- Review
- Open access
- Published:
Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges
Genome Medicine volume 11, Article number: 45 (2019)
Abstract
Tuberculosis (TB) is a global infectious threat that is intensified by an increasing incidence of highly drug-resistant disease. Whole-genome sequencing (WGS) studies of Mycobacterium tuberculosis, the causative agent of TB, have greatly increased our understanding of this pathogen. Since the first M. tuberculosis genome was published in 1998, WGS has provided a more complete account of the genomic features that cause resistance in populations of M. tuberculosis, has helped to fill gaps in our knowledge of how both classical and new antitubercular drugs work, and has identified specific mutations that allow M. tuberculosis to escape the effects of these drugs. WGS studies have also revealed how resistance evolves both within an individual patient and within patient populations, including the important roles of de novo acquisition of resistance and clonal spread. These findings have informed decisions about which drug-resistance mutations should be included on extended diagnostic panels. From its origins as a basic science technique, WGS of M. tuberculosis is becoming part of the modern clinical microbiology laboratory, promising rapid and improved detection of drug resistance, and detailed and real-time epidemiology of TB outbreaks. We review the successes and highlight the challenges that remain in applying WGS to improve the control of drug-resistant TB through monitoring its evolution and spread, and to inform more rapid and effective diagnostic and therapeutic strategies.
Background
Mycobacterium tuberculosis is the causative agent of tuberculosis (TB), which is most often spread person-to-person via cough aerosols. Although many individuals who are exposed to M. tuberculosis never develop active disease, the World Health Organization (WHO) estimated 10 million new cases of active TB and 1.3 million deaths in 2017 alone [1].
Since its initial documentation in the 1940s [2], drug-resistant TB has threatened public health control efforts. In 2016, there were an estimated 490,000 new cases of multidrug-resistant (MDR) TB, which is defined by phenotypic resistance to both isoniazid and rifampicin [3]. Approximately 10% of MDR-TB cases globally can be classified as extensively drug-resistant (XDR), indicating that there is concomitant resistance to quinolones (such as the fluoroquinolones, levofloxacin, and moxifloxacin) and to a second-line injectable agent (amikacin, kanamycin, or capreomycin) [3]. As expected, drug-resistance patterns predict treatment outcome; in 2015, TB treatment success overall was 83%, whereas the success rate was 54% for MDR-TB or rifampicin-resistant-TB (RR-TB) and only 30% for XDR-TB [4].
Culture-based techniques remain the current reference standard for both diagnosis and drug-susceptibility testing of TB, but these processes are time-consuming and require specialized laboratory capacity. More recently, the use of rapid molecular tests for the diagnosis of TB has increased globally, particularly the use of Xpert MTB/RIF (Cepheid, Sunnyvale, CA), a PCR-based assay that simultaneously detects the presence of M. tuberculosis and resistance to rifampicin.
Current recommendations for the treatment of drug-susceptible TB include a 6-month course of a multi-drug regimen of rifampicin, isoniazid, pyrazinamide, and ethambutol. Historically, treatment of MDR- or XDR-TB involved the long-term use of second-line drugs, including injectable agents [5]. More recently, the MDR-TB treatment landscape has changed with the introduction of multiple novel second-line drugs that can be administered orally (Table 1). In 2012, bedaquiline, a diarylquinolone, became the first TB drug from a novel drug class to receive US Food and Drug Administration (FDA) approval in over 40 years [48, 49] (Table 1). Another oral agent, delamanid, a nitro-dihydro-imidazooxazole derivative, has also shown promise for TB treatment [50, 51].
In 2018, the WHO published updated treatment guidelines for MDR/RR-TB [47], recommending fully oral MDR regimens for many patient groups. Recommended treatment strategies include both shorter, standardized MDR regimens (for 9 to 12 months) and longer, individualized treatment regimens (for 18 to 20 months). The updated guidelines group antitubercular drugs on the basis of how they should be combined to create individualized, longer MDR-TB regimens [47] (Table 1).
Despite advances in both diagnostics and therapeutics for TB, challenges remain. Obstacles for rapid M. tuberculosis diagnosis include: (i) the imperfect sensitivity of molecular tests for the detection of this pathogen, particularly in the case of paucibacillary TB (where there is a lower bacterial burden); (ii) lack of comprehensive molecular assays due to incomplete knowledge of all resistance mutations in TB; and (iii) technical limitations to the numbers of mutations that can be included on diagnostic molecular platforms. Furthermore, the rollout of rapid diagnostic platforms to low-resource settings has been a challenge. Remaining treatment challenges include: prolonged treatment courses, leading to greater drug exposure, toxicity, and non-compliance; unacceptable side-effect profiles; logistics of drug access; and re-infection [52].
The dawning of the new age of genome sequencing began to revolutionize our approach to human diseases, including TB. In 1998, Cole et al. [53] reported the complete genome sequence of the M. tuberculosis reference strain H37Rv, which was approximately 4.41 million base pairs in length and encoded approximately 4000 genes. The first sequencing of a clinical reference strain, CDC1551, quickly followed [54]. An accompanying editorial optimistically stated: “After several decades in the slow lane of classical microbiology, M. tuberculosis is once again at the cutting edge of science” [55]. However, even at the time of these breakthroughs, there was recognition that translating these genomic data into clinical benefit would prove challenging [55]. Despite these challenges, it is clear, more than 20 years later, that M. tuberculosis genomic data have been remarkably useful in improving our understanding of how drug-resistant TB evolves and spreads and in helping to inform diagnostics and therapies.
In this review, we discuss the molecular epidemiologic and diagnostic advances made by sequencing M. tuberculosis, with a focus on drug-resistant TB. We do not review the practice of whole-genome sequencing (WGS) of M. tuberculosis as this has been reviewed recently [56]. Key findings that are discussed include the use of WGS to identify drug-resistance determinants in M. tuberculosis and to elucidate the evolution and spread of drug-resistant organisms, and the clinical applications of this technology (Table 2).
