kumamoto, Japan. The docking score and the ACE inhibitory activity data of the compound (3a) with respect to the standard drug Lisinopril is provided in Table 2.
Table 2: In vitro ACE inhibitory activity of the Compound (3a) and standard drug Lisinopril
| Compound | Concentration
(μg/mL) |
%ACE Inhibition
(Mean ± SD) |
IC50
(μg/mL) |
Docking Score |
|
Lisinopril |
1 | 46.22 ± 0.32* |
0.99 |
-11.36 |
| 2 | 57.12 ± 0.11* | |||
| 4 | 66.92 ± 0.06* | |||
| 8 | 80.86 ± 0.16* | |||
|
Compound (3a) |
1 | 13.75 ± 0.11* |
4.40 |
-6.54 |
| 2 | 31.97 ± 0.12* | |||
| 4 | 59.64 ± 0.12* | |||
| 8 | 73.45 ± 0.19* |
*p < 0.0001; n = 3.
The in vitro ACE inhibitory activity revealed that the compound (3a) had IC50 value of 4.40 μg/mL, wherein the standard drug Lisinopril had IC50 value of 0.99 μg/mL. These results were also supported by the results of molecular modelling, wherein it was evident that the compound (3a) (Figure 3) did not bind effectively with the active site of the ACE enzyme as compared to the standard drug Lisinopril (Figure 2). This may be due to the fact that the compound (3a) does not have free functional groups that aid in the binding with the active site of the ACE enzyme. It is possible that the incorporation of free amino groups and free carboxylic acid groups in the structure of the compounds (3a-3l) may provide better and more potent ACE inhibitors. This finding is in accordance to our previously reported work.7
Conclusion
Based on the molecular modelling results and the in vitro enzymatic ACE inhibitory activity data, it is evident that the chemical structures of the title compounds need to be modified in order to increase their potency as inhibitors of the ACE enzyme. The incorporation of free amino group and free carboxylic acid groups in the chemical structures of the title compounds is recommended. Accordingly, further studies are in progress in our laboratory.
Conflict of interest
The authors declare no Conflict of Interest.
References
- Gaetano, S. Epidemiology of cardiovascular disease in the 21st century: Updated numbers and updated facts. J. Cardiovasc. Dis. 2013; 1(1): 1-2.
- Al-Sieni, A.I., Baghdadi, M,A,, Al-Abbasi, F.A. Country wide statistical assessment of smoking induced atherosclerotic and metabolic risk factors in Saudi population. Asian J. Pharm. Health Sci. 2014; 4(1): 916-20.
- El-Bcheraoui, C., Memish, Z.A., Tuffaha, M., Daoud, F., Robinson, M., Jaber, S., Mikhitarian, S., Al-Saeedi, M., Al-Mazroa, M.A., Mokdad, A.H., Al-Rabeeah, A.A. Hypertension and its associated risk factors in the Kingdom of Saudi Arabia, 2013: A national survey. Int. J. Hypertens. 2014; 1-8: Article ID 564679, http://dx.doi.org/10.1155/2014/564679.
CrossRef - Aljefree, N., Ahmed. F. Prevalence of cardiovascular disease and associated risk factors among adult population in the Gulf region: A systematic review. Adv. Public Health. 2015; 1-23: Article ID 235101,
http://dx.doi.org/10.1155/2015/235101.
CrossRef - Temilolu, O.A., Miller, M,. Cardiovascular disease: A global problem extending into the developing world. World J. Cardiol. 2009; 1(1): 3-10.
CrossRef - Reddy, K.S. Cardiovascular diseases in the developing countries: Dimensions, determinants, dynamics and directions for public health action. Public Health Nutr. 2002; 5(1A): 231-7.
CrossRef - Imran, M., Nayeem, N. Synthesis and antihypertensive activity of some novel pyridazinones. Orient. J. Chem., 2016; 32(1): 267-74.
CrossRef - Brown, N.J., Vaughan, D.E. Angiotensin-converting enzyme inhibitors. Circulation, 1998; 97: 1411-20.
CrossRef - Natesh, R., Schwager, S.L.U., Sturrock, E.D., Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature, 2003; 421: 551-4.
CrossRef - Loving, K., Salam, N.K., Sherman, W. Energetic analysis of fragment docking and application to structure-based pharmacophore hypothesis generation. J. Comput. Aided Mol. Des. 2009; 23: 541-554.
CrossRef - Lam, L.H., Shimamura, T., Manabe, S., Ishiyama, M., Ukeda H. Assay of angiotensin I-converting enzyme-inhibiting activity based on the detection of 3-hydroxybutyrate with water-soluble tetrazolium salt. Anal. Sci. 2008; 24(8): 1057-60.
CrossRef - Bang, T.H., Suhara, H., Doi, K., Ishikawa, H., Fukami, K., Parajuli, G.P., Katakura, Y., Yamashita, S., Watanabe, K., Adhikari, M.K., Manandhar, H.K., Kondo, R., Shimizu, K. Wild mushrooms in Nepal: Some potential candidates as antioxidant and ACE-Inhibition sources. Evid. Based Complement. Alternat. Med. 2014; 1-11: Article ID 195305, http://dx.doi.org/10.1155/2014/195305.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





