Published online Dec 26, 2019. doi: 10.12998/wjcc.v7.i24.4172
Peer-review started: September 18, 2019
First decision: October 14, 2019
Revised: November 27, 2019
Accepted: December 13, 2019
Article in press: December 13, 2019
Published online: December 26, 2019
Colorectal cancer (CRC) has been designated a major global problem, especially due to its high prevalence in developed countries. CRC mostly occurs sporadically (75%-80%), and only 20%-25% of patients have a family history. Several processes are involved in the development of CRC such as a combination of genetic and epigenetic alterations. Epigenetic changes, including DNA methylation play a vital role in the progression of CRC. Complex interactions between susceptibility genes and environmental factors, such as a diet and sedentary lifestyle, lead to the development of CRC. Clinical and experimental studies have confirmed the beneficial effects of dietary polyunsaturated fatty acids (PUFAs) in preventing CRC. From a mechanistic viewpoint, it has been suggested that PUFAs are pleiotropic agents that alter chromatin remodeling, membrane structure and downstream cell signaling. Moreover, PUFAs can alter the epigenome via modulation of DNA methylation. In this review, we summarize recent investigations linking PUFAs and DNA methylation-associated CRC risk.
Core tip: Polyunsaturated fatty acids, including ω-3 (eicosapentaenoic acid and docosahexaenoic acid) may have a potential preventive role in colorectal cancer (CRC) by changing DNA methylation. In this review after summarizing the latest knowledge regarding changes in the DNA methylation pattern and its association with CRC, we aim to highlight the link between polyunsaturated fatty acids and DNA methylation in CRC, which is currently an interesting field of research.
- Citation: Moradi Sarabi M, Mohammadrezaei Khorramabadi R, Zare Z, Eftekhar E. Polyunsaturated fatty acids and DNA methylation in colorectal cancer. World J Clin Cases 2019; 7(24): 4172-4185
- URL: https://www.wjgnet.com/2307-8960/full/v7/i24/4172.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i24.4172
Cancer is one of the leading causes of death worldwide, creating a global health problem[1]. The incidence of colorectal cancer (CRC) ranks third, it is among the most commonly diagnosed cancers, and the second leading cause of mortality[2]. More than 700000 people die annually due to CRC[3]. The incidence of CRC corresponds with age and a high risk of CRC is also associated with life in developed regions[4]. CRC develops through the synergistic effect of several genetic and epigenetic changes that lead to transformation of the normal intestinal epithelium into invasive adenocarcinoma[5]. Various genetic mechanisms are involved in the development of CRC. Defective DNA mismatch repair, presenting with microsatellite instability phenotypes and chromosomal instability, are the most important mechanisms. Today, the role of epigenetic alterations, such as the CpG island methylator phenotype (CIMP) is an important factor in the development of CRC[6]. Tumor suppressor genes (TSGs) can be influenced by abnormal de novo methylation in CRC, which is identified as a major epigenetic mechanism[7]. Aberrant hypermethylation, usually affects multiple loci in colorectal tumors, and it is referred to as the CIMP[8]. The correlations between CIMP status, clinical outcome and response to chemotherapy in CRC patients, have been documented[9,10]. In addition, several other molecular mechanisms for the resistance of CRC to chemotherapy have been reported[11,12]. Three active forms of DNA methyltransferases (DNMTs) including DNMT1, DNMT3A, and DNMT3B are responsible for the generation and maintenance of DNA methylation[13]. The functions of DNMT3A and DNMT3B enzymes are known as de novo methyltransferases[14]. However, DNMT1, is responsible for the maintenance of methyltransferase activity, which plays a role in the transmission of methylation patterns to daughter cells through cell divisions[15]. It is reported that various tumor tissues, such as breast and hepatocellular carcinomas, as well as cell lines, have elevated levels of DNMTs expression[16,17]. Various studies have documented the correlation between elevated expression of DNMTs in aberrant DNA methylation, and CIMP-associated colon cancer. Moreover, there is a significant association between DNMTs overexpression and CpG island hypermethylation, in CRC[15,18,19]. Unhealthy diet is an important environmental risk factor, which is involved in different types of cancers, especially the development of CRC[20]. Different food groups, including high meat consumption are associated with CRC incidence. Numerous studies have documented that high meat intake can increase the risk of CRC[21-23]. In contrast, a large body of evidence supports the protective effect of fish consumption against CRC risk. This is due to high levels of vitamin D and ω-3 fatty acids. Moreover, the positive role of other food groups, such as fruits and vegetables against CRC has been reported. Various studies show that this protective effect is related to the presence of polyphenolic compounds, such as flavonoids, and fiber[4]. Dietary fatty acids are important nutrients, which participate in human health and the prevention of diseases[24]. Polyunsaturated fatty acids (PUFAs) are a small group of dietary unsaturated fatty acids[25]. The role of PUFAs in the alleviation of tumor progression and cancer outcomes, especially CRC has been documented[26]. Recent evidence indicates that PUFAs, such as docosahexaenoic acid (DHA, 22:6Δ4,7,10,13,16,19), and eicosapentaenoic acid (EPA, 20:5Δ5,8,11,14,17) can significantly affect the epigenome status of cells[27]. It was also established that in CRC cells, PUFAs affect the activity of certain microRNAs via the associated alteration in promoter DNA methylation[28,29]. The exact mechanisms, explaining the effect of PUFAs on epigenetic modifications, and gene expression in human normal and cancer cells, are not well understood. In this review, after a brief description of the biology of DNA methylation, and the structure and metabolic role of PUFAs, we will focus on the pathophysiological mechanisms of DNA methylation involved in CRC, as well as linking PUFAs with DNA methylation in CRC, which is currently an interesting field of research.
The enzymatic transfer of a methyl group (CH3) to the 5-position of cytosine is called DNA methylation. This process is carried out by a family of DNMTs. In eukaryotes, cytosine residues are the only residues, in which DNA methylation occurs[30]. Five types of methyltransferases have been recognized: DNMT1, 2, 3A, 3B and DNMT-related protein (DNMT3L). With the exception of DNMT2 and DNMT3L, all have enzyme activity[31]. DNMT1, is recognized as a maintenance methyltransferase and participates in passing the epigenetic information through the exact transmission of methylation patterns to daughter cells, through cell generations[30]. DNMT3A and DNMT3B are involved in the maintenance of DNA methylation patterns through de novo methylation, and participate in various processes, such as correcting errors left by DNMT1 after DNA replication and dynamic demethylation; thus, DNMTs allow transient regulation of gene transcription[32-34].
DNMTs act on specified sequences of DNA, which are known as CpG dinucleotides. Generally, in mammalian cells, the DNA methylation of CpG dinucleotides occurs in normal cells, but the majority of CpGs are usually unmethylated, and designated in clusters known as CpG islands. Approximately, half of the genes in mammalian genomes consist of short CpG islands, which are important as they constitute less than 1% of the total genomic DNA[32,35]. CpG islands may be abnormally methylated in different types of cancer such as CRC[36]. In comparison with normal tissues, it has been rigorously established that DNMT1, DNMT3A, and DNMT3B are misdirected or up-regulated to different degrees in some malignancies, and there is a meaningful association between overexpression of DNMTs with CpG island hypermethylation in CRC tumors[16,17]. Conversely, many studies have shown the controversial role of increased expression of DNMTs in aberrant DNA methylation, and CIMP-associated colon cancer. However, our recent study indicated that no significant association was found between overexpression of DNMTs with specific CpG island hypermethylation in CRC cell lines[37].