Identifying M. tuberculosis drug-resistance determinants
Drug resistance in M. tuberculosis is the result of chromosomal mutations in existing genes that are passed along through vertical descent, that is, passed from mother to daughter cells. Unlike many other bacterial pathogens, M. tuberculosis rarely recombines via lateral exchange of DNA [83] and also lacks plasmids. Many of the resistance determinants were discovered before the sequencing of the M. tuberculosis genome was completed. By 1998, resistance mechanisms had already been discovered for classical first- and second-line TB drugs including isoniazid (alterations in genes katG and inhA); rifampicin (in rpoB); streptomycin (in rrs and rpsL); pyrazinamide (in pncA); ethambutol (in embB); quinolones (in gyrA); and kanamycin (in rrs) (reviewed in Ramaswamy and Musser [84]) (Table 1). However, the targeted amplification and sequencing of known or suspected resistance genes revealed that these mechanisms were insufficient to explain all phenotypic resistance [85, 86], and resistance mechanisms for several newer drugs—including pretomanid, bedaquiline, and delamanid—were discovered over the next eight years during a period when WGS was becoming routine. Together, in the past 20 years, WGS-based approaches, focused on both laboratory-derived and naturally circulating populations of drug-resistant M. tuberculosis, have provided a more complete account of the genomic features that cause treatment resistance, enabling the identification of novel resistance mechanisms for existing drugs, and the determination of the mechanisms of action of newly discovered drugs.
Identifying resistance determinants in laboratory-derived mutants
Drug-resistant mutants can be derived in vitro by growing drug-susceptible M. tuberculosis strains in drug-containing media, and selecting for mutants that are able to grow in the presence of the drug. Sequencing laboratory-derived resistant mutants has played a critical role in identifying both the mechanism of action of new TB drug classes, including diarylquinolines (for example, bedaquiline) [19] and nitroimidazopyrans (for example, PA-824) [19, 29], and rare resistance mechanisms for established antitubercular drugs, including ethambutol [15], pyrazinamide [13], carbapenems [30], cycloserine [87], clofazimine, and bedaquiline [20]. For example, WGS of laboratory mutants identified drug efflux as a mechanism of resistance to clofazimine and bedaquiline [20,21,22], and this approach continues to be a mainstay for identifying the mechanism of action of compounds that are in development for TB [88].
Although laboratory-derived mutants are helpful in elucidating novel resistance mechanisms, mutations that have evolved in laboratory settings do not always match those in clinical isolates of drug-resistant M. tuberculosis [89, 90], for reasons that are largely unknown. Studies by Ford et al. [91, 92] suggested that these mismatches could not be explained by differences in the mutation rate in these settings, because the in vitro mutation rate of M. tuberculosis correlates well with the in vivo mutation rate in both humans and in a macaque model. Differences in the relative fitness of specific mutants grown in in vitro compared to in vivo conditions are probably responsible for these mismatches, but more work is needed. Regardless of the reason, if the goal is to identify a full complement of resistance mutations on which to base molecular diagnostics, isolates from clinical collections must be studied as these bacteria have evolved their resistance within the host.
Quantifying and identifying resistance determinants in clinical strains
Among the larger studies exploring resistance in natural populations, Walker et al. [58] analyzed the genomes of 3651 drug-resistant and -susceptible M. tuberculosis isolates for associations between phenotypic resistance to eight first- and second-line drugs, and then predicted genotypic resistance on the basis of a compiled catalog of 232 resistance mutations in 23 candidate resistance genes. Resistance to most drugs could be predicted accurately, with a mean sensitivity of 92% and specificity of 98%, suggesting that the majority of resistance—particularly for first-line drugs—is explained by known mechanisms and mutations (Table 1). Numerous other studies have found similar results using smaller datasets [7, 25, 57, 69, 93, 94]. This result was echoed in a more recent study by the Comprehensive Resistance Prediction for Tuberculosis (CRYPTIC) Consortium and the 100,000 Genomes Project that focused solely on first-line drugs, which included analysis of 10,209 globally diverse M. tuberculosis isolate genomes against a database of mutations identified in a literature search [60]. Notably, predictions for mutations that are associated with resistance to pyrazinamide were greatly improved over earlier predictions; this study achieved 91.3% sensitivity in predicting resistance to this drug compared to 57% sensitivity in Walker et al. [58]. Although the news has been encouraging with respect to completing the catalog of mutations that cause resistance to first-line drugs, few studies have attempted to predict resistance to second-line drugs [95]. Some of these drugs, such as D-cycloserine, pyrazinamide, and para-aminosalicylic acid (PAS), are more difficult to assay because they have been reported to have variable drug phenotypes in clinical microbiology laboratories [96] (discussed later).
To fill gaps in the catalog of drug-resistance mechanisms, genome-wide association study (GWAS) approaches, originally designed for use on human genomic data, have been adapted for non-recombining microbes such as M. tuberculosis and used to predict novel resistance mechanisms [97, 98] (Table 3). The majority of GWAS predictions remain experimentally unverified, but several new resistance-associated genotypes have been validated. Farhat et al. [7] sequenced 116 M. tuberculosis genomes and developed a phylogenetic convergence test, ‘PhyC’, to identify resistance associations. They identified a mutation in ponA1 (c.1095G>T) and showed that it conferred a minimum inhibitory concentration (MIC) to rifampicin that was twofold higher than that of wild-type bacteria. Zhang et al. [57] sequenced 161 genomes from China and searched for genes that appeared to be under positive selection and more frequently mutated in drug-resistant isolates. Resistance-associated polymorphisms in two intergenic regions upstream of the known resistance genes thyA-Rv2765 and thyX-hsdS.1 were shown to cause increased gene expression of a lacZ construct in Mycobacterium smegmatis, suggesting that these mutations may mediate PAS resistance through the overexpression of downstream genes.
Desjardins et al. [25] used a combination of the correlated evolution test [104] (to test for correlated evolution of genotype and phenotype) and a simple GWAS framework to search for novel drug-resistance mechanisms in 498 genomes from South Africa and China. Of note, they combined all variants within each gene that were predicted to inactivate gene function, and used these combinations as the input into the association test to increase statistical power in the detection of genomic features that are associated with resistance. They found that loss-of-function mutations in ald (Rv2780), which is predicted to encode an alanine dehydrogenase, correlated with unexplained resistance [25]. They also confirmed experimentally that these mutations conferred increased resistance of laboratory and clinical isolates to D-cycloserine [25], a key drug in MDR- and XDR-TB regimens that has severe psychiatric and central nervous system toxicities.
