Abstract
The objective of this study is to demonstrate through empirical evaluation the potential of a number of computer vision (CV) methods for sex determination from human skull. To achieve this, six local feature representations, two feature learnings, and three classification algorithms are rigorously combined and evaluated on skull regions derived from skull partitions. Furthermore, we introduce for the first time the application of multi-kernel learning (MKL) on multiple features for sex prediction from human skull. In comparison to the classical forensic methods, the results in this study are competitive, attesting to the suitability of CV methods for sex estimation. The proposed approach is fully automatic.
Similar content being viewed by others
1 Introduction
Sex determination from human skull plays an essential role in forensic anthropology. In the literature, morphological assessment and morphometric analysis are the two main approaches, which have historically been demonstrated for capturing sexually dimorphic characteristics from cranial regions. Morphological assessment involves some procedural steps where a forensic expert visually examines the anatomical regions of the skull (such as the glabella, mastoid, nuchal crest, orbital, and mental eminence), reports the observed variations on the skull with standard semantic terms, quantifies the descriptions on an ordinal scale, and eventually uses discriminant function analysis (DFA) to predict the sex of the skull. Morphometric analysis, on the other hand, requires forensic experts to annotate and measure the distance between anatomical landmarks. These measurements are considered as input to a DFA model to determine the sex of the skull.
Though both techniques are generally acceptable concepts founded on well-grounded principles for forensic examination, they exhibit some limitations. For instance, both estimation techniques are based on manual derivation of estimation parameters. Morphological assessment being a method that relies on visual perception is highly influenced by subjectivity of the observer. Besides, it necessitates a certain level of experience as well as familiarity of the forensic expert with the cranial samples in a population group that is being studied. Whereas, the process of morphometric analysis is laborious as it requires ample amount of time for accurate and precise landmark annotation. Moreover, variation in the shape of the skull is another limitation of morphometric analysis, which inhibits the generalization ability of the method to diverse population groups.
This paper presents an automatic estimation method which eschews the need for human manual assessment. We demonstrate with experimental evidence the potential of computer vision (CV) methods for cranial sex estimation. Of particular interest is application of different 3D local shape descriptors, feature learning, and classification methods. In addition, for the first time, we present multi-kernel learning on multiple features for cranial sex estimation. 3D local descriptors have been used in several CV and medical applications such as 3D object recognition [1, 2], face recognition [3], gender recognition [4, 5], diagnosis of cranial deformity, and detection of anatomical landmarks [6–8]. However, to the best of our knowledge, there are no related works on their application to 3D representation of skulls for sex determination.
The remainder of the paper is organized as follows. Section 2 presents the related works on cranial sex estimation. In Section 3, we describe the experimental dataset used for validating our work. Section 4 presents the CV framework for cranial estimation which includes sub-stages of pre-processing, 3D local shape representation, feature learning, and classification. In section 5, we demonstrate the usefulness of the proposed CV methods by the prediction results attained. Finally, discussion and conclusion are given in Section 6 to summarize this paper.
2 Related works
Morphological assessment and morphometric analysis are the two main techniques employed by forensic anthropolgists for sex determination [9]. The earliest studies using morphological assessment were reported in [10–12]. Studies in the 90’s have established standardized quantification methods using ordinal scale to aid visual assessment of sexually dimorphic characteristics from five anatomical sites of the skull [13]. These traits include the robusticity of the nuchal crest, size of the mastoid process, sharpness of the supraorbital margin, prominence of the glabella, and projection of the mental eminence. A forensic expert would examine how the dimorphic characteristics are expressed in those five anatomical sites and their visual similarity to the diagram presented in [13]. With various ordinal scoring methods, forensic experts have been able to achieve estimation performance between 83 and 90% [12, 14]. It has also been demonstrated that specific cranial regions such as the shape of supraorbital margin [15] can be assessed for sex determination with prediction rate of 70%. This is made possible by forming the contour shape of the supraorbital with plasticine impression, which is then visually assessed and quantified on a 7-point ordinal scale. A modification of such technique has been presented in [16] replacing manual assessment with computer-aided method by using 2D wavelet transform on 3D reconstruction of the scanned supraorbital impression to study its shape variation.
Alternative sex estimation technique is based on morphometric analysis, which involves linear or geometric measurement of anatomical landmarks. The choice of landmarks to annotate varies among the methods reported in literature. Franklin et al. [17] reported using eight measurements of the 3D landmarks to perform morphometric analysis with discriminant function analysis, which yielded prediction rates between 77 and 80%. The author discovered high sexual dimorphism in the facial width (bizygomatic breadth) and the length and height of the cranial vault. Bigoni et al. [18] conducted a study on 139 cranial samples of the Central European population, where 82 ecto-cranial landmarks were annotated from seven sub-regions (the configuration of the neurocranium, cranial base, midsagittal curve of vault, upper face, orbital region, nasal region, and palatal region) of the skull. Generalized procrustes analysis (GPA) was adopted for analyzing the shape configuration, and no sex difference were noticed in the sample set when landmarks from the whole cranium were used. However, through partial shape examinations on each of the seven regions, there were indications of strong sexual dimorphism in the midsagittal curve, the upper face, the orbital region, the nasal region, and the palatal region, but no sex variation in the cranium base and the neurocranium configuration. Another study has compared two discriminant function analysis methods on 17 craniometric variables from 90 Iberian skulls [19]. The authors observed higher metric variables in male samples than in female samples [19]. Luo et al. [20] presented statistical analysis of the holistic shape of the frontal part of the skull using principal component analysis (PCA) and linear discriminant analysis (LDA) on Chinese samples.
While morphological assessment and morphometric analysis appear to be technically simple, they have been demonstrated to be effective for cranial sex estimation. Moreover, they are concepts which have been established on well-grounded principles and universally acceptable as evidence in court cases and forensic investigation [21]. Nevertheless, there are a number of weaknesses in the two techniques that are worth mentioning:
-
Subjective perception of the observer (forensic expert) affects the confidence of prediction [18, 22]. This is a natural phenomenon associated with visual assessment and verbal description of the observed variation, especially in a situation where the descriptions connoted from a particular group is unable to generalize to other groups due to discrepancies in the perception of the observers.
