Abstract
Purpose
To compare breast cancer subtyping with the three centrally assessed microarray-based assays BluePrint, MammaPrint, and TargetPrint with locally assessed clinical subtyping using immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH).
Methods
BluePrint, MammaPrint, and TargetPrint were all performed on fresh tumor samples. Microarray analysis was performed at Agendia Laboratories, blinded for clinical and pathological data. IHC/FISH assessments were performed according to local practice at each institution; estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) assessments were performed on 132 samples, and Ki-67 on 79 samples.
Results
The concordance between BluePrint and IHC/FISH subtyping was 94 % for the Luminal-type, 95 % for the HER2-type, and 94 % for the Basal-type subgroups. The concordance of BluePrint with subtyping using mRNA single gene readout (TargetPrint) was 96 % for the Luminal-type, 97 % for the HER2-type, and 98 % for the Basal-type subgroups. The concordance for substratification into Luminal A and B using MammaPrint and Ki-67 was 68 %. The concordance between TargetPrint and IHC/FISH was 97 % for ER, 80 % for PR, and 95 % for HER2.
Conclusions
The implementation of multigene assays such as TargetPrint, BluePrint, and MammaPrint may improve the clinical management of breast cancer patients. High discordance between Luminal A and B substratification based on MammaPrint versus locally assessed Ki-67 or grade indicates that chemotherapy decisions should not be based on the basis of Ki-67 readout or tumor grade alone. TargetPrint serves as a second opinion for those local pathology settings where high-quality standardization is harder to maintain.
Similar content being viewed by others
Breast cancer is no longer considered to be a single disease but rather a diverse and heterogeneous group of diseases. Simple hierarchical clustering of breast tumors according to their gene expression patterns has led to the identification of molecular subtypes.1 The different molecular subtypes of breast cancer were found to have different prognoses and response to treatment regimens even though this classification system had been developed without consideration for patient survival rates. Luminal, human epidermal growth factor receptor 2 (HER2), and Basal-type subgroups are distinguished by the differential expression of the estrogen receptor (ER), progesterone receptor (PR), and HER2. Luminal-type tumors have a more favorable outcome, whereas Basal-type and HER2-type subgroups are more sensitive to chemotherapy. As a result, the management of breast cancer needs to take into consideration the different molecular subtypes to properly assess the likely prognosis and determine the most appropriate therapy.2 Molecular classification is thus becoming increasingly important in guiding treatment decisions.
At present, the methodology for molecular subtyping is not standardized, and the methodology and interpretation of results varies between different laboratories.3 , 4 Breast cancer subtype identification is being assessed clinically by immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) or molecularly by gene expression profiling.1 , 5 – 7 One of the more recently available molecular profiles is BluePrint. This molecular subtyping profile has been developed using a rational-based model to ensure a robust and reproducible profile with concordant IHC/FISH and mRNA (TargetPrint) assessed samples for ER, PR, and HER2. BluePrint is a molecular subtyping profile that determines the mRNA levels of 80 genes that best discriminate between three breast cancer subtypes and was validated using four independent validation cohorts with 784 patients.8 A further stratification of the Luminal group into types A and B is important in determining the need for chemotherapy and is thought to be based on the expression of markers of tumor grade and/or proliferation, such as Ki-67. However, risk stratification by multigene assays such as MammaPrint or Oncotype Dx are preferred for making this distinction, whereby the MammaPrint Low-Risk patients are Luminal A and MammaPrint High-Risk patients are equivalent to Luminal B.9
TargetPrint is an additional test that is an alternative measurement of ER, PR, and HER2 to IHC/FISH assessment and can be used in conjunction with MammaPrint and BluePrint. TargetPrint measures mRNA levels to provide a quantitative result. The mRNA readout has been shown to be robust and reproducible in a number of studies.10 , 11 Further, it can serve as a reliable second pathologic assessment for locally assessed parameters, especially because it is part of a multiprofile platform for breast cancer treatment management.12
Taken together, the diagnostic multigene array featuring >4,500 genes is a platform enabling multiple gene expression profiles to be analyzed simultaneously from one tumor specimen. MammaPrint is a 70-gene profile for prognostic and predictive tumor analysis; TargetPrint offers a microarray-based readout of ER, PR, and HER2 (single gene readout) and provides results comparable to IHC and FISH for providing objective and quantitative assessment of tumor receptor status. BluePrint is an 80-gene profile (four genes overlapping with MammaPrint), developed with a rational-based method, specifically to discriminate between the molecular subtypes Luminal, HER2, and Basal. Molecular subtyping is currently being used in clinical trials to help stratify patients, anticipating that such information will have a beneficial effect on patient outcomes. The scalability of the platform ensures the simultaneous detection and measurement of hundreds of normalization and control genes. It also exemplifies the versatility of gene expression profiling in the diagnostic setting, enabling the addition of many more profiles (such as future drug response profiles).