Hicks et al. [105] used the algorithm phyOverlap to perform a GWAS on 549 clinical isolates from China, in which they identified mutations that disproportionately occurred in isoniazid-resistant isolates. In addition to known resistance and compensatory mutations for first- and second-line drugs, they identified an association with prpR (Rv1129c). They then went on to characterize prpR as a transcriptional regulator of propionate metabolism which, instead of drug resistance, confers tolerance to multiple antibiotics in a macrophage model of infection.
In one of the largest GWAS published to date, Coll et al. [59] combined PhyC with a GWAS approach within a mixed-regression framework to detect determinants of resistance to 14 drugs in a large dataset of 6465 global M. tuberculosis clinical isolates. Although no functional experiments were performed to validate the predictions, new resistance-associated mutations were identified, including new codons in ethA (a gene known to activate ethionamide, which is a prodrug) that are associated with ethionamide resistance, and mutations in the thyX promoter associated with PAS resistance. Mutations in the promoter of thyX have been previously shown to upregulate thyX [43, 57, 106].
Predicting susceptibility and drug resistance in M. tuberculosis
As the list of suspected resistance determinants grows, there has been a need to establish well-curated databases of drug-resistance single nucleotide polymorphisms (SNPs) [107]. Initially, SNP databases, including TBDB [108] and PATRIC [109], were created to bring together genome annotation data and other functional data. Unfortunately, some of the pioneering databases of drug-resistance-associated mutations in M. tuberculosis, including TBDReamDB [110], have not been maintained to include emerging data.
Software and web-based tools have also been developed to enable the community to infer TB drug resistance from WGS data. These tools include CASTB [111], KVarQ [112], MyKrobe Predictor TB [113], PhyResSE [114], TBProfiler [115], and TGS-TB [116]. Studies have compared the sensitivity and specificity of these tools in predicting drug resistance [117,118,119], and have found that they tend to perform quite well for first-line drugs but less optimally for second-line drugs. In addition to tools, there have been improvements to databases, including the creation of the Relational Sequencing TB Database Platform (ReSeqTB) [120, 121] and efforts from the CRyPTIC Consortium [122], which seeks to develop a curated database of clinically relevant drug-resistance mutations.
Continued refinement of these drug-resistance databases and prediction tools is necessary. Miotto et al. [123] performed a systematic review in which they assigned a confidence level to associations of individual and groups of mutations with phenotypic drug resistance. Importantly, they identified that certain mutations that are included in current commercial diagnostic tests, including eis c-2a, do not have a convincing association with drug resistance. Input from ongoing large sequencing projects will be needed to optimize the inference of resistance phenotypes from sequence data, especially for mutations that are present at low frequency in natural populations.
Challenges in uncovering the remaining resistance elements
Although WGS approaches have been successful in identifying resistance mechanisms, there are computational and experimental challenges that hamper efforts to complete the catalog of TB drug resistance. For example, for non-recombining organisms such as M. tuberculosis, interpretation of GWAS output can be complicated because non-causal variation can be tightly linked to causal variation [124]. Furthermore, as a result of frequent multidrug resistance, resistance mutations for one drug can appear to be highly associated with phenotypic resistance to multiple drugs [25], and confirmatory wet lab studies, which are non-trivial in M. tuberculosis, are often necessary to identify causal resistance mutations correctly. In addition, genotype–phenotype associations are largely dependent on accurate phylogenies, and phylogenetic reconstruction can be challenging in M. tuberculosis because of its slow rate of evolution [92, 125,126,127,128], which gives rise to relatively few SNPs in clinical isolates.
When defining phenotypic resistance, different studies often use different drug concentration cutoffs and test in different media, complicating the meta-analysis of multiple datasets. In addition, phenotypic resistance testing of some antitubercular drugs, including pyrazinamide and D-cycloserine, is notoriously challenging and unreliable [129], introducing phenotypic inaccuracies that can confound analyses. Furthermore, the dichotomous classification of phenotypic resistance as ‘resistant’ or ‘susceptible’ will fail to identify drug-resistance mutations that result only in minimal increases in MIC, and there is emerging evidence that such mutations may be clinically relevant. TB relapse following treatment has been found to occur more commonly in individuals who harbored M. tuberculosis isolates that were susceptible to, yet had minimally increased MIC values for, either isoniazid or rifampicin [130]. Future study designs that address phenotypic resistance as a spectrum, rather than a binary value, will be needed to identify such mutations.
Heteroresistance, defined as the coexistence of pathogen populations that have differing nucleotides at a specific drug-resistance locus [131], can also confound genotype–phenotype comparisons [132,133,134]. A bacterial culture in which only a small fraction of the population is resistant can appear to be resistant when tested on media containing a drug, yet when grown on drug-free media for genome sequencing, the sensitive fraction can dominate, resulting in a genotypic prediction of sensitivity [132]. The problem of heteroresistance seems to be particularly common with fluoroquinolone resistance [135].
Last, innate characteristics of the M. tuberculosis genome—namely, highly repetitive DNA sequences and the high guanine-cytosine (GC) content of the genome (65.6%) [53]—present technical difficulties for both WGS and bioinformatic analyses. GC-rich regions can be troublesome for library PCR amplification and sequencing, and reads that represent highly repetitive regions of the genome can confound alignments by mapping to multiple regions of the genome and hampering accurate de novo assemblies. In addition, approximately 10% of the coding regions in M. tuberculosis are dedicated to two repetitive protein families that are unique to mycobacteria (the PE and PPE families), which have conserved Pro-Glu (PE) and Pro-Pro-Glu (PPE) motifs [53]. Even with WGS investigation [136], the function of the PE and PPE genes has remained elusive, although recent studies have suggested that they may play a role in virulence [137]. Their association with drug resistance remains largely unexplored because bioinformatic studies of M. tuberculosis often exclude these genes [138, 139]. In the future, long-read sequencing technology may allow these regions to be sequenced successfully in order to assess if they have a role in drug resistance.