-
Inter-observer variability is commonly experienced when observers select landmarks in the case of morphometric analysis. Furthermore, this influence of population difference affects the accuracy and precision of the traits used for identification. As a result, an estimation method used in a specific population may not generalize to other populations [23]
-
Inaccurate and incomplete landmarks is another limitation of morphometric analysis. In fact, it has been shown that the inter-observer error is approximately 10% for most measurements [21]. Accurate annotation of anatomical landmarks requires ample amount of time and it often needs expensive, specialized anthropometric equipment. In addition, forensic expert usually face the challenge of performing measurements that capture subtle variations among cranial traits that are easy to see but very difficult to measure [14].
2.1 Main contributions
The main objective of this paper is to demonstrate the potential of computer vision methods for sex determination from human skull. Though it is accurate that the 3D feature descriptors adopted in this paper are well known, from a holistic point of view, a framework integrating several stages of pre-processing—feature extraction, multiregion representation, and classification—has not been reported in the literature (to the best of our knowledge), which indicates the novelty of this work. Such pipeline provides an incentive for forensic anthropologists to look at sex estimation problem from a totally different perspective. Moreover, the proposed approaches are completely automatic. We therefore summarize the contributions of the paper as follows:
-
This paper proposes using computer vision approaches based on local feature representation, feature learning, and classification for sex prediction from human cranial data obtained from CT scans. The proposed method advances the conventional forensic methods as it does not rely on manually configured estimation parameters.
-
We propose to partition the skull along the axial, coronal, and sagittal axes, extract 3D local shape features, and aggregate those features into compact representations that possess discriminative capabilities. To segment the skulls into smaller regions, we constrained the partitioning to the X-, Y-, and Z-axes, where accurate distribution of features in each local sub-region can be generated.
-
Furthermore, comprehensive performance analysis of different combination of local features and feature learning methods with different number of regions from the three planes is presented.
-
Finally, we introduce the concept of multi-kernel learning on multiple features for cranial sex prediction. To the best of our knowledge, this is the first study to approach the problem of skull sex estimation from this novel perspective.
3 Application of computer vision methods
This section introduces the proposed framework for sex classification, which is composed of four main stages: 3D data pre-processing, 3D feature representation, multi-region feature representation, and classification.
3.1 Experimental dataset
The experimental data is a 100 sample set of post-mortem computed tomography (PMCT) scan slices obtained from the Hospital Kuala Lumpur (HKL). There are 54 males and 46 females between the ages of 5 and 85 years from South East Asia. Seventy percent of the data are Malaysian (Malay, Chinese, Indian) and 30% non-Malaysian. The scanning device is a Toshiba CT scanner with scanning settings of 1.0 slice thickness and 0.8 slice interval. The data contain slices belonging to the head region with resolutions between 512 × 512 × 261 and 512 × 512 × 400, depending on the size of the skull of each subject. Legal consent for the use of the dataset was obtained prior to commencing this research.
3.2 3D data pre-processing
The consecutive 2D CT slices of each subject are stacked vertically to obtain 3D volumetric data, which are then filtered to reduce noise, local irregularities, and roughness using a discretized spline smoothing method [24]. The 3D smoothing technique utilizes 3D discrete cosine transform-based penalized least square regression (DCT-PLS) on equally spaced high-dimension data. The main idea of the algorithm is to reformulate PLS regression problem with DCT, where the data are expressed in the form of cosine functions oscillating at different frequencies [24].
Afterward, the denoised 3D volumetric data is reconstructed into 3D surface using marching cubes algorithm [25] as illustrated in Fig. 1. The reconstruction is achieved using an iso-value of (150) to obtain the regions containing hard tissues. As the reconstructed 3D surface is of high dimension (>650,000 vertex points), mesh simplification is performed to reduce the surface to 13% of the original size [26]. The resulting downsampled skulls have <130,000 vertices, with well-preserved structural details of the surface after downsampling, as shown in Fig. 1.
3.2.1 Background object removal
After surface reconstruction, we noticed some dynamic background objects scanned with the subjects, with the same isovalue as the hard tissue, around some skull samples (∼27 samples). In order to remove these background objects, we propose a method based on online sequential least square (OLS) with Gaussian mixture model (GMM) sample initialization. We are able to design this method using 27 noisy samples; however, the same approach can be improved further when there is availability of larger noisy samples. Initially, a training data matrix is prepared by randomly selecting eight clean and eight noisy skull samples. For each noisy skull, we manually annotate and cut out the background objects. The set of background objects which have been manually segmented are considered to form the negative training samples, while the clean skulls are regarded as the positive training samples, as illustrated in Fig. 2. Therefore, the training set is composed of eight clean and eight segmented background objects resulting in ∼ 750,000 training vertex points for the two classes. Due to the size of the training set, we used a sequential learning method where each time the training is performed in batches. However, the initial batch of data for training is initialized using a GMM to fit a mixture model composing of K components. Consider a training set A = {v 1,…v i }, where A is a matrix consisting of clean and segmented background objects and v i is a vertex point; GMM is used to cluster the set A into K clusters. Then, the OLS training process is initialized by choosing a cluster as the initial batch A 0. After selecting the initial batch, the following minimizing problem is solved:
where x 0 are the coefficients, λ is the regularization term, and b 0 = {+1,−1} are the labels (+ 1 for clean skull, −1 for the background object) of the initial subsets. The initial solution x 0 can be obtained analytically using:
The coefficients x 0 are referred to as the least square solution to the minimization problem in Eq. (1). The inverse matrix \(M_{0}^{-1}\) can be obtained as: \( M_{0}^{-1} ~=~\left (A_{0}^{\top } A_{0} + {\lambda \textsc {I}}\right)^{-1}\) and the output coefficients \(x_{0}~=~M_{0}^{-1} A_{0}^{\top } b_{0}\).