With standardization still pending, a comparison of the different methods for molecular classification of early stage breast cancer is of increased interest. The tumor samples for which different methodologies are discordant are of particular interest because they could indicate which method is more appropriate for diagnosing different patient groups and could potentially change prognosis and treatment. The added value of using a rational-based profile such as BluePrint is that the BluePrint profile captures the downstream processes involved in the specific subtype, rather than just the initiating gene or protein, which could be present without being functional (Fig. 1).
Methods for measuring ER activity: The added value of using a rational-based profile such as BluePrint is that the BluePrint profile captures the downstream processes involved in the specific subtype, rather than just the initiating gene or protein, which could potentially be present (and therefore positive as measured by IHC and TargetPrint) without being functional. ER estrogen receptor, mRNA messenger RNA, IHC immunohistochemistry
The objective of this study was to compare subtyping results obtained with the three centrally assessed microarray-based assays BluePrint, MammaPrint, and TargetPrint with locally assessed clinical subtyping using IHC/FISH and to investigate potential discordances between the different techniques.
Patients and Methods
Patients
Samples from patients with a tumor size ≤5 cm, up to three positive lymph nodes, and stage T1-4, N0-3 disease were eligible for inclusion. All patients provided written informed consent for their tumor samples to be evaluated in the study.
Tumor Samples
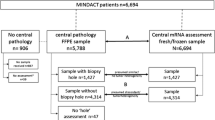
Fresh tumor samples were obtained from surgical specimens (minimum 3 mm3 tumor tissue) or core needle biopsies (two cores of tumor tissue from a 14-gauge needle or one core from a 10–12-gauge needle). Fresh tissue samples were placed directly into RNARetain (Asuragen, Austin, TX, USA) preservative. Tumor samples were prospectively obtained from 135 breast cancer patients between December 2008 and December 2011 at 11 institutions in the United States and Europe.
Molecular and Clinical Subtyping
BluePrint, MammaPrint, and TargetPrint were all performed on fresh tumor samples. Microarray analysis (RNA labeling, microarray hybridization, and scanning) for obtaining the profiles was performed at the centralized Agendia Laboratories blinded for clinical and pathological data. RNA was cohybridized with a standard reference to the custom-designed diagnostic chip, each containing oligonucleotide probes for the profiles in triplicate or more.13 IHC/FISH assessments were performed according to local practice at each institution. The threshold for ER and PR was set at 1 % positive staining. The threshold for HER2 3+ was 10 % or more positive staining and HER2 2+ cases were assessed by FISH. HER2 3+ cases were considered positive. FISH was performed on HER2 2+ by IHC (n = 11). The cutpoint of Ki-67 at 14 % was recommended by the St. Gallen’s expert consensus panel and was derived from comparison with gene-array data as a prognostic factor.9 , 14
Pathological ER, PR, and HER2 assessments were performed on 132 samples and Ki-67 on 79 samples (Ki-67 was not regularly performed in all participating centers). A clinical subtype classification was possible for 85 patients.