Understanding the evolution and spread of drug resistance in M. tuberculosis
Prior to WGS, the diversity and epidemiology of resistant M. tuberculosis were examined using DNA fingerprinting techniques, like IS6110 restriction fragment length polymorphism (RFLP) typing [140], spoligotyping (spacer oligonucleotide typing, a method of typing strains according to the distinct hybridization patterns of their spacer oligonucleotides) [141], and mycobacterial interspersed repetitive units-variable number of tandem repeats (MIRU-VNTR) typing [142,143,144,145]. These techniques enabled assessments of the diversity of resistant strains in specific geographic regions [146,147,148,149] and, when combined with the genetic profiling of resistance mutations, allowed strain-level monitoring of patients on TB therapy [150].
The dramatic increase in resolution afforded by WGS has extended the sensitivity and resolution with which the diversity and evolution of drug-resistant M. tuberculosis can be assessed. This has resulted in the more confident identification of cases of recent transmission [151] and re-infection [152], and has provided insights into the evolution of resistance within individual patients and across populations. WGS has also enabled more sensitive differentiation of de novo acquisition of resistance (where resistance mutations emerge within a host) from person-to-person transmission of resistance, a critical capability given that these two scenarios require different health-system responses in order to stem resistance.
Within patient evolution of drug resistance
Despite the slow evolutionary rate of M. tuberculosis, estimated at 0.3–0.6 SNPs/genome/year [69, 92, 125,126,127,128], experimental data suggest that drug resistance can evolve within an individual patient during TB treatment. Eldholm et al. [61] described the first documented case of XDR evolution of M. tuberculosis from a fully susceptible ancestor within a single patient, by sequencing nine serial isolates collected over a 42-month period. During this time, seven known resistance mutations emerged in a stepwise fashion after the clinical use of each corresponding drug, revealing how TB drug pressures can rapidly shape M. tuberculosis populations in vivo.
However, the evolution of drug resistance within a host is not always linear, and instead can involve a complex interplay of heterogeneous M. tuberculosis populations [153, 154]. In particular, transient genetic diversity can exist before a dominant clone emerges. In addition, as the size of the transmission bottleneck (the number of bacteria transmitted during an infection event) in M. tuberculosis is not well understood [155], it is difficult to estimate the relative contribution of diversity that is transmitted to the patient versus diversity that evolves within the patient. Numerous WGS studies, performed either on isolates or directly on DNA extracted from serially collected sputum samples, have revealed substantial transient genetic diversity in pathogen populations within patients, particularly within resistance genes [61, 62, 106, 156,157,158,159]. This diversity was observed to endure months before a single variant became fixed in the population (the situation when only a single variant remains). In the study by Eldholm et al. [61] mentioned above, the seven resistance-conferring mutations that eventually dominated were from amongst 35 mutations observed in total throughout the sampling period [61, 160]. They joined eight other mutations that were not resistance-associated but that also became fixed in the population, probably as the result of a phenomenon called ‘hitchhiking’ in which non-adaptive mutations are selected because of their linkage and physical proximity to consequential mutations.
The relative fitness cost of drug-resistance mutations often determines which mutations become fixed within a host. While multiple mutations that confer resistance to a specific drug can evolve repeatedly, mutations conferring no or little fitness cost are typically selected, resulting in fixed dominant mutations [61, 156]. Compensatory mutations (discussed in more detail later), which serve to counterbalance the deleterious effects of acquired resistance, have also been shown to emerge during treatment [156].
WGS has also revealed how combination chemotherapy effectively prevents the emergence of drug resistance during treatment for TB. In a study of very deep WGS of serial sputum specimens from patients receiving treatment for TB, Trauner et al. [62] demonstrated that the combined action of multiple active drugs prevented transient mutants from fixing within a population and becoming dominant. The fewer the drugs that were applied, the more likely it was that resistance would develop and become fixed.
Population views of drug-resistance evolution
A number of careful WGS studies have empirically established SNP-based criteria to discriminate cases of recent transmission from unrelated infections—usually using the criterion that recently transmitted strains differ by fewer than 6–12 total SNPs across the M. tuberculosis genome [63, 125, 126, 161]. In a 2016 review, Nikolayevskyy and colleagues [63] systematically compared WGS to fingerprinting techniques for detecting transmission, including a meta-analysis of 12 studies published between 2005 and 2014. They concluded that results from WGS studies not only have higher discriminatory power, but they also enable more sensitive detection of transmission events that may have been missed by epidemiologic methods.
Although traditional spoligotyping analyses suggested that drug-resistant strains were diverse, WGS of clinical isolates began to reveal the full breadth of diversity in resistant M. tuberculosis. The TB epidemic in South Africa over the past two decades has been well-studied in this regard. In an early WGS investigation, Ioerger et al. [64] examined 14 phenotypically diverse strains within the Beijing lineage and showed that resistance mutations arose independently multiple times, and that XDR isolates may be less fit and less able to transmit. WGS studies across larger sets of strains from the same region in South Africa suggested that, although de novo resistance is indeed common, highly resistant strains (including MDR and XDR strains) have the ability to spread broadly by person-to-person transmission. This includes the ongoing transmission of a circulating XDR clone in South Africa that is linked to the infamous Tugela Ferry XDR outbreak [162] that brought XDR-TB to the world stage in 2005. A more recent large-scale study confirmed that XDR strains have been broadly transmitted person-to-person in KwaZulu-Natal [65].
The patterns observed in South Africa hold for many other parts of the world. Recent studies have shown that patterns of both de novo evolution and person-to-person spread of drug resistance in M. tuberculosis also occur in Belarus, Russia, England, and Malawi [73, 139, 159, 163, 164]. In a composite analysis of over 5000 M. tuberculosis isolates from patients from around the globe, Manson et al. [66] confirmed that both de novo evolution and person-to-person transmission are important factors for the rise and spread of drug-resistant TB worldwide. The emergence of MDR and XDR M. tuberculosis was found to be a frequent occurrence that is distributed fairly evenly across the globe [66]. This analysis also predicted that 37% of MDR isolates in this study had spread person-to-person, which is probably a vast underestimate of how frequently MDR is transmitted once evolved [66].