Many studies have indicated that promoter-specific DNA hypermethylation of TSGs, and genomic global DNA hypomethylation, occur early in malignant, premalignant or precursor lesions. These gene modifications have potential clinical use as biomarkers, especially for the detection and screening of CRC[38]. For example, LINE-1 hypomethylation, as well as hypermethylation of TSGs, such as Vimentin and SEPT9 are the best hypermethylation DNA signatures for recognition of CRC[39-42]. These findings, including specific promoter hypermethylation and global DNA hypomethylation, are usual features, and help in understanding the importance of these abnormalities in cancer pathogenesis[43]. It has been established that epigenetic alteration machinery such as regional hypermethylation and global DNA hypomethylation, are accepted processes and hallmarks of cancer cells that may lead to modifications, including loss of imprinting, and are antecedents to the classical primary transforming events such as chromosomal instability, and mutations in tumor suppressors and proto-oncogenes[44]. Moreover, DNA hypermethylation of TSGs, involved in cell-cycle regulation, DNA repair, apoptosis, angiogenesis, adhesion, and invasion, is the most common change in tumorigenesis, causing gene silencing at the transcription level, and a failure of typical cellular functions[13,45-48]. On the other hand, DNA hypermethylation of CpG islands often affects transcriptional silencing of tumor suppressor or DNA repair genes, although there are exceptions[49-51]. Hypermethylation of TSGs is introduced as a general mechanism, which participates in tumor suppressor inactivation in cancer, and loss of tumor suppressor protein function has been reported in many tumor types[52,53]. Although methylation of CpG islands, adjacent to the transcription start sites of TSGs, is related to gene silencing, methylation of gene bodies is associated with activation of gene expression[52]. Furthermore, gene expression is not associated with methylation of the downstream gene sequences[54]. Recently, the role of epigenetic changes in cancer has been fully established. Cancer epigenome studies indicated that 1%-10% of CpG islands are abnormally methylated, suggesting that hundreds of genes might be aberrantly methylated in the CRC genome[35]. For example, hypermethylation of diverse TSGs, such as CDKN2A/p16, MLH1 and CDH1 (E-cadherin) and p14ARF, has been documented in the pathogenesis of CRC[37,55,56].
Global genomic DNA hypomethylation refers to loss of DNA methylation in various regions of the whole genome[57]. DNA hypomethylation is a hallmark, and one of the important features of cancer cell lines, such that 85% of cell lines are globally hypomethylated in different types of cancer, including CRC[57,58]. The significant role of global DNA hypomethylation in tumorigenesis is established, and it can occur at various genomic sequences, including repetitive elements, retrotransposons, CpG poor promoters, introns, and gene deserts[59]. In addition, growth stimulating genes, including R-Ras and MAPSIN in gastric cancer, S-100, IGF2, and repetitive sequences in CRC, and MAGE in melanoma can be activated by DNA hypomethylation[60-62]. Moreover, in colorectal tumors an increase, decrease or no change in global methylation status was reported, in comparison with their adjacent normal tissues. Alternative epigenetic progression pathways in tumors are controversial, as global hypomethylation is highly variable in tumor cells, and partially inversely correlated with microsatellite instability[58,63]. Furthermore, the carcinogenic effect of DNA hypomethylation may involve genomic instability, up-regulation of particular genes such as oncogenic microRNAs, translocations, facilitation of illegitimate mitotic recombination, and may also permit the transcription of parasitic sequences, including virus DNA and transposon elements that have been merged with the DNA[64,65].
Fatty acids are recognized as hydrocarbon chains with a methyl group at one end, and a carboxyl group at the other end. In saturated fatty acids, the carbon atoms connect with each other only by carbon-carbon single bonds. While, unsaturated fatty acids (UFAs) have one (monounsaturated fatty acid) or two or more (PUFAs) double bonds in their chain in the cis configuration.
Chemically, PUFAs are categorized as simple lipids. The ω-3 (EPA and DHA) and ω-6 linoleic acid (LA) are two main members of essential PUFAs, which have very different biochemical roles. Mammalian cells cannot synthesize LA and alpha-linolenic acid (α-LNA), due to the lack of required desaturase enzymes (∆12 and ∆15). Hence, these two PUFAs are categorized as essential, and need to be ingested via the diet. LA is present in vegetable seeds and oils, whereas, sources of α-LNA are dark green leafy plants and blackcurrant seed oils[66]. In addition, cold water-derived oily fish, particularly mackerel, salmon and sardines are dietary sources of EPA, and DHA[67,68]. Production of EPA and DHA from α-LNA occurs in the human body. However, endogenous production of the ω-3 PUFAs (EPA and DHA) from α-LNA by humans is very small, and almost non-significant (< 5%-10% for EPA and 2%-5% for DHA).
Dietary intake and various sources of fatty acids can influence the complex metabolism of PUFAs. In addition, in the endoplasmic reticulum, desaturase enzymes (∆6- and ∆5- desaturase) encoded by fatty acid desaturase 2 (FADS2) and FADS1, as well as elongase enzymes are encoded by ELOV5 and ELOV2, and are involved in metabolism[69]. Arachidonic acid and EPA are the first products, which are produced in the metabolic pathway. In the plasma membranes, PUFAs play a role as substrates for enzymes, including cyclooxygenase (COX) and lipooxygenase (LOX), and are converted into an eicosanoid. Irrespective of the mechanism, eicosanoids are highly biologically active hormone-like compounds that affect numerous metabolic activities such as inflammation, hemorrhage, blood pressure, platelet aggregation, immune responses and both vasoconstriction and vasodilatation[70]. It has been suggested that ingestion of EPA and DHA from fish oil and their metabolites, have a competitive function and replace arachidonic acid in phospholipids of the cell plasma membrane, and cause production of prostanoids and leukotrienes with various effects such as anti-inflammatory, anti-chemotactic and anti-tumor[71]. Moreover, the beneficial roles of ω-3 PUFAs have been established in cardiovascular diseases, myocardial infarction, inflammatory bowel disease, diabetes, rheumatoid arthritis, optimal brain function and neurodegenerative diseases[72-75].
The molecular mechanisms of LA, EPA, and DHA are not fully understood. However, many studies have indicated that these fatty acids show pleiotropic effects, and are major modulators of many genes[76]. It has been suggested that PUFAs and their derivatives change gene expression that leads to changes in membrane composition by precisely governing the activity of nuclear transcription factors such as peroxisome proliferator-activated receptor (PPAR)α, PPARβ, and PPARγ[77]. In addition to PPARs, different transcription factors have been recognized as targets for fatty acid regulation, such as hepatic nuclear factor-4α, sterol regulatory element-binding protein, liver X receptors, retinoid X receptors, thyroid hormone receptors (TR-α, TR-β) and nuclear factor-kappaB (NF-kB)[78-81]. For example, it is well established that treatment of human CRC cell lines with ω-3 PUFAs leads to increased membrane fluidity and lipid peroxidation, by acting as substrates for second messengers. On the other hand, these PUFAs reduced vascular endothelial cell growth factor (VEGF), β-catenin, BCL-2, and matrix metalloproteinase (MMP) gene expression levels, by activation of transcription factors such as PPARs. In addition, PUFAs reduced extracellular signal-regulated kinase-1/2 (ERK1/2) signaling[82,83].
Different mechanisms are involved in the beneficial effects of ω-3 PUFAs in cancer[84,85]. In vivo model experiments showed that low consumption of marine-derived ω-3 PUFAs and high intake of ω-6 PUFAs, elevate the risk of breast cancer in women[86,87]. Other studies indicated the crucial role of saturated fat from animal food sources in the increased risk of pancreatic cancer[88]. There is substantial evidence that ω-3 PUFAs (usually a mix of EPA and DHA as fish oil) have a potential role in the treatment and prevention of CRC, and it has been established that there is an inverse association between consumption of ω-3 PUFAs and the risk of CRC[89]. As mentioned above, sufficient consumption of PUFAs play a role as structural constituents of cellular membranes, and are involved in metabolism, inflammation, cell signaling, and regulation of gene expression[90]. Moreover, both EPA and DHA participate in the suppression of angiogenesis and the antineoplastic activity of ω-3 PUFAs is associated with negative regulation of stromal-epithelial cell signaling[91]. Single nucleotide polymorphisms (SNPs) are involved in the metabolism of PUFAs, and abnormal PUFA metabolism due to genetic variation plays a role in increasing cancer risk. It is possible that personalized-diets may be a therapeutic approach to provide specified intakes of PUFAs, based on an individual’s metabolic capability and physiological needs[92]. In FADS1 and FADS2, 330 and 942 identified SNPs have been found, respectively. According to these results, it has been suggested that these SNPs are related to the physiological levels of PUFAs, proposing that genetic variation participates in PUFA metabolism and is probably a cancer risk[69].
