Abstract
Background
Bloom syndrome helicase (BLM) is overexpressed in multiple types of cancers and its overexpression may induce genomic instability. This study aimed to determine the function of BLM expression in pancreatic cancer.
Methods
BLM messenger RNA (mRNA) expression was analyzed using public datasets to determine its relationship with pancreatic cancer prognosis. Overall, 182 patients with pancreatic cancer who underwent radical resection at our institution were enrolled. BLM expression was evaluated by immunohistochemistry (IHC). We explored the effect of BLM on the proliferation, invasion, migration, and chemoresistance of pancreatic cancer cells via small-interfering RNAs and performed pathway analysis using gene set enrichment analysis.
Results
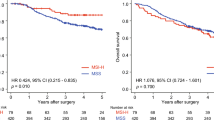
BLM mRNA expression was higher in tumor tissue than in normal tissue and had a prognostic effect on overall survival (OS) and recurrence-free survival. The same results were validated by IHC. Multivariate analysis showed that high BLM expression was an independent poor prognostic factor for OS (hazard ratio [HR] 1.678, p = 0.029). In subgroup analysis, the effect of high BLM expression was more significant on OS in patients with younger age (HR 2.27, p = 0.006), male sex (HR 2.39, p = 0.002), high cancer antigen 19-9 level (HR 2.44, p = 0.001), advanced tumor stage (HR 2.25, p = 0.001), lymph node metastasis (HR 2.51, p = 0.001), nerve invasion (HR 2.07, p = 0.002), and adjuvant chemotherapy (HR 2.66, p < 0.001). In vitro, BLM suppression resulted in reduced tumor proliferation, invasion, migration, and chemoresistance. Mechanistically, BLM expression may be associated with E2F1 and E2F2.
Conclusion
BLM expression is a prognostic factor for patients with pancreatic cancer, especially in those with advanced malignancies and receiving chemotherapy.








Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34.
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-386.
Gillen S, Schuster T, Meyer Zum Buschenfelde C, Friess H, Kleeff J. Preoperative/neoadjuvant therapy in pancreatic cancer: a systematic review and meta-analysis of response and resection percentages. PLoS Med. 2010;7(4):e1000267.
Winter JM, Brennan MF, Tang LH, et al. Survival after resection of pancreatic adenocarcinoma: results from a single institution over three decades. Ann Surg Oncol. 2012;19(1):169–75.
Oettle H, Neuhaus P, Hochhaus A, et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310(14):1473–81.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Sharma S, Doherty KM, Brosh RM Jr. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem J. 2006;398(3):319–37.
Brosh RM Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat Rev Cancer. 2013;13(8):542–58.
Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–52.
Cheok CF, Bachrati CZ, Chan KL, Ralf C, Wu L, Hickson ID. Roles of the Bloom’s syndrome helicase in the maintenance of genome stability. Biochem Soc Trans. 2005;33(Pt 6):1456–9.
Kaur E, Agrawal R, Sengupta S. Functions of BLM helicase in cells: is it acting like a double-edged sword? Front Genet. 2021;12:634789.
Ledet EM, Antonarakis ES, Isaacs WB, Lotan TL, Pritchard C, Sartor AO. Germline BLM mutations and metastatic prostate cancer. Prostate. 2020;80(2):235–7.
Ruan Y, Xu H, Ji X, Zhao J. BLM interaction with EZH2 regulates MDM2 expression and is a poor prognostic biomarker for prostate cancer. Am J Cancer Res. 2021;11(4):1347–68.
Alzahrani FA, Ahmed F, Sharma M, et al. Investigating the pathogenic SNPs in BLM helicase and their biological consequences by computational approach. Sci Rep. 2020;10(1):12377.
Votino C, Laudanna C, Parcesepe P, et al. Aberrant BLM cytoplasmic expression associates with DNA damage stress and hypersensitivity to DNA-damaging agents in colorectal cancer. J Gastroenterol. 2017;52(3):327–40.
Arora A, Abdel-Fatah TM, Agarwal D, et al. Transcriptomic and protein expression analysis reveals clinicopathological significance of bloom syndrome helicase (BLM) in breast cancer. Mol Cancer Ther. 2015;14(4):1057–65.
Mills CC, Kolb EA, Sampson VB. Development of chemotherapy with cell-cycle inhibitors for adult and pediatric cancer therapy. Cancer Res. 2018;78(2):320–5.
Slupianek A, Gurdek E, Koptyra M, et al. BLM helicase is activated in BCR/ABL leukemia cells to modulate responses to cisplatin. Oncogene. 2005;24(24):3914–22.