Geographic movement of people is also an important consideration with regard to person-to-person transmission. Further examination of the MDR clades from Manson et al. [66] revealed that they included widespread international, and even intercontinental, dissemination of strains that were separated by as few as four SNPs, probably due to spread via international travel [67]. Even within a single province in South Africa, Nelson et al. [68] showed, using genomic sequence data and global positioning system coordinates, that many cases of person-to-person transmission (with ≤ 5 SNPs) of XDR-TB occur between people living a median of 108 km apart, pointing to migration between urban and rural areas as a driver of TB spread. Collectively, these studies reinforce the idea that the geographic movement of people must be taken into consideration in any strategy for controlling the spread of TB resistance.
Ordering of the acquisition of resistance and compensatory mutations
Recent WGS studies have helped to illuminate the steps or ‘fitness landscape’ through which M. tuberculosis develops and compensates for drug resistance. Several studies [66, 69, 70] have shown that the order of acquisition of drug-resistance mutations in complex resistance cases is partly constrained in clinical M. tuberculosis. For example, in MDR-TB, isoniazid resistance (most often involving a katG S315T mutation) overwhelmingly evolves prior to resistance to rifampicin and second-line drugs. This was first shown using regional datasets from South Africa [69] and Argentina [70], and recently confirmed by Manson et al. [66] using a global dataset of 5310 strains. In the study by Manson et al. [66], this ordering was shown to hold true over 95% of the time, even for distinct global regions and time frames, including times when both rifampicin and isoniazid were in use, suggesting that the earlier introduction of isoniazid in the 1950s was not the major contributor to this effect. It was also shown that inhA promoter mutations that confer isoniazid resistance (such as those observed by Perdigão et al. [165] in Portugal) were acquired earlier than rifampicin mutations, although the number of samples harboring these mutations was much smaller. Further studies are necessary to determine whether isoniazid preventive monotherapy, which is one of the treatments for latent tuberculosis, may account for some of this effect, as this could result in a background level of increased isoniazid monoresistance.
Compensatory mutations that potentially ease fitness effects caused by resistance often occur after the evolution of primary resistance. This phenomenon was reviewed by Fonseca et al. [71], and examples include mutations in the ahpC promoter region and the rpoC/rpoA genes, which act as compensatory mutations for isoniazid and rifampicin resistance, respectively. Newer WGS work has pointed to several novel compensatory mutations in M. tuberculosis, particularly for rifampicin resistance. Comas et al. [72] identified a set of compensatory mutations in the rpoB gene that conferred high competitive fitness in vitro and were also found frequently in clinical populations. In a large-scale analysis of 1000 strains from Russia, Casali et al. [73] examined strains with primary resistance mutations in rpoB and identified accompanying compensatory mutations in rpoA and rpoC. Cohen et al. [69] identified putative rifampicin compensatory mutations that are present in South African strains by searching for rpoA, rpoB, and rpoC mutations that evolved only after or concurrent with rifampicin resistance-conferring mutations. A recent study of highly resistant M. tuberculosis strains from Central Asia confirmed that the presence of compensatory mutations, particularly those compensating for the fitness cost of mutations that confer rifampicin resistance, is associated with transmission success and higher drug-resistance rates [74]. Beyond rifampicin resistance compensation, Coll et al. [59] identified mutations in pncB2 that may compensate for pyrazinamide resistance conferred by pncA, and similarly, mutations in thyX-hsdS.1 (the thyX promoter) that may compensate for PAS resistance conferred by thyA; however, experimental validation of these potential compensatory relationships is needed. Even fewer studies have identified stepping-stone mutations in M. tuberculosis, which emerge prior to higher-level resistance mutations. Cohen et al. [69] found that ubiA mutations emerge in a stepping-stone fashion prior to more classic embB mutations that confer ethambutol resistance. Safi et al. [15] also showed in vitro that multi-step selection involving ubiA, aftA, embB, and embC is required to achieve the highest levels of ethambutol resistance.
The challenge of mixed infections
Although WGS approaches have great sensitivity in detecting cases of recent transmission, reconstructing the details of transmission networks with WGS [166,167,168] can be difficult. Transmission network mapping is highly dependent on sampling density and studies rarely, if ever, comprehensively sample an outbreak or the extent of within-host diversity. It is also becoming clear, from the prevalence of very close relationships between isolates from patients who have no other direct epidemiological connections, that transmission may largely result from casual contact in community settings [169]. In addition, the phylogenetic reconstruction of transmission networks can be especially challenging, particularly because of the very close relationships between strains and the slow rate of evolution of M. tuberculosis [92, 125,126,127,128].
Mixed infections represent a major challenge for understanding drug-resistance evolution within patients [153, 158, 159]. It can be straightforward to disambiguate co-infections of strains from different lineages, but mixed infections involving strains that have few genetic differences can also occur, making these strains difficult to distinguish. Köser et al. [75] used WGS for rapid drug-susceptibility testing of a patient with XDR-TB, and determined that the patient carried two different XDR-TB Beijing strains with differing resistance mutations. In a study by Liu et al. [76], three dominant sub-clones differing by 10–14 SNPs were detected within a single patient, each with different resistance patterns and probably different anatomical distributions. Also, co-infection by strains with differing resistance patterns may yield misleading composite views of resistance; for example, co-infection with two MDR-TB strains—one with quinolone resistance and the other with aminoglycoside resistance—may be mistaken for infection with a single XDR-TB strain.
Furthermore, newer data suggest that there can be genetic heterogeneity among M. tuberculosis isolates from different parts of the body, potentially leading to incomplete views of drug resistance within a patient (Fig. 1). In a study by Lieberman et al. [77], the authors observed evidence for both within-host evolution and mixed infection by piecing together the genetic variation observed among M. tuberculosis isolates from multiple post-mortem biopsies from the same patient. Another recent study by Dheda et al. [78] showed that drug concentrations at seven body sites were inversely correlated with the MIC of the bacteria isolated from these sites. Sequencing and comparison to pre-treatment and serial sputum samples suggested ongoing acquired resistance and differential evolution across sites [78]. These findings underscore the limitations of diagnosing or studying the evolution of drug-resistant M. tuberculosis using a single patient specimen. However, they also show the promise of WGS for informing interventions related to drug delivery, dosing, and diagnostics, thereby helping to prevent the development of acquired resistance within a patient. More research in this area is needed to determine the breadth and scope of mixed infections among patients with active TB, their contribution to changing drug-resistance patterns over time, and the role of spatial heterogeneity in the evolution of drug resistance.