3.2.2 Updating x k+1
Having obtained the initial solution, the remaining subset of the data can be trained batch-by-batch without keeping the previously trained set to minimize computational time and complexity. Thus, it is necessary to use an efficient approach to update the initially learned coefficients, while solving new linear systems minimization problem [27]. A well-recognized method to sequentially update x 0 is the Sherman-Morrison Woodbury inverse formula [28]. When new sample subsets \(A_{k+1}~=~\{v^{k+1}_{1},\ldots,v^{k+1}_{i}\}\) are learned in batches, we can update x 0 from Eq. (2) as:
where x k is the output coefficients in the previous step, b k + 1 is the newly collected labels and \( M_{k+1}^{-1} = \left (A^{T}_{k+1} A_{k+1} + \lambda \textsc {I}\right)^{-1}\). Once x k+1 has been learned, each new skull sample containing noise, which were not included in the training phase are used to test the model. The results obtained are depicted in Fig. 2.
Afterward, skull alignment is performed using an iterative closest point (ICP) algorithm [29].
3.3 Skull representation with 3D local shape features
3.3.1 Mesh local binary pattern (MeshLBP)
MeshLBP is a local shape representation method for directly extracting local binary patterns from 3D mesh. Basically, the algorithm forms a set of ordered ring facets (ORF) [30, 31] and calculates the primitive functions such as mean or Gaussian curvature for each facet in the ring. Then, the binary patterns are obtained by thresholding the primitive function of neighboring facets by that of the centre facet. Given the primitive function p(f), defined on a mesh, represents the Gaussian curvature. The MeshLBP can be derived as follows:
where r is the ring number and m is the number of facets available in the ring.
These two parameters in the Eq. (5) regulate the radial resolution and azimuthal quantization, while the discrete function α(k) is a weight which enables computation of other variants of LBP. Using α 1(k) = 1 gives binary pattern in the range [0–12], while α 2(k)=2k gives patterns in the range [0–4096]. In order to represent the skull with MeshLBP, ten-ring neighbors are computed for each individual facet (this is chosen to provide a trade-off between fairly covering a large local neighbourhood and less computational time), m = 12 and α 2(k). Then, the MeshLBP is extracted by comparing the Gaussian curvature of center facet to the neighboring facets. An example of MeshLBP feature representation is illustrated in Fig. 3. The output of MeshLBP is V-by-10 matrix for each skull sample, with V denoting the number of vertex points.
Example of MeshLBP feature distribution by mapping the binary patterns to different color map. For illustration, we display the binary pattern of ring (r = 2). Red color indicates areas of small binary pattern (0) due to small variation in the curvature of the local regions. Blue represents patterns with values (1–16), green denotes patterns with values (32–256), and cyan represents patterns with values (512–4096), due to large variation in the curvature of the local regions. For instance, sharp curvature variation can be observed around the local regions of the orbital, nasal bone, and mandible, while the parietal bone exhibits mild variation in curvature
3.3.2 Spin image
Spin image [1] is a point-based descriptor which is invariant to rotation and translation commonly used for object retrieval. Spin image generates 2D histogram for mesh point containing the representation of object geometry. To begin with, oriented points are initially computed for each point on the mesh according to location of the vertex p and its surface normal n, which results in a 2D basis (p,n). Then, to obtain the coordinate system, a tangent plane P passing through p is formed which is perpendicular to the normal n and line L through p that is parallel to n, resulting in a cylindrical coordinate system (α,β), where α is a non-negative distance to L and β is signed distance perpendicular to P. Finally, a spin map S m is formed by projecting the 3D points x on a mesh to 2D coordinates (α,β), which are accumulated into discrete points that are updated incrementally.
The size (i max,j max) of a spin image is determined by the size of the bin and the maximum size of the object expressed in the spin map coordinates.
In this paper, a cell size of 10 and histogram bin of 10 are used for extracting the features, which result in 10 × 10 spin image for each vertex.
3.3.3 Local depth SIFT (LD-SIFT)
LD-SIFT [2] is an extension of SIFT [32] to mesh surface. The process of extracting the LD-SIFT features involves detecting a number of interest points representing the local maxima from difference of Gaussian (DoG) operation on the mesh. From each interest, a sphere support is constructed to cover the neighboring region. LD-SIFT then creates a 2D array that emanates from estimating a tangent plane T within the support region and computing the distance from each point in that region to the plane. Then, it makes the feature scale invariant by setting the viewport size to match the feature scale, as detected by the DoG detector. Further, it computes the principal component analysis (PCA) of the points surrounding the interest point and use their dominant direction as the local dominant angle. Similar to the standard SIFT, the depth map are rotated to a canonical angle with respect to the dominant angle to make the LD-SIFT rotation invariant. From the resulting depth maps, the standard SIFT feature descriptors are computed to create the LD-SIFT feature descriptor. The features represent 8-bin gradient histograms distributed in the local cells of 4 × 4 depth map. As a result, the dimensional of the final feature vector is 128 (4 × 4 × 8) for each detected vertex (interest) point.
3.3.4 Scale invariant heat kernel signature (SIHKS)
Heat kernel signature (HKS) is a type of spectral shape representation method which uses deformable shape analysis to create the point signature of a specific point [33]. The representation follows the concept of modeling shapes as Riemannian manifold and using their heat conduction characteristics as a descriptor.
Initially, heat kernel signature was proposed in [33] for shape representation, which is invariant to isometry and has gained popularity in shape retrieval application. However, the limitation of HKS is that the descriptor is not scale invariant. As a result, Bronstein et al. [34] introduced a method to remove the scale effect by sampling each point logarithmically in time (t = α τ) and then computing the derivative based on the scale to undo the additive function with respect to the scale. They further utilized Fourier transform to remove the shift variation. In order to extract SIHKS, we used a logarithmic scale-space with base α = 2 ranging from τ = 1:20 with increments of 1/5. We then chose the first ten frequencies for feature representation. Each feature point in the 10-length vector denotes the heat kernel signature at a particular scale.