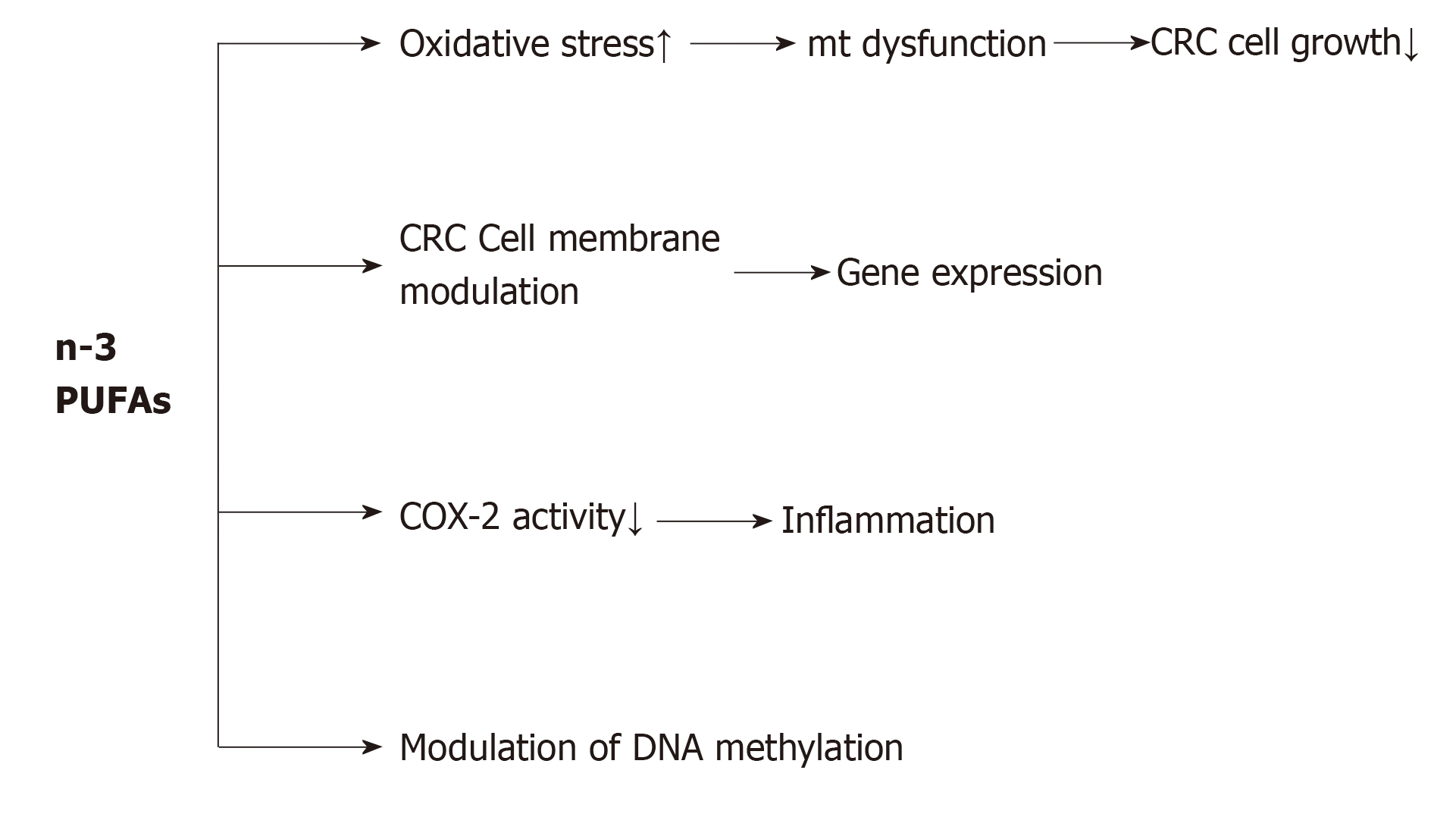
A large body of evidence supports the beneficial effects of dietary PUFAs, including EPA and DHA, for reducing cell proliferation and angiogenesis, and increasing apoptosis, which lead to a reduction in CRC risk. As shown in Figure 1, PUFAs induce an antineoplastic effect against CRC via four general mechanisms: (1) By increasing oxidative stress in colon cancer cells. Increased reactive oxygen species and malondialdehyde level, as end-products of PUFA peroxidation may be related to the progression and pathogenesis of CRC. From a mechanistic viewpoint, it has been suggested that increased reactive oxygen species and malondialdehyde level consequently leads to CRC cell growth inhibition through mitochondrial dysfunction[93]; (2) By changing the biochemical properties of cancer cell membranes such as fluidity, compressibility, fusion and protein function[94]. For example, in mammalian cells, PUFAs by interfering with membrane-associated Ras signaling could modulate gene expression, as well as DNA methylation. Another study indicated that in rats fed with a high-fat ω-3 PUFA diet, the total Ras protein and membrane-bound Ras levels decreased, but the protein levels of cytosolic Ras increased in colon tumors, in comparison with rats fed with a high-fat corn oil diet[95]. This shows that ω-3 PUFAs interfere with Ras activation by reducing its membrane localization. In addition, ω-3 PUFAs can protect against both the initiation and post-initiation stages of carcinogenesis. Davidson et al[96] concluded that consumption of ω-3 PUFAs has a protective effect on the initiation of CRC, and has a promotional effect on apoptosis and aberrant crypt foci levels. Furthermore, the role of fish oil supplementation in colonic apoptosis in rats has been established, conferring resistance to alkylation and oxidation-induced DNA damage[97]. Calviello et al[98] showed a more obvious reduction in cell number with EPA than with DHA, and indicated that both EPA and DHA reduced VEGF and COX-2 expression and prostaglandin E2 (PGE2) levels in CRC cells. (3) By reducing inflammation. Inflammation is one of the hallmarks of tumorigenesis[99]. Various investigations have revealed the anti- and pro-inflammatory mechanisms of PUFAs in cancer promotion and progression. For example, the anti-inflammatory mechanism of EPA is in competition with its ω-6 isomer for metabolism by COX and LOX enzymes, thus reducing the synthesis of pro-inflammatory PGE2 and LTB4. Subsequently, EPA metabolism by the COX pathway gives rise to PGE3, while the LOX pathway results in LTB5. Unlike the actions of PGE2, PGE3 is not involved in cancer cell proliferation, and instead down-regulates the expression of COX-2[100]. Moreover, EPA and DHA play an important role in the suppression of inflammatory transcription factors such as NF-kB; and (4) By modulation of DNA methylation. Many studies have confirmed the role of PUFAs in promoter methylation of different genes. It has been suggested that PGE2 modulates cancer progression via epigenetic modification. In CRC cells, PGE2, which acts in part via the PG receptor EP4, was found to increase the expression of DNMT1 and DNMT3, which led to hypermethylation of the promoter regions, and reduced the mRNA and protein expression levels of TSGs[101].
Increasing evidence suggests that dietary PUFAs affect cell function by modifying the epigenome, especially, DNA methylation[27]. There is a two-way correlation between PUFAs and epigenetics. This means that PUFAs can change the processes of epigenetics, and these epigenetic processes play an important role in the biosynthesis of PUFAs. For example, Niculescu et al[102] reported that DNA methylation of the promoter of the FADS2 gene, increased in the liver of the offspring of mice consuming a diet supplemented with α-LNA during pregnancy[102]. Furthermore, DNA hypermethylation of FADS2, and the levels of histone methylation in placentae, and in adipose tissue, were stimulated after increased fish oil intake. Additionally, nutritional ω-3 PUFA supplementation in pregnant women affected offspring DNA methylation through induction of genomic global DNA methylation in cord blood leukocytes, and a high-fat diet altered the DNA methylation status of target genes[103].