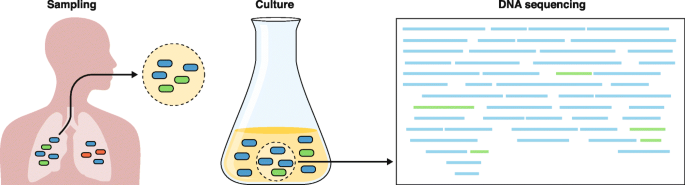
Challenges to predicting drug resistance accurately from clinical specimens using current culture-dependent molecular diagnostics. The left panel depicts an expectorated sputum sample, which may not accurately represent the microbiologic diversity within the source patient. Culturing this sample (center panel) introduces further biases between faster- and slower-growing strains, such that faster-growing strains are over-represented within the cultured sample. Genomic DNA that is isolated and sequenced is input to computer algorithms that determine the genomic content, including the identification of drug-resistance mutations. However, disambiguating samples that contain mixed strains or detecting heteroresistance remains a computational challenge. The left panel was adapted from Ford et al. [170], with permission from Elsevier
From bench to bedside: promise and challenges
Given that the failure to identify and treat patients who have drug-resistant TB leads to increased mortality, spread of resistant strains, and gain of additional drug resistance [171], there is a critical need to diagnose resistant M. tuberculosis in patients rapidly. Several important molecular diagnostic platforms have been established for the identification of M. tuberculosis and drug resistance within this organism, but they are limited to the identification of a defined subset of resistance mutations [172], do not always include the earliest-arising mutations that precede MDR [66], and do not provide knowledge that is useful in determining whether a patient has been re-infected, whether the patient has a recurrent or mixed infection, or whether a particular infection represents a transmission event. WGS holds significant potential to modernize the TB laboratory and improve upon TB management [173], and this topic has been reviewed previously [173, 174]. To date, WGS has been primarily applied as a clinical tool to achieve two goals: first, to detect M. tuberculosis within a clinical sample, and second, to detect resistance mutations and predict resistance patterns so that appropriate treatment can be provided. In order to provide clinically useful information, a diagnostic platform must be rapid. Historically, WGS has relied upon an input of pure mycobacterial cultures, which is time-consuming (requiring multiple weeks) and therefore of less clinical utility. Several investigations have attempted to address this issue by using earlier culture inputs or by attempting culture-independent, direct sequencing from clinical specimens [80, 82, 175]. In a rapid, yet still culture-dependent method, Pankhurst et al. [80] prospectively compared real-time WGS of “early positive liquid cultures” to routine M. tuberculosis diagnostics, and found that WGS achieved a faster time to diagnosis at a lower cost.
Although the advances achieved using WGS are promising, several hurdles must be overcome before it can be put into practical use in the clinic (Fig. 1). Requirements for costly equipment, technical expertise, and substantial computational resources present challenges to implementation [173]. Direct sequencing of patient samples has revealed that the vast majority of DNA present is from the patient or from non-mycobacterial prokaryotes, with variable quantities of mycobacterial DNA present. Doughty et al. [81] performed a pilot study demonstrating the feasibility of direct sequencing using a benchtop sequencer (Illumina MiSeq, San Diego, CA) and sputum samples from eight patients. Although they were able to identify the presence of M. tuberculosis, the low depth of sequencing coverage of the genome (0.002 to 0.7x) prevented drug-susceptibility prediction. Separately, Brown et al. [176] performed an enrichment step with biotinylated RNA baits prior to direct sequencing of sputum, resulting in higher quality data (> 20x depth and > 90% coverage) that allowed the identification of resistance mutations.
Using a targeted DNA enrichment strategy to study 43 individuals with active pulmonary TB, Doyle et al. [177] compared WGS directly from sputum with mycobacterial growth indicator tube (MGIT) WGS. Although direct sputum sequencing was able to identify drug resistance much more rapidly than MGIT WGS, only 74% of sputum samples yielded interpretable WGS data (vs 100% from MGIT); thus, additional optimization of these methods is needed to increase the sensitivity of this approach. Similarly, in a recent study, the use of pyrosequencing of a concentrated sputum sediment (rather than from sputum directly), dramatically shortened the time to initiation of an MDR-treatment regimen [178].
One promising technology that could change clinical WGS is long-read sequencing using the Oxford Nanopore Technologies (ONT; Oxford, UK) platform. An advantage of ONT is the ability to allow sequencing to continue until sufficient coverage of the genome has been obtained, potentially solving the problem of low or variable amounts of M. tuberculosis in clinical samples [82]. Early ONT studies have shown promise in identifying antimicrobial-resistance genes in different bacterial species [179]. Unfortunately, at present, both the high error rate of ONT MinION and potential difficulties with GC-rich regions limit the utility of this technology; thus, improvements in accuracy are necessary to enable the identification of resistance associated with point mutations [179]. ONT metagenomic sequencing has been successfully applied to improve pathogen detection and antimicrobial-resistance testing in other clinical settings [180]; however, to date, applications of this technology to M. tuberculosis have been limited to pre-clinical research [82].
Despite these challenges, WGS offers several advantages over the technologies that are currently employed for diagnosis and epidemiological monitoring of TB. Using WGS directly on patient sputum could reduce the turnaround time for diagnosis and determination of antibiotic-resistance status from weeks to hours [61, 159], and would prevent the introduction of culture-induced biases. The depth of information provided by WGS could be used to identify whether an individual harbors multiple co-infecting strains [106, 160] and to distinguish recurrent infection as either relapse or re-infection [174, 181]. In addition, WGS could provide real-time epidemiological information that could be useful for understanding patterns of drug resistance and for establishing chains of transmission [174]. Encouragingly, the high levels of concordance observed between the genotypes and phenotypes of clinical samples indicate that WGS can provide high accuracy for both diagnosing TB and informing treatment options [113]. Finally, WGS of patient samples would provide a high level of convenience, by combining diagnosis, resistance profiling, and epidemiological analysis into a single test [85]. Given these advantages, the WHO has recently published a technical guide for the implementation of next-generation sequencing (NGS) technologies for the detection of drug resistance in M. tuberculosis [182].