Clinical characteristics are shown in Table 1. According to BluePrint subtyping, 114 patients were Luminal-type, 6 HER2-type, and 15 Basal-type. Luminal-type patients were further substratified by MammaPrint as Low Risk (n = 50, equivalent to Luminal A) or High Risk (n = 64, equivalent to Luminal B).
Statistical Analysis
Agreement measurements between binary microarray and IHC classifications were based on two-way contingency table analysis and included overall concordance and Cohen’s κ coefficient scores; a score of >0.75 is considered excellent, 0.40–0.75 is fair to good, and <0.40 is poor agreement. All measurements were associated with a 95 % confidence interval (CI).
Results
Comparison of Subtyping with BluePrint and IHC/FISH
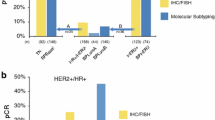
The results of BluePrint and IHC/FISH subtyping for 85 patients are listed in Table 2. Of 68 patients classified by BluePrint as Luminal-type, 65 were Luminal (IHC hormone receptor positive [HR+], HER2−), two were HER2 (IHC/FISH HR−, HER2+), and one was Basal (IHC/FISH HR−, HER2−) according to clinical subtyping. All six patients classified by BluePrint as HER2 type were HER2 (IHC/FISH HR−, HER2+) according to clinical subtyping. Of 11 patients classified by BluePrint as Basal, two were Luminal (IHC HR+, HER2−), two were HER2 (IHC/FISH HR−, HER2+), and seven were Basal (IHC/FISH HR−, HER2−) according to clinical subtyping. The concordance between BluePrint and IHC/FISH subtyping was 94 % for the Luminal-type subgroup, with a κ score of 0.82 (95 % CI 0.67–0.97); 95 % for the HER2-type subgroup, with a κ score of 0.73 (95 % CI 0.46–0.99); and 94 % for the Basal-type subgroup, with a κ score of 0.70 (95 % CI 0.45–0.96).
Concordance Between Molecular Subtyping Profile BluePrint, Single Gene mRNA Readout by TargetPrint, and Locally Assessed IHC/FISH
The concordance between BluePrint, TargetPrint, and IHC/FISH is shown in Fig. 2. Of 114 patients classified by BluePrint as Luminal-type, 110 were ER+ and/or PR+ and four were HER2+ according to TargetPrint. According to clinical subtyping, 109 patients were ER+ and/or PR+, two were HER2+, and one was ER−, PR−, and HER2−. All six patients classified by BluePrint as HER2 type were also HER2+ according to TargetPrint and clinical subtyping. Of 15 patients classified by BluePrint as Basal-type, 13 were TargetPrint triple negative, and two were TargetPrint ER+ and/or PR+. Clinical subtyping classified eight patients as triple negative, five were ER+ and/or PR+, and two were HER2+. The concordance of BluePrint with IHC/FISH subtyping was 93 % for the Luminal-type subgroup, with a κ score of 0.76 (95 % CI 0.61–0.92), 96 % for the HER2-type subgroup, with a κ score of 0.74 (95 % CI 0.48–0.99), and 93 % for the Basal-type subgroup, with a κ score of 0.64 (95 % CI 0.39–0.88). The concordance of BluePrint with TargetPrint was slightly higher for all three subgroups: 95 % for the Luminal-type, with a κ score of 0.84 (95 % CI 0.71–0.96); 97 % for the HER2 type, with a κ score of 0.74 (95 % CI 0.48–0.99); and 98 % for the Basal-type, with a κ score of 0.92 (95 % CI 0.81–1.00).
Substratification of Luminal-Type into Luminal A and B Based on MammaPrint or Ki-67 IHC
The results for substratification of the Luminal-type subgroup using MammaPrint and Ki-67 (n = 65) are shown in Table 2. Of 35 Luminal-type patients classified by MammaPrint as Low Risk, 26 were clinically Luminal A (IHC HR+, HER2−, Ki-67 low [<14 %]) and nine were clinically Luminal B (IHC HR+, HER2−, Ki-67 high). Of 30 Luminal-type patients classified by MammaPrint as High Risk, 12 were Luminal A (IHC HR+, HER2−, Ki-67 low [<14 %]) and 18 were Luminal B (IHC HR+, HER2−, Ki-67 high) according to clinical subtyping. The concordance between MammaPrint and Ki-67 was 68 %, with a κ score of 0.35 (95 % CI 0.11–0.58).