3.3.5 ShapeDNA
ShapeDNA initially proposed by Reuter et al. [35] computes the fingerprint of any 3D object by deriving the eigenvalues of Laplace-Beltrami operator of the shape model. It is isometry invariant and insensitive to noise. Normally, eigenvalues λ and eigenfunctions u are the solutions to the laplacian eigenvalue problem △u = −λ u, where △u:=div(grad(u)),div is the divergence of the underlying Riemanian manifold and grad represents the gradient. The first normed N eignevalues 0 < λ 1 < λ 2,…λ n are chosen as the shape descriptor. In this paper, we selected the first 100 eigenvalues as the shape descriptor. Figure 4 depicts some examples of ShapeDNA representation.
3.3.6 Area of skull regions
Finally, the surface area of each local sub-region of the skull is computed as additional features. The skull is initially partitioned in smaller regions along the X, Y, or Z, and the corresponding area is computed following the approach in [36]
3.4 Multiple-region feature representation
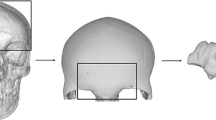
The common practice in computer vision applications is to directly concatenate features extracted from keypoints into long-tailed vector or, as an alternative, use the bag-of-word model to construct a compact representation. Nevertheless, we discovered that these approaches result in high possibility of the local features from a particular class to possess dissimilar representation. This inherently makes features from two different classes to exhibit the same or similar feature values, which reduces the informative characteristics of such methods. Hence, in this work, we employed a heuristic partitioning method, where each skull is divided into several regions along a particular orientation (axis). In each region, we stack the representations from each point on each other and aggregate the features into compact representation as shown in Fig. 5. It is essential to note that the partitioning could have been performed following the anatomic structure of the skull. However, such partitioning is itself a research topic, and there are currently no reported methods for anatomic partitioning in the literature. We intend to investigate this research aspect in our future works. Thus, such exploration would go beyond the scope of the current paper.
Assuming spin image of 10×10 size is extracted from V = 10,000 vertices, we stack the spin image from each vertex point on one another and calculate the aggregate, which results in a final spin image of 10 × 10 descriptor size and feature vector of 100 dimensions, as shown in Fig. 5. This makes the representation more distinctive than the long-tailed concatenated or bag-of-word representation. Similar approach is used for LD-SIFT by stacking the gradient features from the keypoints and taking aggregate, which results in a final feature vector of 128 dimensions. For MeshLBP, the descriptor extracted is 10 scalar values from 10 rings for each vertex point, making a V × 10 descriptor matrix; thus, a 32-histogram bin is aggregated for each ring along the vertices and the final feature vector is the concatenation of the histograms from the 10 rings (32 × 10 = 320 feature descriptor). We used 32 bins to keep the dimension of the feature vector to a reasonable length. Also, SIHKS is compactly represented in this fashion, using 32-bin histogram bins to represent each frequency, which yields 32 × 10 = 320 descriptor. To obtain discriminant representation from the extracted features, we used the Kernel principal component analysis (KPCA) [37] which is a dimensionality reduction method that generalizes the standard PCA to non-linear feature representation and K-SVD [38], a dictionary learning technique.
Furthermore, to make our evaluation comprehensive, the partition is performed along the X-, Y-, and Z-axes, as depicted in Fig. 6, and the number of regions N examined in each axis is N = {1:2:99}. The results attained from the regions in each axis are illustrated in Section 4.
3.5 Multi-kernel learning (MKL) on multiple features
Besides testing the performance ability of the six local shape descriptors described in Section 4.2, we examine the potential of MKL to fuse multiple features with different representation properties. MKL has attracted significant amount of attention in CV research domain. In this paper, the soft margin MKL algorithm introduced by Xu et al. [39] is adopted, where a kernel slack variable is first introduced for each of the base kernels when learning the kernel. This approach is advancement over the MKL framework generally regarded as the hard margin MKL [40], which imposes sparsity on a category of features and selects the features that best optimize the object function. In fact, it has been pointed out in [39] that the hard margin MKL is a method which only selects the base kernels with minimum objective. This could easily lead to overfitting problem, particularly in a situation where the base kernels contain noisy features. Following the notion of standard hard margin SVM, it is believed that data from two classes can be separated by a hard margin. However, to enable usability of SVM in real applications the slack variables were introduced to the hard margin SVM, which allows some training errors to be incorporated to the training data, thereby minimizing the overfitting problem [39]. This concept inspired the development of soft margin MKL, which instead introduces kernel slack variable for each of the base kernels [39].
Futhermore, by conducting independent experiments on each local feature, we are able to figure out the best feature set in Section 4.1, where it turns out that MeshLBP > Spin Image > LD-SIFT > SIHKS > ShapeDNA > Region Area. Hence, the point of using MKL in this paper is to try to induce information from more than one feature source by different combination, which is why the soft margin MKL is more suitable for this purpose. As a result, it makes it unnecessary to select the best feature again or highlight the best set of features. Moreover combination of all features exhibited poor performance indicating that information cannot be induced from all features at the same time.
The motivation behind MKL is to combine or fuse multiple kernels or features rather than using a single feature representation to make prediction, with expectation that such combination leads to potential gain in performance. Assuming, we have a training set M consisting of \({(h_{i},g_{i},w_{i})}^{M}_{i = 1}\) where h i is a type of local features of the ith sample and g i is a another type of features of the same sample and w = (−1,+1) are the class labels. We can map the features to the reproducing kernel hilbert space (RKHS) with a kernel function k(·,·) defined over each of the two feature types. Without loss of generality, this notion applies to more than two feature types.