Aslibekyan et al[104] showed an association between numerous biologically important epigenetic markers, including regions on chromosomes 3, 10 and 16, and long-term consumption of seafood-derived ω-3 PUFAs. Another study demonstrated that treatment of U937 leukemic cells with EPA (100 µmol/L), increased the mRNA expression of CCAAT/enhancer-binding proteins C/EBP-β, C/EBP-ẟ, and c-Jun, which was accompanied by single specific locus demethylation of the C/EBP-ẟ promoter[105]. Moreover, the addition of DHA diminished the global level of dimethyl forms of H3K4, H3K9, H3K27, H3K36 and H3K79 in M17 neuroblastoma cells[106]. Furthermore, the beneficial effect of ω-3 PUFAs has been reported in the prevention of inflammatory disorders via epigenetic modulation of the immune system. For example, Lee et al[107] reported that ω-3 PUFA supplementation may modulate methylation levels in LINE1 repetitive elements, IFNγ and IL13, during pregnancy. Another investigation by Boigues et al[28] showed that the role of PUFAs in the modulation of gene promoters was linked with lipid metabolism, and regulation of the activity of specific microRNAs.
The anti-tumor effects of dietary PUFAs, especially EPA and DHA, on the reduced risk of CRC has been established. For advanced CRC, 5-fluorouracil-based chemotherapy is the first-line treatment[108]. Vasudevan et al[109] demonstrated that EPA by itself or in combination with other agents, including 5-fluorouracil could be a potential preventive strategy for recurring CRC, in both in vitro and in vivo models. Huang et al[110] demonstrated that treatment of a CRC rat model with ω-3 PUFA was accompanied by decreased tumor incidence and tumor size and a close correlation was found between the anticancer effects of ω-3 PUFA, and increased genomic DNA hydroxymethylation. Moreover, it has been suggested that PUFAs have a role in modulation of gene silencing by affecting gene promoter methylation. For instance, treatment of colon cancer cells with fish oil and pectin increased apoptosis, which was accompanied by increased methylation in the Bcl-2 promoter[111]. Ceccarelli et al[112] indicated that EPA directly regulates and demethylates DNA in hepatocarcinoma cell lines. Furthermore, Serini et al[94] showed that during treatment of the inflammatory response of the large bowel with ω-3 PUFAs altered M2 macrophage polarization, and may exert beneficial effects on gene expression through epigenetic modification. Overall, these data suggest that alterations in epigenetic pathways may be involved in the anticancer properties of ω-3 PUFAs.
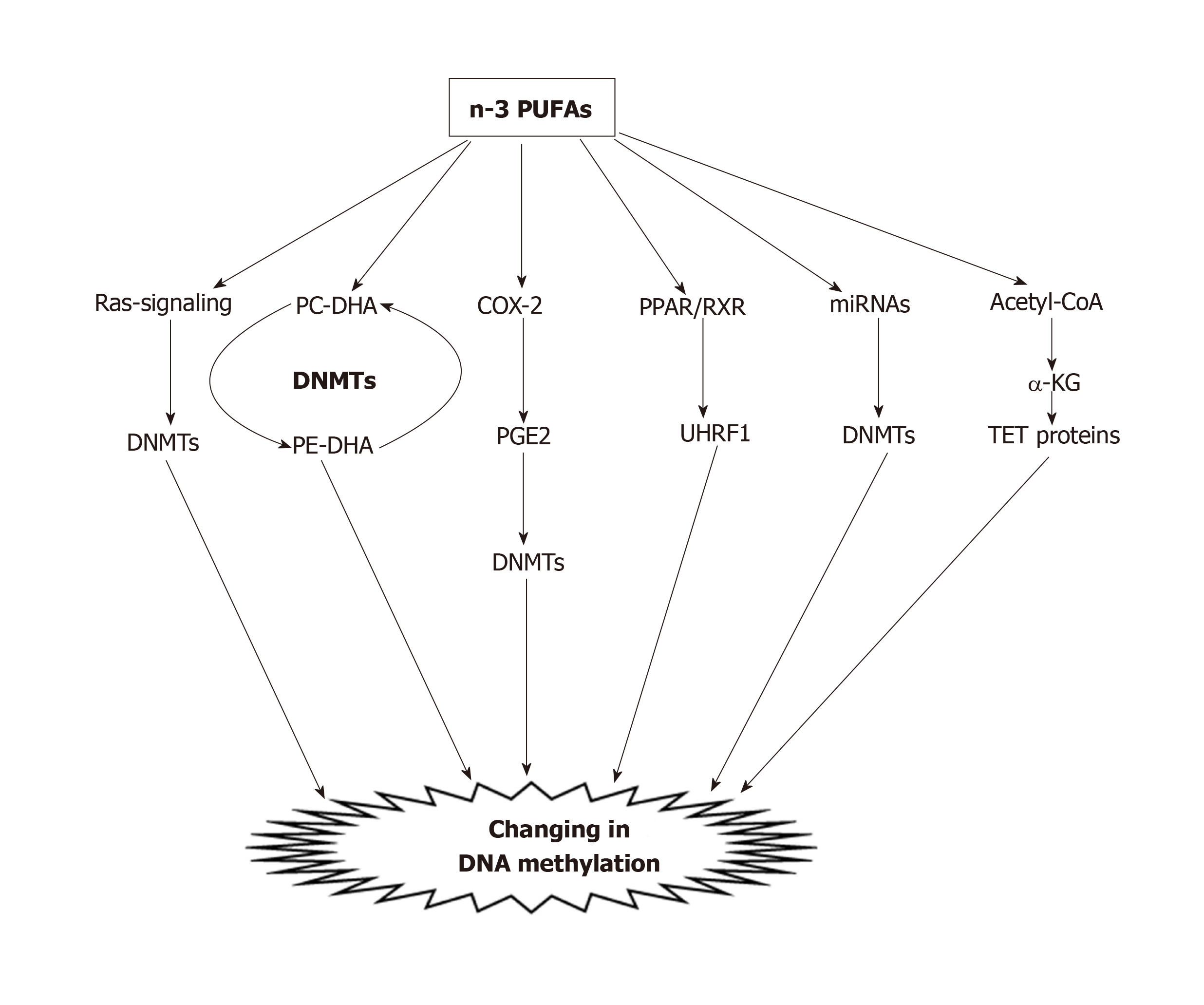
To date, many studies have demonstrated the effects of PUFAs on the DNA methylation processes of CRC in vitro. Our previous investigation showed that PUFA treatment, caused decreased methylation of COX-2, p16INK4a, PPARγ, CDH1, PTEN and MMP2 in CRC cell lines[113]. Other studies indicated that treatment of HCT116 colon cancer cells with DHA (50 µmol/L) or LA, was significantly accompanied by a reduction of both genomic global DNA methylation and DNA methylation of apoptosis-related genes, including Bcl2, Cideb, Dapk1, Ltbr, and Tnfrsf25[114]. The mechanism by which PUFAs alter the epigenome is currently unclear. Experimental studies revealed that there are different possible mechanisms (Figure 2) in the reduction of DNA methylation by PUFAs, including: (1) Via the Ras-signaling pathway. It has been shown that lipid rafts modulate signals generating from the cell membrane and linking DNA methylation and chromatin dynamics[115]. Activation of Ras stimulates DNMT1 mRNA transcription, and increasing levels of DNMT1 can affect hypermethylation of certain genes[116-118]. In our previous study, we observed different impacts of PUFAs on DNMT expression levels in five CRC cell lines. We found that EPA, DHA, and LA significantly reduced the expression levels of DNMT1 and DNMT3B in LS180 and HCT116 CRC cell lines, whereas DNMT1 expression was significantly induced in SW742 and HT29/29 cell lines. Moreover, with regard to DNMT3A we found a trend for coordinating a significant decrease in expression in five CRC cell lines, except the HT29/219 cell line[113]. The results of this study suggest that PUFAs can alter global and gene-specific DNA methylation, as well as the expression of DNMTs in a cell-type specific manner in CRC cells[113]; (2) By transferring methyl groups from phosphatidylethanolamine-DHA (PE-DHA) to phosphatidylcholine-DHA[119]. From a mechanistic viewpoint, methyl groups from S-adenosyl methionine are required for the conversion of PE-DHA to PC-DHA; therefore, a lack of cellular DHA leads to a deficiency of PE-DHA and the resulting excess of methyl groups will be available for other transmethylation reactions of DNA by DNMTs[29,119]; (3) By inhibiting COX-2 enzyme. COX-2 inhibition causes aberrant methylation and disturbs epigenetic regulation. PUFAs affect DNA methylation by inhibiting COX-2, and by decreasing the production of pro-inflammatory eicosanoids, and the risk of colon cancer[84,120]. Tsujii[121] showed that COX-2 induced hypermethylation of TSGs through increased DNMT expression via the production of PGE2. In another study by Tsujii[121], it was reported that COX-2 reduced DNMT activity, and increased global hypomethylation through an increase in demethylase expression and promoter hypomethylation of oncogenes. In addition, COX-2-derived PGE2 increases gene-specific and global DNA methylation through increased DNMT3A expression. These effects of PGE2 are tissue-specific, and enhancement of DNMT3A expression was facilitated by PGE2 signaling through its E prostanoid 2 receptors. In addition, COX-2-derived PGE2 elevated DNMT1 and DNMT3B protein expression, upregulated CpG island methylation, and stimulated intestinal tumor growth in APC -/+ mice. Moreover, it has been observed that DNMT1 and DNMT3B protein expression increased due to administration of PGE2 in colonic tumor epithelial cells, and elevated the number and size of intestinal polyps via DNA methylation[101]; (4) PUFA can bind to intracellular PPAR receptors. PPARs, such as PPARγ, bind as a heterodimer to the retinoic acid X receptor (RXR) and after heterodimerization of PPARγ with RXR, binds to the promoters of different genes and mediators such as UHRF1, which ultimately binds to the promoter regions of their target genes, and changes their expression by altering DNA methylation[122,123]. Our previous study indicated that n-3 PUFAs could modulate the expression of PPARα and DNMT3b in rat liver and colon tissues[124]. These findings suggest that this association could be created due to the interaction between the PUFAs and the epigenome, which causes reprogrammed epigenetic marks; (5) By modulating the gene expression of microRNAs (miRNAs). Zhang et al[125] demonstrated that in pregnant and lactating mice fed a high-fat diet, the expression of key miRNAs significantly reduced in the adult offspring. It has been documented that miRNAs can modify chromatin remodeling and DNA methylation by altering the activity of DNMTs, and histone methyltransferase[126,127]. Moreover, our previous study indicated that DHA (100 µmol/L) decreased DNA methylation of the miR-126 gene promoter, and inhibited its VEGF protein target level in a cell-type specific manner in CRC cell lines[29]. Thus, miRNAs represent a possible mechanism by which PUFAs modify the epigenome. However, such mechanisms await empirical evidence; and (6) By activation of ten-eleven translocation (TET) proteins. TET proteins are involved in the demethylation of 5-methylcytosine by sequential oxidation, and it has been well documented that TET proteins are positively activated by α-ketoglutarate[128]. The current findings clearly indicate that the activity of TET proteins might be enhanced by the metabolism of PUFAs. Increased dietary fatty acids may lead to β-oxidation of fatty acid. Accordingly, increased flux of acetyl-CoA derived from β-oxidation to the Krebs’s cycle, will give rise to an α-ketoglutarate concentration that may lead to up-regulation of TET enzymes, and DNA demethylation[129]. Based on these mechanisms, it has been suggested that PUFAs could induce alterations of DNA methylation by changing the activities of DNMTs and TET enzymes.