Nakagawa S, Okabe H, Sakamoto Y, et al. Enhancer of zeste homolog 2 (EZH2) promotes progression of cholangiocarcinoma cells by regulating cell cycle and apoptosis. Ann Surg Oncol. 2013;20(Suppl 3):S667-675.
Okabe H, Beppu T, Ueda M, et al. Identification of CXCL5/ENA-78 as a factor involved in the interaction between cholangiocarcinoma cells and cancer-associated fibroblasts. Int J Cancer. 2012;131(10):2234–41.
Nakao Y, Nakagawa S, Yamashita YI, et al. High ARHGEF2 (GEF-H1) expression is associated with poor prognosis via cell cycle regulation in patients with pancreatic cancer. Ann Surg Oncol. 2021;28(8):4733–43.
Du X, Zhang C, Yin C, et al. High BLM expression predicts poor clinical outcome and contributes to malignant progression in human cholangiocarcinoma. Front Oncol. 2021;11:633899.
Morkel M, Wenkel J, Bannister AJ, Kouzarides T, Hagemeier C. An E2F-like repressor of transcription. Nature. 1997;390(6660):567–8.
Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23(24):4709–16.
Ren B, Cam H, Takahashi Y, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16(2):245–56.
Xie L, Li T, Yang LH. E2F2 induces MCM4, CCNE2 and WHSC1 upregulation in ovarian cancer and predicts poor overall survival. Eur Rev Med Pharmacol Sci. 2017;21(9):2150–6.
Liang B, Zhao J, Wang X. Clinical performance of E2Fs 1–3 in kidney clear cell renal cancer, evidence from bioinformatics analysis. Genes Cancer. 2017;8(5–6):600–7.
Sun CC, Li SJ, Hu W, et al. Comprehensive analysis of the expression and prognosis for E2Fs in human breast cancer. Mol Ther. 2019;27(6):1153–65.
Zhan L, Huang C, Meng XM, et al. Promising roles of mammalian E2Fs in hepatocellular carcinoma. Cell Signal. 2014;26(5):1075–81.
Ma L, Tian X, Wang F, et al. The long noncoding RNA H19 promotes cell proliferation via E2F–1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2016;17(10):1051–61.
Schild C, Wirth M, Reichert M, Schmid RM, Saur D, Schneider G. PI3K signaling maintains c-myc expression to regulate transcription of E2F1 in pancreatic cancer cells. Mol Carcinog. 2009;48(12):1149–58.
Yamazaki K, Yajima T, Nagao T, et al. Expression of transcription factor E2F–1 in pancreatic ductal carcinoma: an immunohistochemical study. Pathol Res Pract. 2003;199(1):23–8.
Yao Z, Chen Q, Ni Z, et al. Long non-coding RNA differentiation antagonizing nonprotein coding RNA (DANCR) promotes proliferation and invasion of pancreatic cancer by sponging miR-214-5p to regulate E2F2 expression. Med Sci Monit. 2019;25:4544–52.
Sun FB, Lin Y, Li SJ, Gao J, Han B, Zhang CS. MiR-210 knockdown promotes the development of pancreatic cancer via upregulating E2F3 expression. Eur Rev Med Pharmacol Sci. 2018;22(24):8640–8.
Lin C, Hu Z, Yuan G, et al. MicroRNA-1179 inhibits the proliferation, migration and invasion of human pancreatic cancer cells by targeting E2F5. Chem Biol Interact. 2018;291:65–71.
Cam H, Balciunaite E, Blais A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16(3):399–411.
Acknowledgment
The authors would like to acknowledge TCGA, GEO, Kaplan–Meier Plotter, GSEA, and GEPIA2 databases, which were used free of charge. They would also like to thank Editage (www.editage.cn) for English language editing, and Yoko Ogata, Noriko Yasuda, and Lingfeng Fu for their technical assistance.
Funding
This work was supported by the Youth Project of the University-level Scientific Research Development Fund of North Sichuan Medical College (grant no. XJ2021007001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
Chuan Lan, Yo-ichi Yamashita, Hiromitsu Hayashi, Shigeki Nakagawa, Katsunori Imai, Kosuke Mima, Takayoshi Kaida, Takashi Matsumoto, Masataka Maruno, Zhao Liu, Xiyu Wu, Feng Wei, and Hideo Baba have declared no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lan, C., Yamashita, Yi., Hayashi, H. et al. High Expression of Bloom Syndrome Helicase is a Key Factor for Poor Prognosis and Advanced Malignancy in Patients with Pancreatic Cancer: A Retrospective Study. Ann Surg Oncol 29, 3551–3564 (2022). https://doi.org/10.1245/s10434-022-11500-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-11500-9