3.5.1 SVM-based MKL
Suppose we have a set of base kernels K = {K 1,…K N } for our training set, the objective of the standard problem solved by the MKL introduced in [39] is:
where, S V M{K n ,α}=−1/2(α⊙w)′ K n (α⊙w), α i are the coefficients of the samples, and μ=[μ 1,…μ N ]′ 1 are the coefficients measuring the importance of the nth base kernel. \(\mathcal {A} = \{ \alpha | \alpha ' \mathbf {1} = 1, \alpha ' w = 0, 0 \leq \alpha \leq C \} \) is the domain for α and \(\mathcal {M} = \{ \alpha | \mathbf {0} \leq \mu, \sum ^{N}_{n=1} \mu = 1 \} \) is the domain for μ. For the solution in Eq. 6, a hard margin is constructed resulting in selection of only the most important base kernel, which eventually defeats the purpose of finding interaction between different feature sets. Therefore, in this work, we used the soft margin formulation, where a slack variable which is the difference between the target margin τ and the objective is introduced as:
The loss incurred by the kernel slack can be expressed as
where ℓ(·) is a hinge loss function ℓ(·)=max(0,ℓ n ).
Besides using MKL to induce information from different features, it will be interesting to explore the interaction and joint contribution of different regions using MKL. However, as it itself is another research topic and our current focus is on evaluating the performances of different types of features and their combinations for sex identification of the cranial, we hope to include this direction in our future research.
4 Experimental results
In this section, two different experiments are reported to evaluate the described CV methods. Firstly, we test the effect of partition orientation on the four local descriptors, meaning that the prediction results attained are compared based on the regions in X-,Y-, and Z-axes. In addition to that the effect of number of partitions (regions) the skull is divided is tested. Secondly, we evaluate the performance of MKL for various feature combinations, similarly with respect to the three partition axes.
4.1 Sex determination using 3D local features
To evaluate the performance of the four local descriptors for sex determination, we have selected three baseline predictive models: (1) support vector machine (SVM) [41], (2) kernel extreme learning machine (KELM) [42], and (3) sparse representation classifier (SRC) [43]. The standard protocol used throughout this paper is that the input dataset is divided into 60 and 40% as training and testing set, respectively, by random sampling without replacement. In the training stage, a five-fold cross-validation is performed to derive the best regularization value C (between 2−10 and 210) for the three classifiers. Once training is completed, the skulls are classified into male or female using the separate unused testing set. The experiments are repeated 10 times, and the average is computed as the sex prediction rate.
4.1.1 Effect of number of regions and partition orientation on sex prediction
We now simultaneously demonstrate how the orientation of partition and the number of skull partition influence the results of sex prediction. These evaluations are divided into two categories, where the first evaluation involves using ordinary 3D local descriptors to train the three predictive models and the second evaluation involves using KPCA and KSVD to learn compact representation from the feature vectors before classification. In the first evaluation, the skulls are initially partitioned into different regions, and the local features are extracted, aggregated, and finally concatenated from all regions before serving as an input to the predictive models. It can be noticed in Fig. 7 that partitions derived from the z-axis generally provided the best prediction rates, particularly using MeshLBP, as these results remain consistent across the three predictive models. When the MeshLBP features extracted from partitions in the z-axis are used to train the KELM classifier, the highest prediction rate obtained is 80.25%; SRC classifier showed similar performance with a prediction rate of 80%, while with SVM classifier, the highest prediction rate is 82%. Moreover, in all experiments, MeshLBP showed better performance than the region area, ShapeDNA, SI-HKS, LD-SIFT, and spin image local descriptors.
Whereas, the Y-axis in some cases showed comparable trend in performances, especially using LD-SIFT and Spin image local descriptors, which also remain consistent across the three classifiers. For instance the prediction results of spin image are 76.25, 73.5, and 73% using KELM, SRC, and SVM classifiers respectively.
However, for MeshLBP, it can be seen that the prediction results achieved from partitions in X- and Y-axes are lower than those from Z-axis. Besides that, we observed that the effect of increase in number of partitions is not so evident in all experiments, except for MeshLBP where the prediction results peaked at five regions before deteriorating as the number of partitions approach 99 regions. With region area, ShapeDNA, and SI-HKS, the performances are neither influenced by orientation of partition nor by number of regions as the prediction rate is generally less than 70% in the three predictive models, thus leading to the conclusion that the SI-HKS descriptor is not suitable for sex prediction from human skulls.
In the second evaluation, we used KSVD and KPCA to learn compact representations from the local features prior to classification, which significantly reduce the dimensionality of the feature vectors. Using KSVD, we noticed a slight improvement in the performance of the local descriptors. For instance, MeshLBP increased from 82 to 83% using KELM classifier, while the results using SRC and SVM also increase by at least 1%. Similarly, the prediction rates of LD-SIFT and spin image increased across the three classifiers. Meanwhile, a similar trend in prediction results with respect to the orientation of partitions can be observed from the results in Fig. 8, as the Y-axis tends to exhibit comparable performance to the Z-axis partitions.
Using KPCA, we further observed improvement in prediction results, especially in the case of spin image and MeshLBP where the prediction rates increased to 85.5 and 86% respectively, as depicted in Fig. 9. In addition, it is obvious that the partitions derived from the Z-axis provided better results than X- and Y-axes. Also, increasing the number of partitions resulted in reduced prediction results for MeshLBP; however, the trend is only slightly evident for spin image and LD-SIFT. Overall, SVM classifier showed better prediction performance than SRC and KELM.