DNA methylation is a hallmark of cancer cells, and plays a pivotal role in the progression of CRC. DNA methylation can influence different pathways such as chromatin remodeling, gene expression, signaling transduction, and other signaling pathways, and could be a key step in CRC tumorigenesis. There is substantial evidence that PUFAs can modify the epigenome, and the beneficial properties associated with ω-3 PUFAs might be explained by DNA methylation. It has been suggested that PUFAs can induce alterations in DNA methylation by different mechanisms, including modulation of cancer cell membrane, and activities of intracellular PPARs, COX-2, noncoding-RNAs, DNMTs and TET proteins. However, there is a key limitation to progress in this field, which is due to the small changes in DNA methylation and their effects on gene expression, and the effect of PUFAs on less accessible tissues, including liver or brain. Further research is needed to provide potential novel insights into the mechanisms related to the influence of PUFAs on DNA methylation and CRC risk.
Manuscript source: Unsolicited manuscript
Manuscript source: Invited manuscript
Specialty type: Medicine, Research and Experimental
Country of origin: Iran
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Li Y, Qiu YD, Vynios D S-Editor: Zhang L L-Editor: Webster JR E-Editor: Ma YJ
| 1. | Grosso G, Bella F, Godos J, Sciacca S, Del Rio D, Ray S, Galvano F, Giovannucci EL. Possible role of diet in cancer: systematic review and multiple meta-analyses of dietary patterns, lifestyle factors, and cancer risk. Nutr Rev. 2017;75:405-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 269] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 2. | Zargar P, Ghani E, Mashayekhi FJ, Ramezani A, Eftekhar E. Acriflavine enhances the antitumor activity of the chemotherapeutic drug 5-fluorouracil in colorectal cancer cells. Oncol Lett. 2018;15:10084-10090. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 3. | Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3058] [Cited by in F6Publishing: 2876] [Article Influence: 410.9] [Reference Citation Analysis (1)] |
| 4. | Baena R, Salinas P. Diet and colorectal cancer. Maturitas. 2015;80:258-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 124] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 5. | Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116:762-774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 97] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 6. | Alwers E, Jia M, Kloor M, Bläker H, Brenner H, Hoffmeister M. Associations Between Molecular Classifications of Colorectal Cancer and Patient Survival: A Systematic Review. Clin Gastroenterol Hepatol. 2019;17:402-410.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 7. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3299] [Cited by in F6Publishing: 3278] [Article Influence: 192.8] [Reference Citation Analysis (0)] |
| 8. | Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681-8686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1834] [Cited by in F6Publishing: 1803] [Article Influence: 72.1] [Reference Citation Analysis (0)] |
| 9. | Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer. 2013;109:1004-1012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 10. | Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup? Int J Clin Oncol. 2008;13:498-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Naghibalhossaini F, Shefaghat M, Mansouri A, Jaberi H, Tatar M, Eftekhar E. The Impact of Thymidylate Synthase and Methylenetetrahydrofolate Reductase Genotypes on Sensitivity to 5-Fluorouracil Treatment in Colorectal Cancer Cells. Acta Med Iran. 2017;55:751-758. [PubMed] [Cited in This Article: ] |
| 12. | Eftekhar E, Jaberie H, Naghibalhossaini F. Carcinoembryonic Antigen Expression and Resistance to Radiation and 5-Fluorouracil-Induced Apoptosis and Autophagy. Int J Mol Cell Med. 2016;5:80-89. [PubMed] [Cited in This Article: ] |
| 13. | Moradi Sarabi M, Ghareghani P, Khademi F, Zal F. Oral Contraceptive Use May Modulate Global Genomic DNA Methylation and Promoter Methylation of APC1 and ESR1. Asian Pac J Cancer Prev. 2017;18:2361-2366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 14. | Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem. 2012;287:33116-33121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 126] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 15. | Ibrahim AE, Arends MJ, Silva AL, Wyllie AH, Greger L, Ito Y, Vowler SL, Huang TH, Tavaré S, Murrell A, Brenton JD. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60:499-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 16. | Girault I, Tozlu S, Lidereau R, Bièche I. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res. 2003;9:4415-4422. [PubMed] [Cited in This Article: ] |
| 17. | Oh BK, Kim H, Park HJ, Shim YH, Choi J, Park C, Park YN. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int J Mol Med. 2007;20:65-73. [PubMed] [Cited in This Article: ] |
| 18. | Nosho K, Shima K, Irahara N, Kure S, Baba Y, Kirkner GJ, Chen L, Gokhale S, Hazra A, Spiegelman D, Giovannucci EL, Jaenisch R, Fuchs CS, Ogino S. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15:3663-3671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 19. | Schmidt WM, Sedivy R, Forstner B, Steger GG, Zöchbauer-Müller S, Mader RM. Progressive up-regulation of genes encoding DNA methyltransferases in the colorectal adenoma-carcinoma sequence. Mol Carcinog. 2007;46:766-772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Willett WC. Diet and cancer: one view at the start of the millennium. Cancer Epidemiol Biomarkers Prev. 2001;10:3-8. [PubMed] [Cited in This Article: ] |
| 21. | Aykan NF. Red Meat and Colorectal Cancer. Oncol Rev. 2015;9:288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 22. | Carr PR, Jansen L, Walter V, Kloor M, Roth W, Bläker H, Chang-Claude J, Brenner H, Hoffmeister M. Associations of red and processed meat with survival after colorectal cancer and differences according to timing of dietary assessment. Am J Clin Nutr. 2016;103:192-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 23. | Zhao Z, Feng Q, Yin Z, Shuang J, Bai B, Yu P, Guo M, Zhao Q. Red and processed meat consumption and colorectal cancer risk: a systematic review and meta-analysis. Oncotarget. 2017;8:83306-83314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 68] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 24. | Park WJ. The Biochemistry and Regulation of Fatty Acid Desaturases in Animals. In: Burdge GC. Polyunsaturated Fatty Acid Metabolism. Elsevier 2018; 87-100. [DOI] [Cited in This Article: ] |
| 25. | Venegas-Calerón M, Sayanova O, Napier JA. An alternative to fish oils: Metabolic engineering of oil-seed crops to produce omega-3 long chain polyunsaturated fatty acids. Prog Lipid Res. 2010;49:108-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 150] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 26. | Gerber M. Omega-3 fatty acids and cancers: a systematic update review of epidemiological studies. Br J Nutr. 2012;107 Suppl 2:S228-S239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 27. | Burdge GC, Lillycrop KA. Fatty acids and epigenetics. Curr Opin Clin Nutr Metab Care. 2014;17:156-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 28. | Hernando Boigues JF, Mach N. The effect of polyunsaturated fatty acids on obesity through epigenetic modifications. Endocrinol Nutr. 2015;62:338-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Moradi Sarabi M, Zahedi SA, Pajouhi N, Khosravi P, Bagheri S, Ahmadvand H, Shahryarhesami S. The effects of dietary polyunsaturated fatty acids on miR-126 promoter DNA methylation status and VEGF protein expression in the colorectal cancer cells. Genes Nutr. 2018;13:32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 30. | Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1287] [Cited by in F6Publishing: 1327] [Article Influence: 147.4] [Reference Citation Analysis (0)] |