Of the Luminal patients, 66 % were grade 2. Six Luminal MammaPrint Low-Risk patients were grade 3 and 11 Luminal MammaPrint High-Risk patients were grade 1, resulting in a 52 % discordance between MammaPrint and grade, with a κ score of 0.07 (95 % CI 0.00–0.39).
Comparison of Centrally Assessed Single Gene mRNA Readout of ER, PR, and HER2 by TargetPrint and Locally Assessed IHC/FISH
Side by side TargetPrint single gene mRNA readout and local IHC/FISH data for ER, PR, and HER2 was available for 132 patients (Table 3). Of 116 patients classified as ER+ by TargetPrint, 115 were ER+ and one was ER− by IHC. Of 16 patients classified as ER− by TargetPrint, three were ER+ and 13 were ER− by IHC. Of 94 patients classified as PR+ by TargetPrint, 83 were PR+ and 11 were PR− by IHC. Of 38 patients classified as PR− by TargetPrint, 16 were PR+ and 22 were PR− by IHC. Of 10 patients classified as HER2+ by TargetPrint, seven were HER2+ and three were HER2− by IHC/FISH. Of 122 patients classified as HER2− by TargetPrint, three were HER2+ and 119 were HER2− by IHC/FISH. The concordance between TargetPrint and IHC was 96 % for ER, with a κ score of 0.85 (95 % CI 0.70–0.99), 79 % for PR with a κ score of 0.48 (95 % CI 0.31–0.66), and 95 % for HER2, with a κ score of 0.68 (95 % CI 0.42–0.93).
Discussion
Apart from high concordance between BluePrint and clinical subtyping by IHC/FISH for the Luminal-type, HER2-type, and Basal-type subgroups, the discordance for the substratification into Luminal A and B was considerable. The distinction between Luminal A versus Luminal B is considered to be the threshold for advising treatment with adjuvant chemotherapy and is thus critical in the diagnosis of breast cancer. Both Ki-67 and grade are commonly used surrogate markers for this Luminal A/B substratification. The current study compared these surrogate markers to MammaPrint, the preferred method of substratification. Locally assessed Ki-67 was discordant in 33 % of patients and tumor grade was discordant in 48 % of patients. This indicates that even though Ki-67 and/or grade are assumed to be fairly reliable measures of proliferation, they cannot substitute for a multigene assay such as MammaPrint in tailoring chemotherapy treatment decisions.
The high concordance between BluePrint and clinical subtyping (93 % for the Luminal-type subgroup, κ score of 0.76, 95 % CI 0.61–0.92; 96 % for the HER2-type, κ score of 0.74, 95 % CI 0.48–0.99; and 93 % for the Basal-type, κ score of 0.64, 95 % CI 0.39–0.88) implies that high-quality, locally assessed IHC/FISH can, to an extent, be used in the absence of molecular subtyping. However, the discordant cases will ultimately provide insight into the optimal methods for identifying specific prognostic and treatment response information for individual patients. A previous study reported 0.6 and 8 % of clinically ER+ tumors to be Basal-like or HER2-like, respectively.6 Also, the concordance of subtyping with TargetPrint is higher in comparison to locally assessed IHC/FISH, indicating that IHC/FISH assessment could still be improved. The observed subtype discrepancies reveal a potentially important impact on treatment decision making.