4.2 Sex determination using MKL on multiple features
In this experiment, we used the linear kernel function for mapping each local feature, which are then combined in the MKL framework. We focus on LD-SIFT, spin image, and MeshLBP since these three feature representation methods showed better performance than region area, ShapeDNA, and SI-HKS in Section 4.1. Within this framework, four different sets of combination are performed with respect to the orientation of partition (X, Y, Z) and number of regions (1:2:99). Particularly, we have examined the integration of:
-
LD-SIFT + MeshLBP
-
Spin Image + MeshLBP
-
Spin Image + LD-SIFT
-
Spin Image + LD-SIFT + MeshLBP
The results attained are illustrated in Fig. 10. Learning a multi-kernel representation for LD-SIFT + MeshLBP, the best prediction rates attained are 72.5% (X-axis), 71.5% (Y-axis), and 78% (Z-axis). With spin image + MeshLBP, the prediction rates are 72% (X-axis), 70% (Y-axis), and 86% (Z-axis), while the results of spin image + LD-SIFT are 79.5% (X-axis), 72.5% (Y-axis), and 85.5% (Z-axis). Similar to the case of single descriptor, it can be noted that the partitions derived from the Z-axis provided better performance than X- and Y-axes in all experiments.
Moreover, the influence of number of regions is slightly evident on the results of spin image + LD-SIFT and Spin + MeshLBP, as the peak recognition rate is achieved at five regions before receding as the partitions approach 99 regions. However, we observed no significant impact of number of region on the prediction rates of LD-SIFT + MeshLBP. Finally, we attempted to combine spin image + LD-SIFT + MeshLBP, but the prediction results decreased on the three orientations of skull partition, indicating less compatibility among the three features. Quite interestingly, spin + MeshLBP produced the most compatible combination as the prediction rate of 86% is comparable to the benchmark attained with single descriptor. We denote from these experiments that MKL is useful for sex prediction from human skulls albeit the selection of the most compatible 3D local descriptor seems to be necessary. Unlike the soft margin MKL which tries to induce information from multiple sources, we used the hard margin MKL based on primal formulation, which imposes penalty on the features and selects those that best optimize the objective. However, the results as shown in Fig. 11 are not so promising compared to the soft margin MKL. This can be attributed to the fact that the hard margin MKL tends to be dependent on the discriminative ability of the base kernels [44]. Thus, in a case where the single features are already discriminant, the hard margin MKL will be unable to exhibit any better performance than the single features. On the other hand, this indicates that if the single features are not discriminant such as heat kernel signature or ShapeDNA, their combination will have a negative impact on the performance unless they can be singled out.
4.3 Comparison with forensic approach
For the sake of completeness, we compared the performance of the proposed CV methods with conventional forensic estimation method [45] as shown in Table 1. Prior to describing the forensic method, it is worth emphasizing that the objective of this paper is to demonstrate the potential of computer vision methods for sex determination from human skull. Despite the fact that the 3D feature descriptors adopted in this paper are commonly used in the domain of computer vision, from a general perspective, a framework which integrates several stages of pre-processing—feature extraction, multiregion representation, and classification—has not been reported in the literature. This differentiates the proposed framework from the existing forensic methods, and such framework serves as an incentive for forensic anthropologists to approach sex estimation problem from a completely different perspective.
The conventional forensic method [45] is based on traditional morphometrics, where 22 estimation parameters are measured covering anatomical locations of 87 cranial samples (45 males and 42 females). These 87 samples are drawn the same dataset we obtained from the Hospital Kuala Lumpur (HKL). The measurements include the maximum cranial length, maximum cranial breadth, cranial base length, basion-bregma height, bizygomatic breadth, maximum frontal breadth, minimum frontal breadth, basion prosthion, upper facial height, nasal height, nasal breadth, orbital height of the right eye, orbital height of the left eye, orbital breadth of the right eye, orbital breadth of the left eye, biorbital breadth, auriculo-bregmatic height of the right side, auriculo-bregmatic height of the left side, naso-occipital length, mastoidal bregma height of the right side, mastoidal bregma height of the left side, and nasion bregma height. Through independent t test, the authors were able to deduce the difference between male and female categories. Discriminant function analysis(DFA) is then used to generate equations for modeling the measurements and regression analysis to classify the samples into their respective classes. The prediction result from the study obtained through cross-validation is between 78.2 and 86.2%.
5 Discussion and conclusion
Sex determination from human skull is a very essential aspect of forensic examination. In the past few decades, forensic anthropologists have suggested two main approaches for determining sexual dimorphic characteristics from human skull, which are morphological assessment and morphometric analysis. These two methods either visually assess specific cranial sites that possess sexual dimorphism or perform inter-distance measurement between anatomical landmarks that have been carefully annotated.
Probing from a different perspective and diverging completely from the conventional forensic approaches, we suggest a possible framework for sex determination from human skull based on computer vision techniques, which include automatic representation of human skulls with advanced local shape features and learning of compact and discriminant representation from the extracted features. We introduced multi-region-based representation that are derived from partitioning the skulls along three main axes (X, Y, Z) which are anatomically equivalent to the sagittal, coronal, and axial axes. Inclusive of this is the aggregation of several region descriptors into compact features that represent each region with better discriminant capabilities. Also, we examine the influence of increase in number of regions and orientation of partition on prediction results.
From a general perspective, our experimental results give indication that these CV methods are suitable for sex determination with the best prediction rate of 86% through KPCA subspace representation of compact MeshLBP features from five sub-regions of the skull. Intuitively, we discovered that orientation of partition have significant influence on the results of MeshLBP, as the difference between the results obtained from using partitions derived from Z-axis are superior to those from X- and Y-axes. However, this observation is not so evident in the case of LD-SIFT and spin image because the results of X-axis are quite competitive with those obtained from Z-axis. We mainly attribute this to the fact that spin image are LD-SIFT descriptors embodying rotation invariance property. It is also discovered from the experiments that increase in number of partition does not affect the performance of LD-SIFT and spin image, but MeshLBP seems to be sensitive to increase in number of regions. Furthermore, we presented the application of MKL on multiple feature set for cranial sex estimation. The framework provided possibility of integrating two or more 3D local shape descriptors with differing representation properties. It was discovered from the experiments that MKL is similarly suitable for cranial sex estimation with the fusion of spin image + MeshLBP, revealing the most compatibility with a prediction rate of 86%. Quite interestingly, similar trends observed from previous experiments with respect to orientation of partition and number of regions remain valid in the MKL framework.