Routine whole-genome sequencing of mycobacterial isolates
In 2017, England became the first nation to launch routine WGS of all prospectively identified M. tuberculosis clinical isolates [183]. Sponsored by Public Health England (PHE), prospective WGS is being performed on all positive mycobacterial cultures. Within 5–7 days of receipt of the culture from the reference lab, data will be provided on the mycobacterial species, the predicted drug susceptibility, and the molecular epidemiology of the strains. If, from the sequence analysis, a strain is predicted to be fully susceptible to first-line antitubercular drugs, phenotypic drug-susceptibility testing (DST) will no longer be performed routinely. However, if drug resistance to any first-line drug is identified, then phenotypic DST will follow. Beyond drug-susceptibility prediction, these efforts will have profound implications for TB control because WGS data can be used for real-time molecular epidemiology in this context.
Given the high sensitivity of WGS in detecting drug resistance to first-line TB drugs [60], similar algorithms utilizing WGS to predict susceptibility (rather than to identify drug resistance) for first-line drugs, in lieu of phenotypic DST, have been endorsed in the Netherlands and in New York [60]. It seems highly likely that these kinds of efforts would be helpful in higher-burden TB settings than those mentioned here, but the feasibility of this approach has not yet been established, from either a practical or economic standpoint, in settings where the numbers of drug-resistant TB cases are high.
Conclusions and future directions
Since the first applications of WGS to M. tuberculosis in 1998, WGS techniques have greatly accelerated our understanding of drug-resistance mechanisms in this pathogen. Importantly, WGS studies now indicate that, for many drugs, the vast majority of resistance is explained by known mutations. The increasing availability of whole-genome sequences from phenotypically diverse M. tuberculosis, combined with improved GWAS algorithms, is enabling the discovery of the remaining determinants of unexplained resistance. In addition, WGS has provided valuable insight into how resistance mutations evolve and spread. It is clear that both de novo acquisition of resistance mutations and clonal transmission are critical factors in the spread of drug-resistant TB.
Furthermore, WGS investigations have revealed that there is a specific order in which drug-resistance mutations are acquired: isoniazid resistance is almost always acquired before rifampicin resistance, which has significant implications for the design of diagnostic tests. Within individual patients, WGS studies have highlighted that mixed infections are common, and often represent important intermediates in the evolution of drug resistance.
WGS also holds great promise for revolutionizing the rapid clinical diagnosis of TB in the future. Although there are still substantial technical hurdles, WGS can be used to diagnose the presence of M. tuberculosis rapidly, as well as to pinpoint appropriate antibiotic treatment regimens by identifying the complement of M. tuberculosis drug-resistance mutations that are present within a clinical sample. Indeed, improvements in the prediction of drug susceptibility with WGS may obviate the need for phenotypic culture methods, especially for first-line drugs.
Although WGS offers many benefits, targeted NGS, in which sequence data are obtained from only a focused panel of genes or genetic regions rather than from the entire genome, is gaining momentum [184]. One of the advantages of targeted NGS over WGS is that it can be performed directly on clinical specimens and is, therefore, faster than culture-based WGS. Other advantages include reduction in both labor and computational efforts and reduced costs. The potential offered by the application of targeted NGS to the prediction of drug resistance from genomic data is self-evident. Nevertheless, it seems that WGS would have greater discriminatory power than targeted NGS for molecular epidemiology purposes.
Ultimately, the use of WGS is expected to continue to advance our understanding of M. tuberculosis drug resistance. Furthermore, its practical use in clinical settings holds great potential to improve public health through real-time molecular epidemiology tracking, to identify global hotspots of drug-resistance emergence, and to facilitate the development of improved approaches for the diagnosis and treatment of drug-resistant TB.
Abbreviations
- DST:
-
Drug-susceptibility testing
- GWAS:
-
Genome-wide association study
- MDR:
-
Multidrug-resistant
- MIC:
-
Minimum inhibitory concentration
- NGS:
-
Next generation sequencing
- ONT:
-
Oxford Nanopore Technologies
- PAS:
-
Para-aminosalicylic acid
- RR-TB:
-
Rifampicin-resistant-TB
- SNP:
-
Single nucleotide polymorphism
- TB:
-
Tuberculosis
- WGS:
-
Whole-genome sequencing
- WHO:
-
World Health Organization (WHO)
- XDR:
-
Extensively drug-resistant
References
World Health Organization. Global Tuberculosis Report 2018. Geneva: WHO; 2018. http://apps.who.int/iris/bitstream/handle/10665/274453/9789241565646-eng.pdf?ua=1. Accessed 9 July 2019
Streptomycin in Tuberculosis Trials Committee. Streptomycin treatment for pulmonary tuberculosis. Br Med J. 1948;2:769–82 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2091872/pdf/brmedj03701-0007.pdf. Accessed 9 July 2019.
World Health Organization. Global Tuberculosis Report 2016. Geneva: WHO; 2016. https://apps.who.int/iris/bitstream/handle/10665/250441/9789241565394-eng.pdf?sequence=1. Accessed 9 July 2019
World Health Organization. Global Tuberculosis Report 2017. Geneva: WHO; 2017. https://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf. Accessed 9 July 2019
World Health Organization. WHO treatment guidelines for drug-resistant tuberculosis. 2016 update. Geneva: WHO; 2016. https://apps.who.int/iris/bitstream/handle/10665/250125/9789241549639-eng.pdf?sequence=1. Accessed 9 July 2019
Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston MJ, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–50.
Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45:1183–9.
Heym B, Alzari PM, Honoré N, Cole ST. Missense mutations in the catalase-peroxidase gene, katG, are associated with isoniazid resistance in Mycobacterium tuberculosis. Mol Microbiol. 1995;15:235–45.
Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98–102.
Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263:227–30.
Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–40.
Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, et al. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–3.
Zhang S, Chen J, Shi W, Liu W, Zhang W, Zhang Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect. 2013;2:e34.
Shi W, Zhang X, Jiang X, Yuan H, Lee JS, Barry CE, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333:1630–2.
Safi H, Lingaraju S, Amin A, Kim S, Jones M, Holmes M, et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-β-D-arabinose biosynthetic and utilization pathway genes. Nat Genet. 2013;45:1190–7.
Alcaide F, Pfyffer GE, Telenti A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob Agents Chemother. 1997;41:2270–3.