| 31. | Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1792] [Cited by in F6Publishing: 1681] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 32. | Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805-811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 556] [Cited by in F6Publishing: 523] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 33. | Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, Benes V, Jeltsch A, Gannon F, Salbert G. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 680] [Cited by in F6Publishing: 634] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 34. | Eftekhar E, Rasti M, Nahgibalhossaini F, Sadeghi Y. The Study of DNA Methyltransferase-3B Promoter Variant Genotype among Iranian Sporadic Breast Cancer Patients. Iran J Med Sci. 2014;39:268-274. [PubMed] [Cited in This Article: ] |
| 35. | Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686-700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 432] [Cited by in F6Publishing: 475] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 36. | Deng G, Chen A, Pong E, Kim YS. Methylation in hMLH1 promoter interferes with its binding to transcription factor CBF and inhibits gene expression. Oncogene. 2001;20:7120-7127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 59] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 37. | Sarabi MM, Naghibalhossaini F. Association of DNA methyltransferases expression with global and gene-specific DNA methylation in colorectal cancer cells. Cell Biochem Funct. 2015;33:427-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 38. | Chan AO, Broaddus RR, Houlihan PS, Issa JP, Hamilton SR, Rashid A. CpG island methylation in aberrant crypt foci of the colorectum. Am J Pathol. 2002;160:1823-1830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 180] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 39. | Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, Pretlow TP, Lutterbaugh J, Kasturi L, Willson JK, Rao JS, Shuber A, Markowitz SD. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124-1132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 267] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 40. | Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, Sontag S, Johnson D, Skoletsky J, Durkee K, Markowitz S, Shuber A. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 181] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 41. | Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, Levin B, Juhl H, Arber N, Moinova H, Durkee K, Schmidt K, He Y, Diehl F, Velculescu VE, Zhou S, Diaz LA, Kinzler KW, Markowitz SD, Vogelstein B. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 42. | Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, Schernhammer ES, Giovannucci EL, Fuchs CS. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734-1738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 276] [Cited by in F6Publishing: 289] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 43. | Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2049] [Cited by in F6Publishing: 1973] [Article Influence: 151.8] [Reference Citation Analysis (0)] |
| 44. | Feinberg AP. Cancer epigenetics is no Mickey Mouse. Cancer Cell. 2005;8:267-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 45. | Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2672] [Cited by in F6Publishing: 2493] [Article Influence: 155.8] [Reference Citation Analysis (0)] |
| 46. | Carmona FJ, Esteller M. Epigenomics of human colon cancer. Mutat Res. 2010;693:53-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 47. | Mokarram P, Zamani M, Kavousipour S, Naghibalhossaini F, Irajie C, Moradi Sarabi M, Hosseini SV. Different patterns of DNA methylation of the two distinct O6-methylguanine-DNA methyltransferase (O6-MGMT) promoter regions in colorectal cancer. Mol Biol Rep. 2013;40:3851-3857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Mokarram P, Kavousipour S, Sarabi MM, Mehrabani G, Fahmidehkar MA, Shamsdin SA, Alipour A, Naini MA. MGMT-B gene promoter hypermethylation in patients with inflammatory bowel disease - a novel finding. Asian Pac J Cancer Prev. 2015;16:1945-1952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 49. | Coppedè F. Epigenetic biomarkers of colorectal cancer: Focus on DNA methylation. Cancer Lett. 2014;342:238-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 50. | Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 314] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 51. | Mokarram P, Shakiba-Jam F, Kavousipour S, Sarabi MM, Seghatoleslam A. Promoter Methylation Status of Two Novel Human Genes, UBE2Q1 and UBE2Q2, in Colorectal Cancer: a New Finding in Iranian Patients. Asian Pac J Cancer Prev. 2015;16:8247-8252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 52. | Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 763] [Cited by in F6Publishing: 771] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 53. | Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891-11896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 700] [Cited by in F6Publishing: 732] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 54. | Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 498] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 55. | Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res. 1998;58:3455-3460. [PubMed] [Cited in This Article: ] |
| 56. | Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, Van Tornout JM, Jones PA. Methylation of the 5' CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531-4535. [PubMed] [Cited in This Article: ] |
| 57. | Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1361] [Cited by in F6Publishing: 1260] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 58. | Estécio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, Tajara EH, Issa JP. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 201] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 59. | Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capellà G, Ribas M, Peinado MA. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462-9468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 226] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 60. | Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138-162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 324] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 61. | Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753-1755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 555] [Cited by in F6Publishing: 505] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 62. | Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159-1161. [PubMed] [Cited in This Article: ] |
| 63. | Huang YW, Kuo CT, Stoner K, Huang TH, Wang LS. An overview of epigenetics and chemoprevention. FEBS Lett. 2011;585:2129-2136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 64. | Nakayama M, Wada M, Harada T, Nagayama J, Kusaba H, Ohshima K, Kozuru M, Komatsu H, Ueda R, Kuwano M. Hypomethylation status of CpG sites at the promoter region and overexpression of the human MDR1 gene in acute myeloid leukemias. Blood. 1998;92:4296-4307. [PubMed] [Cited in This Article: ] |
| 65. | Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol. 2005;83:296-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 168] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 66. | Roynette CE, Calder PC, Dupertuis YM, Pichard C. n-3 polyunsaturated fatty acids and colon cancer prevention. Clin Nutr. 2004;23:139-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 181] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 67. | Davis BC, Kris-Etherton PM. Achieving optimal essential fatty acid status in vegetarians: current knowledge and practical implications. Am J Clin Nutr. 2003;78:640S-646S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 161] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 68. | Burdge G. Alpha-linolenic acid metabolism in men and women: nutritional and biological implications. Curr Opin Clin Nutr Metab Care. 2004;7:137-144. [PubMed] [Cited in This Article: ] |