Comparing the results of CV methods with those of standard forensic approach, our results are within the often reported sex prediction range (70–90%) using morphometric or morphological assessment [14, 46]. This novel perspective has introduced an alternative and efficient approach in forensic anthropology, which could potentially set the path to bridging the semantic gap between visual assessment and perceived dimorphic characteristics. Besides, we are able to discard the challenge of incomplete anatomical landmarks, which affects the performance of morphometric analysis. From our experiments, we can make three fundamental suggestions for potential future studies:
-
Extracting features from multiple regions is important for skull representation as it tends to out-perform holistic representation of the skull.
-
Feature aggregation instead of long-tailed concatenation should be considered to compactly represent each local region of the skull as it makes the features more distinctive with less correlation among features from the same class.
-
Prediction performance with respect to orientation of partition is highly dependent on the properties of the local shape descriptor. For instance, MeshLBP showed great performance along the Z-axis but deteriorated significantly along X- and Y-axes in all experiments, while the results of spin image and LD-SIFT on X- and Z-axes are quite comparable.
Abbreviations
- CV:
-
Computer vision
- DFA:
-
Discriminant function analysis
- GMM:
-
Gaussian mixture model
- KPCA:
-
Kernel principal component analysis
- KSVD:
-
K-singular value decomposition
- KELM:
-
Kernel extreme learning machine
- MKL:
-
Multi-kernel learning
- OLS:
-
Online sequential least square
- RKHS:
-
Reproducing kernel hilbert space
- SVM:
-
Support vector machine
- SRC:
-
Sparse representation classifier
References
Johnson AE, Hebert M (1999) Using spin images for efficient object recognition in cluttered 3D scenes. IEEE Trans Pattern Anal Mach Intell 21(5):433–449.
Darom T, Keller Y (2012) Scale invariant features for 3D mesh models. IEEE Trans Image Process 21(5):1–32. doi:10.1109/TIP.2012.2183142.
Tolga I, Halici U (2012) 3-D face recognition with local shape descriptors. IEEE Trans Inf Forensic Secur 7(2):577–587.
Han X, Ugail H, Palmer I (2009) Gender classification based on 3D face geometry features using SVM In: Proceedings of IEEE International Conference on CyberWorlds, 114–118. doi:10.1109/CW.2009.41.
Ballihi L, Amor BB, Daoudi M, Srivastava A, Aboutajdine D (2012) Boosting 3-D-geometric features for efficient face recognition and gender classification. IEEE Trans Inf Forensic Secur 7(6):1766–1779.
Shapiro LG, Wilamowska K, Atmosukarto I, Wu J, Heike C, Speltz M, Cunningham M (2009) Shape-based classification of 3D head data In: International Conference on Image Analysis and Processing, 692–700.. Springer, Berlin.
Atmosukarto I, Wilamowska K, Heike C, Shapiro LG (2010) 3D object classification using salient point patterns with application to craniofacial research. Pattern Recognit 43(4):1502–1517.
Yang S, Shapiro LG, Cunningham ML, Speltz ML, Birgfeld C, Atmosukarto I, Lee SI (2012) Skull retrieval for craniosynostosis using sparse logistic regression models In: MCBR-CDS, 33–44.
Franklin D, Cardini A, Flavel A, Kuliukas A (2012) The application of traditional and geometric morphometric analyses for forensic quantification of sexual dimorphism: preliminary investigations in a Western Australian population. Int J Legal Med 126(4):549–558. doi:10.1007/s00414-012-0684-8.
Krogman WM (1962) The human skeleton in forensic medicine. Am Assoc Adv Sci. doi:10.1126/science.135.3506.782-a.
Stewart TD, Kerley ER (1979) Essentials of forensic anthropology: especially as developed in the United States. Charles C. Thomas, Springfield.
Konigsberg LW, Hens SM (1998) Use of ordinal categorical variables in skeletal assessment of sex from the cranium. Am J Phys Anthropol 107(1):97–112.
Buikstra JE, Ubelaker DH (1994) Standards for data collection from human skeletal remains In: Proceedings of a seminar at the Field Museum of Natural History (Arkansas Archaeology Research Series 44).
Walker PL (2008) Sexing skulls using discriminant function analysis of visually assessed traits. Am J Phys Anthropol 136(1):39–50. doi:10.1002/ajpa.20776.
Graw M, Czarnetzki A, Haffner HT (1999) The form of the supraorbital margin as a criterion in identification of sex from the skull: investigations based on modern human skulls. Am J Phys Anthropol 108:91–96.
Pinto SCD, Urbanovȧ P, Cesar RM (2016) Two-dimensional wavelet analysis of supraorbital margins of the human skull for characterizing sexual dimorphism. IEEE Trans Inf Forensic Secur 11(7):1542–1548.
Franklin D, Freedman L, Milne N (2005) Sexual dimorphism and discriminant function sexing in indigenous South African crania. HOMO - J Comparative Hum Biol 55(3):213–228. doi:10.1016/j.jchb.2004.08.001.
Bigoni L, Velemínská J, Bružek J (2010) Three-dimensional geometric morphometric analysis of cranio-facial sexual dimorphism in a Central European sample of known sex. HOMO- J Comparative Human Biol 61(1):16–32. doi:10.1016/j.jchb.2009.09.004.
Jiménez-Arenas JM, Esquivel JA (2013) Comparing two methods of univariate discriminant analysis for sex discrimination in an Iberian population. Forensic science international 228:175.e1–175.e4. doi:10.1016/j.forsciint.2013.03.016.
Luo L, Wang M, Tian Y, Duan F, Wu Z, Zhou M, Rozenholc Y (2013) Automatic sex determination of skulls based on a statistical shape model. Comput Math Methods Med 2013:1–7.