There was high concordance between TargetPrint and IHC/FISH for ER (96 %, κ score of 0.85, 95 % CI 0.70–0.99) and HER2 (95 %, κ score of 0.68, 95 % CI 0.42–0.93), and fair concordance for PR (79 %, κ score of 0.48, 95 % CI 0.31–0.66). These results are similar to those reported in other studies, with concordance of approximately 97 % for ER, 94 % for HER2, and 86 % for PR.11 , 15 In the largest TargetPrint comparison to high-quality centralized pathology assessment in the first 800 MINDACT patients, concordance was found to be 98, 96, and 85 % for ER, HER2, and PR, respectively, indicating that TargetPrint provides an objective and quantitative assessment of tumor receptor status and can provide a second pathology assessment for locally assessed parameters.12 In the current study, the high concordance between TargetPrint and IHC/FISH shows the high standard of pathology in the participating hospitals in general. However, the range for ER assessment between hospitals was 88–100 %, indicating that further standardization in local pathology assessment is warranted for optimal diagnosis in early stage breast cancer. The range for PR assessment between hospitals was 63–100 and 75–100 % for HER2 assessment.
In conclusion, there was high concordance between BluePrint and clinical subtyping by IHC/FISH for the Luminal-type, HER2-type, and Basal-type subgroups and between TargetPrint and IHC/FISH for ER and HER2. The implementation of multigene assays such as TargetPrint, BluePrint, and MammaPrint may improve the clinical management of breast cancer patients. High discordance between Luminal A and B stratification based on MammaPrint versus locally assessed Ki-67 indicates that, within the Luminal subgroup of patients, chemotherapy decisions should not be based on Ki-67 readout or tumor grade alone. TargetPrint serves as a second opinion for those local pathology settings where high-quality standardization is difficult to maintain.
References
Perou CM, Sørlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer, 2011. Ann Oncol. 2011;22:1736–47.
Arihiro K, Umemura S, Kurosumi M, et al. Comparison of evaluations for hormone receptors in breast carcinoma using two manual and three automated immunohistochemical assays. Am J Clin Pathol. 2007;127:356–65.
Oyama T, Ishikawa Y, Hayashi M, Arihiro K, Horiguchi J. The effects of fixation, processing and evaluation criteria on immunohistochemical detection of hormone receptors in breast cancer. Breast Cancer. 2007;14:182–8.
Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7(5):e1000279.
Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–32.
Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800.
Krijgsman O, Roepman P, Zwart W, et al. A diagnostic gene profile for molecular subtyping of breast cancer associated with treatment response. Breast Cancer Res Treat. 2012;133:37–47.
Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thürlimann B, Senn HJ. Thresholds for therapies: highlights of the St. Gallen International Expert Consensus on the primary therapy of early breast cancer, 2009. Ann Oncol. 2009;20:1319–29.
Roepman P, Horlings HM, Krijgsman O, et al. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin Cancer Res. 2009;15:7003–11.
Wesseling J, Cusumano G, Tinterri C, et al. High concordance for microarray based determination of ER, PR and HER2 receptor status and local IHC/FISH assessment worldwide in 827 patients (abstract P5-11-09). Cancer Res. 2011;71(24 Suppl.):543 s.
Viale G, Bogaerts J, Slaets L, et al. High concordance of protein (by IHC), gene (by FISH; HER2 only) and microarray readout (by TargetPrint) of ER/PR/HER2: results from the MINDACT trial (abstract P1-07-06). Cancer Res. 2011;71(24 Suppl.):190 s.
Glas AM, Floore A, Delahaye LJ, et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics. 2006;7:278.
Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–50.
Gevensleben H, Göhring UJ, Büttner R, et al. Comparison of MammaPrint and TargetPrint results with clinical parameters in German patients with early stage breast cancer. Int J Mol Med. 2010;26:837–43.
Acknowledgment
Supported in part by the Associazione Ricerca in Campo Oncologico (ARCO), Cremona, Italy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nguyen, B., Cusumano, P.G., Deck, K. et al. Comparison of Molecular Subtyping with BluePrint, MammaPrint, and TargetPrint to Local Clinical Subtyping in Breast Cancer Patients. Ann Surg Oncol 19, 3257–3263 (2012). https://doi.org/10.1245/s10434-012-2561-6
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-012-2561-6