| 69. | Azrad M, Turgeon C, Demark-Wahnefried W. Current evidence linking polyunsaturated Fatty acids with cancer risk and progression. Front Oncol. 2013;3:224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 70. | Benatti P, Peluso G, Nicolai R, Calvani M. Polyunsaturated fatty acids: biochemical, nutritional and epigenetic properties. J Am Coll Nutr. 2004;23:281-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 230] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 71. | Clevers H. At the crossroads of inflammation and cancer. Cell. 2004;118:671-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 393] [Cited by in F6Publishing: 345] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 72. | Roth EM, Harris WS. Fish oil for primary and secondary prevention of coronary heart disease. Curr Atheroscler Rep. 2010;12:66-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 73. | Saravanan P, Davidson NC, Schmidt EB, Calder PC. Cardiovascular effects of marine omega-3 fatty acids. Lancet. 2010;376:540-550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 382] [Cited by in F6Publishing: 387] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 74. | Fetterman JW, Zdanowicz MM. Therapeutic potential of n-3 polyunsaturated fatty acids in disease. Am J Health Syst Pharm. 2009;66:1169-1179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 75. | Moradi Sarabi M, Doosti M, Einollahi N, Hesami SS, Dashti N. Effect of eicosapentaenoic acid on the expression of ABCG1 gene in the human monocyte THP-1 cells. Acta Med Iran. 2014;52:176-181. [PubMed] [Cited in This Article: ] |
| 76. | Deckelbaum RJ, Worgall TS, Seo T. n-3 fatty acids and gene expression. Am J Clin Nutr. 2006;83:1520S-1525S. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 221] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 77. | Jump DB, Clarke SD. Regulation of gene expression by dietary fat. Annu Rev Nutr. 1999;19:63-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 480] [Cited by in F6Publishing: 488] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 78. | Duplus E, Glorian M, Forest C. Fatty acid regulation of gene transcription. J Biol Chem. 2000;275:30749-30752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 274] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 79. | Deckelbaum RJ, Johnson RA, Worgall TS. Unsaturated fatty acids inhibit sterol regulatory element-dependent gene expression: a potential mechanism contributing to hypertriglyceridemia in fat-restricted diets. Proc Soc Exp Biol Med. 2000;225:184-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 80. | Sheng H, Shao J, Morrow JD, Beauchamp RD, DuBois RN. Modulation of apoptosis and Bcl-2 expression by prostaglandin E2 in human colon cancer cells. Cancer Res. 1998;58:362-366. [PubMed] [Cited in This Article: ] |
| 81. | Grygiel-Górniak B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications--a review. Nutr J. 2014;13:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 811] [Cited by in F6Publishing: 751] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 82. | Cockbain AJ, Toogood GJ, Hull MA. Omega-3 polyunsaturated fatty acids for the treatment and prevention of colorectal cancer. Gut. 2012;61:135-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 219] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 83. | Collett ED, Davidson LA, Fan YY, Lupton JR, Chapkin RS. n-6 and n-3 polyunsaturated fatty acids differentially modulate oncogenic Ras activation in colonocytes. Am J Physiol Cell Physiol. 2001;280:C1066-C1075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 107] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 84. | Chapkin RS, McMurray DN, Lupton JR. Colon cancer, fatty acids and anti-inflammatory compounds. Curr Opin Gastroenterol. 2007;23:48-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 100] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 85. | Calviello G, Serini S, Piccioni E. n-3 polyunsaturated fatty acids and the prevention of colorectal cancer: molecular mechanisms involved. Curr Med Chem. 2007;14:3059-3069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 86. | Thiébaut AC, Chajès V, Gerber M, Boutron-Ruault MC, Joulin V, Lenoir G, Berrino F, Riboli E, Bénichou J, Clavel-Chapelon F. Dietary intakes of omega-6 and omega-3 polyunsaturated fatty acids and the risk of breast cancer. Int J Cancer. 2009;124:924-931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 87. | Murff HJ, Shu XO, Li H, Yang G, Wu X, Cai H, Wen W, Gao YT, Zheng W. Dietary polyunsaturated fatty acids and breast cancer risk in Chinese women: a prospective cohort study. Int J Cancer. 2011;128:1434-1441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 88. | Thiébaut AC, Jiao L, Silverman DT, Cross AJ, Thompson FE, Subar AF, Hollenbeck AR, Schatzkin A, Stolzenberg-Solomon RZ. Dietary fatty acids and pancreatic cancer in the NIH-AARP diet and health study. J Natl Cancer Inst. 2009;101:1001-1011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 89. | Chen GC, Qin LQ, Lu DB, Han TM, Zheng Y, Xu GZ, Wang XH. N-3 polyunsaturated fatty acids intake and risk of colorectal cancer: meta-analysis of prospective studies. Cancer Causes Control. 2015;26:133-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 90. | Astorg P. Dietary N-6 and N-3 polyunsaturated fatty acids and prostate cancer risk: a review of epidemiological and experimental evidence. Cancer Causes Control. 2004;15:367-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 91. | Tsuzuki T, Shibata A, Kawakami Y, Nakagawa K, Miyazawa T. Conjugated eicosapentaenoic acid inhibits vascular endothelial growth factor-induced angiogenesis by suppressing the migration of human umbilical vein endothelial cells. J Nutr. 2007;137:641-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 92. | Schaeffer L, Gohlke H, Müller M, Heid IM, Palmer LJ, Kompauer I, Demmelmair H, Illig T, Koletzko B, Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745-1756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 398] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 93. | Cai F, Dupertuis YM, Pichard C. Role of polyunsaturated fatty acids and lipid peroxidation on colorectal cancer risk and treatments. Curr Opin Clin Nutr Metab Care. 2012;15:99-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 222] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 94. | Serini S, Ottes Vasconcelos R, Fasano E, Calviello G. Epigenetic regulation of gene expression and M2 macrophage polarization as new potential omega-3 polyunsaturated fatty acid targets in colon inflammation and cancer. Expert Opin Ther Targets. 2016;20:843-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 95. | Singh J, Hamid R, Reddy BS. Dietary fat and colon cancer: modulating effect of types and amount of dietary fat on ras-p21 function during promotion and progression stages of colon cancer. Cancer Res. 1997;57:253-258. [PubMed] [Cited in This Article: ] |
| 96. | Davidson LA, Nguyen DV, Hokanson RM, Callaway ES, Isett RB, Turner ND, Dougherty ER, Wang N, Lupton JR, Carroll RJ, Chapkin RS. Chemopreventive n-3 polyunsaturated fatty acids reprogram genetic signatures during colon cancer initiation and progression in the rat. Cancer Res. 2004;64:6797-6804. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 116] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 97. | Hong MY, Turner ND, Carroll RJ, Chapkin RS, Lupton JR. Differential response to DNA damage may explain different cancer susceptibility between small and large intestine. Exp Biol Med (Maywood). 2005;230:464-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 98. | Calviello G, Di Nicuolo F, Gragnoli S, Piccioni E, Serini S, Maggiano N, Tringali G, Navarra P, Ranelletti FO, Palozza P. n-3 PUFAs reduce VEGF expression in human colon cancer cells modulating the COX-2/PGE2 induced ERK-1 and -2 and HIF-1alpha induction pathway. Carcinogenesis. 2004;25:2303-2310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 99. | Balkwill F, Coussens LM. Cancer: an inflammatory link. Nature. 