Williams BA, Rogers TL (2006) Evaluating the accuracy and precision of cranial morphological traits for sex determination. J Forensic Sci 51(4):729–735. doi:10.1111/j.1556-4029.2006.00177.x.
Lewis CJ, Garvin HM (2016) Reliability of the walker cranial nonmetric method and implications for sex estimation. J Forensic Sci 61(3):743–751. doi:10.1111/1556-4029.13013.
Guyomarc’h P, Bruzek J (2011) Accuracy and reliability in sex determination from skulls: a comparison of Fordisc 3.0 and the discriminant function analysis. Forensic Sci Int 208:30–35. doi:10.1016/j.forsciint.2011.03.011.
Garcia D (2010) Robust smoothing of gridded data in one and higher dimensions with missing values. Comput Stat Data Anal 54(4):1167–1178. doi:10.1016/j.csda.2009.09.020.
Lorensen WE, Cline HE (1987) Marching cubes: a high resolution 3D surface construction algorithm. ACM SIGGRAPH Comput Graphics 21(4):163–169. doi:10.1145/37402.37422.
Cignoni P, Montani C, Scopigno R (1998) A comparison of mesh simplification algorithms. Comput Graphics 22(1):37–54. doi:10.1016/S0097-8493(97)00082-4.
Do TN, Fekete JD (2007) Large scale classification with support vector machine algorithms In: Proceedings of the 6th International Conference on Machine Learning and Applications, ICMLA, 148–153. doi:10.1109/ICMLA.2007.25.
Golub GH, Van Loan CF (1996) Matrix computations. Phys Today. doi:10.1063/1.3060478.
Besl P, McKay N (1992) A method for registration of 3-D shapes. IEEE Trans Pattern Anal Mach Intell 14(2):239–256. doi:10.1109/34.121791.
Werghi N, Rahayem M, Kjellander J (2012) An ordered topological representation of 3D triangular mesh facial surface : concept and applications. EURASIP J Adv Signal Process 2012(144):1–20.
Werghi N, Berretti S, del Bimbo A (2015) The Mesh-LBP : a framework for extracting local binary patterns from discrete manifolds. IEEE Trans Image Process 24(1):220–235.
Lowe DG (2004) Distinctive image features from scale-invariant keypoints. International Journal of Computer Vision 60(2):91–110.
Sun J, Ovsjanikov M, Guibas L (2009) A concise and probably informative multi-scale signature based on heat diffusion. Eurographics Symposium on Geometry Processing 28(5):1383–1392. doi:10.1111/j.1467-8659.2009.01515.x.
Bronstein AM, Bronstein MM, Ovsjanikov M (2012) Feature-based methods in 3D shape analysis In: 3D Imaging, Analysis and Applications, 185–219.. Springer, London.
Reuter M, Wolter F, Peinecke N (2006) Laplace–Beltrami spectra as ‘Shape-DNA’ of surfaces and solids. Computer-Aided Design 38(4):342–366.
Zhang C, Tsuhan C (2001) Efficient feature extraction for 2D/3D objects in mesh representation In: Proceedings of the 2001 IEEE International Conference on Image Processing, 935–938.
Schölkopf B, Smola A, Müller KR (1998) nonlinear component analysis as a kernel eigenvalue problem. Neural Comput 10(5):1299–1319. doi:10.1162/089976698300017467.
Aharon M, Elad M, Bruckstein A (2013) K-SVD: an algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans Signal Process 54(11):4311–4322. doi:10.1109/TSP.2006.881199.
Xu X, Tsang IW, Xu D (2013) Soft margin multiple kernel learning. IEEE Trans Neural Netw Learn Syst 24(5):749–761. doi:10.1109/TNNLS.2012.2237183.
Gönen M, Alpaydin E (2011) Multiple kernel learning algorithms. J Mach Learn Res 12:2211–2268.
Cortes C, Vapnik V (1995) Support vector networks. Mach Learn 297:273–297.
Huang GB (2014) An insight into extreme learning machines: random neurons, random features and kernels. Cogn Comput 6(3):376–390.
Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y (2009) Robust face recognition via sparse representation. IEEE Trans Pattern Anal Mach Intell 31(2):210–227.
Gehler P, Nowozin S (2009) On feature combination for multiclass object classification In: Proceedings of 2009 IEEE 12th International Conference on Computer Vision, 221–228.
Ibrahim A, Alias A, Nor FM, Swarhib M, Abu Bakar SN, Das S (2017) Study of sexual dimorphism of Malaysian crania: an important step in identification of the skeletal remains. Anat Cell Biol 50:86–92.
Garvin HM, Sholts SB, Mosca LA (2014) Sexual dimorphism in human cranial trait scores: effects of population, age, and body size. Am J Phys Anthropol 154:259–269.
Acknowledgements
This research is sponsored by the eScienceFund grant 01-02-12-SF0288, Ministry of Science, Technology, and Innovation (MOSTI), Malaysia. The project has received full ethical approval from the Medical Research & Ethics Committee (MREC), Ministry of Health, Malaysia (ref: NMRR-14-1623-18717), and from the University of Nottingham Malaysia Campus (ref: IYL170414). Iman Yi Liao would like to thank the National Institute of Forensic Medicine (NIFM), Hospital Kuala Lumpur, for providing the PMCT data and is grateful to Dr. Ahmad Hafizam Hasmi (NIFM) and Ms. Khoo Lay See (NIFM) for their assistance in coordinating the data preparation.
Author information
Authors and Affiliations
Contributions
OAA and IYL designed the proposed method and drafted the manuscript. IYL and NA supervised the work. NA and MH coordinated the collection of the dataset. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Arigbabu, O.A., Liao, I.Y., Abdullah, N. et al. Computer vision methods for cranial sex estimation. IPSJ T Comput Vis Appl 9, 19 (2017). https://doi.org/10.1186/s41074-017-0031-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41074-017-0031-6