2004;431:405-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 492] [Cited by in F6Publishing: 491] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 100. | Bagga D, Wang L, Farias-Eisner R, Glaspy JA, Reddy ST. Differential effects of prostaglandin derived from omega-6 and omega-3 polyunsaturated fatty acids on COX-2 expression and IL-6 secretion. Proc Natl Acad Sci USA. 2003;100:1751-1756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 453] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 101. | Xia D, Wang D, Kim SH, Katoh H, DuBois RN. Prostaglandin E2 promotes intestinal tumor growth via DNA methylation. Nat Med. 2012;18:224-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 129] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 102. | Niculescu MD, Lupu DS, Craciunescu CN. Perinatal manipulation of α-linolenic acid intake induces epigenetic changes in maternal and offspring livers. FASEB J. 2013;27:350-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 103. | Bianchi M, Alisi A, Fabrizi M, Vallone C, Ravà L, Giannico R, Vernocchi P, Signore F, Manco M. Maternal Intake of n-3 Polyunsaturated Fatty Acids During Pregnancy Is Associated With Differential Methylation Profiles in Cord Blood White Cells. Front Genet. 2019;10:1050. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 104. | Aslibekyan S, Wiener HW, Havel PJ, Stanhope KL, O'Brien DM, Hopkins SE, Absher DM, Tiwari HK, Boyer BB. DNA methylation patterns are associated with n-3 fatty acid intake in Yup'ik people. J Nutr. 2014;144:425-430. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 105. | Ceccarelli V, Racanicchi S, Martelli MP, Nocentini G, Fettucciari K, Riccardi C, Marconi P, Di Nardo P, Grignani F, Binaglia L, Vecchini A. Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells. J Biol Chem. 2011;286:27092-27102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 106. | Sadli N, Ackland ML, De Mel D, Sinclair AJ, Suphioglu C. Effects of zinc and DHA on the epigenetic regulation of human neuronal cells. Cell Physiol Biochem. 2012;29:87-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 107. | Lee HS, Barraza-Villarreal A, Hernandez-Vargas H, Sly PD, Biessy C, Ramakrishnan U, Romieu I, Herceg Z. Modulation of DNA methylation states and infant immune system by dietary supplementation with ω-3 PUFA during pregnancy in an intervention study. Am J Clin Nutr. 2013;98:480-487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 113] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 108. | Eftekhar E, Naghibalhossaini F. Carcinoembryonic antigen expression level as a predictive factor for response to 5-fluorouracil in colorectal cancer. Mol Biol Rep. 2014;41:459-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 109. | Vasudevan A, Yu Y, Banerjee S, Woods J, Farhana L, Rajendra SG, Patel A, Dyson G, Levi E, Maddipati KR, Majumdar AP, Nangia-Makker P. Omega-3 fatty acid is a potential preventive agent for recurrent colon cancer. Cancer Prev Res (Phila). 2014;7:1138-1148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 110. | Huang Q, Mo M, Zhong Y, Yang Q, Zhang J, Ye X, Zhang L, Cai C. The Anticancer Role of Omega-3 Polyunsaturated Fatty Acids was Closely Associated with the Increase in Genomic DNA Hydroxymethylation. Anticancer Agents Med Chem. 2019;19:330-336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 111. | Cho Y, Turner ND, Davidson LA, Chapkin RS, Carroll RJ, Lupton JR. A chemoprotective fish oil/pectin diet enhances apoptosis via Bcl-2 promoter methylation in rat azoxymethane-induced carcinomas. Exp Biol Med (Maywood). 2012;237:1387-1393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 112. | Ceccarelli V, Valentini V, Ronchetti S, Cannarile L, Billi M, Riccardi C, Ottini L, Talesa VN, Grignani F, Vecchini A. Eicosapentaenoic acid induces DNA demethylation in carcinoma cells through a TET1-dependent mechanism. FASEB J. 2018;fj201800245R. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 113. | Sarabi MM, Naghibalhossaini F. The impact of polyunsaturated fatty acids on DNA methylation and expression of DNMTs in human colorectal cancer cells. Biomed Pharmacother. 2018;101:94-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 114. | Cho Y, Turner ND, Davidson LA, Chapkin RS, Carroll RJ, Lupton JR. Colon cancer cell apoptosis is induced by combined exposure to the n-3 fatty acid docosahexaenoic acid and butyrate through promoter methylation. Exp Biol Med (Maywood). 2014;239:302-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 115. | Patra SK, Szyf M. DNA methylation-mediated nucleosome dynamics and oncogenic Ras signaling: insights from FAS, FAS ligand and RASSF1A. FEBS J. 2008;275:5217-5235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 116. | Lund P, Weisshaupt K, Mikeska T, Jammas D, Chen X, Kuban RJ, Ungethüm U, Krapfenbauer U, Herzel HP, Schäfer R, Walter J, Sers C. Oncogenic HRAS suppresses clusterin expression through promoter hypermethylation. Oncogene. 2006;25:4890-4903. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 117. | MacLeod AR, Rouleau J, Szyf M. Regulation of DNA methylation by the Ras signaling pathway. J Biol Chem. 1995;270:11327-11337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 148] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 118. | Rouleau J, MacLeod AR, Szyf M. Regulation of the DNA methyltransferase by the Ras-AP-1 signaling pathway. J Biol Chem. 1995;270:1595-1601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 105] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 119. | Kulkarni A, Dangat K, Kale A, Sable P, Chavan-Gautam P, Joshi S. Effects of altered maternal folic acid, vitamin B12 and docosahexaenoic acid on placental global DNA methylation patterns in Wistar rats. PLoS One. 2011;6:e17706. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 157] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 120. | Zhang C, Yu H, Ni X, Shen S, Das UN. Growth inhibitory effect of polyunsaturated fatty acids (PUFAs) on colon cancer cells via their growth inhibitory metabolites and fatty acid composition changes. PLoS One. 2015;10:e0123256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 121. | Tsujii M. Cyclooxygenase, cancer stem cells and DNA methylation play important roles in colorectal carcinogenesis. Digestion. 2013;87:12-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 122. | Kota BP, Huang TH, Roufogalis BD. An overview on biological mechanisms of PPARs. Pharmacol Res. 2005;51:85-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 474] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 123. | Sabatino L, Fucci A, Pancione M, Colantuoni V. PPARG Epigenetic Deregulation and Its Role in Colorectal Tumorigenesis. PPAR Res. 2012;2012:687492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 124. | Maktoobian Baharanchi E, Moradi Sarabi M, Naghibalhossaini F. Effects of Dietary Polyunsaturated Fatty Acids on DNA Methylation and the Expression of DNMT3b and PPARα Genes in Rats. Avicenna J Med Biotechnol. 2018;10:214-219. [PubMed] [Cited in This Article: ] |
| 125. | Zhang J, Zhang F, Didelot X, Bruce KD, Cagampang FR, Vatish M, Hanson M, Lehnert H, Ceriello A, Byrne CD. Maternal high fat diet during pregnancy and lactation alters hepatic expression of insulin like growth factor-2 and key microRNAs in the adult offspring. BMC Genomics. 2009;10:478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 126. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805-15810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1300] [Cited by in F6Publishing: 1241] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 127. | Klein ME, Lioy DT, Ma L, Impey S, Mandel G, Goodman RH. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat Neurosci. 2007;10:1513-1514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 338] [Cited by in F6Publishing: 362] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 128. | Kaelin WG, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 613] [Cited by in F6Publishing: 622] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 129. | Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans. 2013;41:727-740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 174] [Article Influence: 15.8] [Reference Citation Analysis (0)] |