Maruri F, Sterling TR, Kaiga AW, Blackman A, van der Heijden YF, Mayer C, et al. A systematic review of gyrase mutations associated with fluoroquinolone-resistant Mycobacterium tuberculosis and a proposed gyrase numbering system. J Antimicrob Chemother. 2012;67:819–31.
Takiff HE, Salazar L, Guerrero C, Philipp W, Huang WM, Kreiswirth B, et al. Cloning and nucleotide sequence of Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–80.
Andries K, Verhasselt P, Guillemont J, Göhlmann HWH, Neefs J-M, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–7.
Almeida D, Ioerger T, Tyagi S, Li SY, Mdluli K, Andries K, et al. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2016;60:4590–9.
Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58:2979–81.
Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, et al. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0102135.
Hillemann D, Rüsch-Gerdes S, Richter E. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob Agents Chemother. 2008;52:800–1.
Beckert P, Hillemann D, Kohl TA, Kalinowski J, Richter E, Niemann S, et al. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob Agents Chemother. 2012;56:2743–5.
Desjardins CA, Cohen KA, Munsamy V, Abeel T, Maharaj K, Walker BJ, et al. Genomic and functional analyses of Mycobacterium tuberculosis strains implicate ald in D-cycloserine resistance. Nat Genet. 2016;48:544–51.
Cáceres NE, Harris NB, Wellehan JF, Feng Z, Kapur V, Barletta RG. Overexpression of the D-alanine racemase gene confers resistance to D-cycloserine in Mycobacterium smegmatis. J Bacteriol. 1997;179:5046–55.
Feng Z, Barletta RG. Roles of Mycobacterium smegmatis D-alanine:D-alanine ligase and D-alanine racemase in the mechanisms of action of and resistance to the peptidoglycan inhibitor D-cycloserine. Antimicrob Agents Chemother. 2003;47:283–91.
Chen JM, Uplekar S, Gordon SV, Cole ST. A point mutation in cycA partially contributes to the D-cycloserine resistance trait of Mycobacterium bovis BCG vaccine strains. PLoS One. 2012. https://doi.org/10.1371/journal.pone.0043467.
Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, et al. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59:5316–23.
Kumar P, Kaushik A, Bell D, Chauhan V, Xia F, Stevens RL, et al. Mutation in an unannotated protein confers carbapenem resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.02234-16.
Suzuki Y, Katsukawa C, Tamaru A, Abe C, Makino M, Mizuguchi Y, et al. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J Clin Microbiol. 1998;36:1220–5.
Finken M, Kirschner P, Meier A, Wrede A, Böttger EC. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis: alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol. 1993;9:1239–46.
Nair J, Rouse DA, Bai GH, Morris SL. The rpsL gene and streptomycin resistance in single and multiple drug-resistant strains of Mycobacterium tuberculosis. Mol Microbiol. 1993;10:521–7.
Honoré N, Cole ST. Streptomycin resistance in mycobacteria. Antimicrob Agents Chemother. 1994;38:238–42.
Meier A, Kirschner P, Bange FC, Vogel U, Böttger EC. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994;38:228–33.
Douglass J, Steyn LM. A ribosomal gene mutation in streptomycin-resistant Mycobacterium tuberculosis isolates. J Infect Dis. 1993;167:1505–6.
Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HIM, Barry CE. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55:2515–22.
Morlock GP, Metchock B, Sikes D, Crawford JT, Cooksey RC. ethA, inhA, and katG loci of ethionamide-resistant clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2003;47:3799–805.
Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, et al. Activation of the pro-drug ethionamide is regulated in mycobacteria. J Biol Chem. 2000;275:28326–31.
Zheng J, Rubin EJ, Bifani P, Mathys V, Lim V, Au M, et al. Para-aminosalicylic acid is a prodrug targeting dihydrofolate reductase in Mycobacterium tuberculosis. J Biol Chem. 2013;288:23447–56.
Mathys V, Wintjens R, Lefevre P, Bertout J, Singhal A, Kiass M, et al. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2009;53:2100–9.
Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol. 2004;53:275–82.
Fivian-Hughes AS, Houghton J, Davis EO, Hill M, Nw L. Mycobacterium tuberculosis thymidylate synthase gene thyX is essential and potentially bifunctional, while thyA deletion confers resistance to p-aminosalicylic acid. Microbiology. 2012;158:308–18.
Zhang X, Liu L, Zhang Y, Dai G, Huang H, Jin Q. Genetic determinants involved in p-aminosalicylic acid resistance in clinical isolates from tuberculosis patients in northern China from 2006 to 2012. Antimicrob Agents Chemother. 2015;59:1320–4.
Zaunbrecher MA, Sikes RD, Metchock B, Shinnick TM, Posey JE. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2009;106:20004–9.
Maus CE, Plikaytis BB, Shinnick TM. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49:571–7.
World Health Organization. Rapid Communication: key changes to treatment of multidrug- and rifampicin-resistant tuberculosis (MDR/RR-TB). Geneva: WHO; 2018. https://www.who.int/tb/publications/2018/WHO_RapidCommunicationMDRTB.pdf?ua=1. Accessed 9 July 2019
Diacon AH, Donald PR, Pym A, Grobusch M, Patientia RF, Mahanyele R, et al. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother. 2012;56:3271–6.
Diacon AH, Pym A, Grobusch MP, de Los Rios JM, Gotuzzo E, Vasilyeva I, et al. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med. 2014;371:723–32.
Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, et al. OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 2006;3:2131–44.
Gler MT, Skripconoka V, Sanchez-Garavito E, Xiao H, Cabrera-Rivero JL, Vargas-Vasquez DE, et al. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med. 2012;366:2151–60.
Bloom BR, Atun R, Cohen T, Dye C, Fraser H, Gomez GB, et al. Tuberculosis. In: Holmes KK, Bertozzi S, Bloom BR, et al., editors. Major infectious diseases. Washington DC: The International Bank for Reconstruction and Development/The World Bank; 2017. https://www.ncbi.nlm.nih.gov/books/NBK525174/. Accessed 9 July 2019.
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44.
Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–90.
Young DB. Blueprint for the white plague. Nature. 1998;393:515–6.
Cabibbe AM, Walker TM, Niemann S, Cirillo DM. Whole genome sequencing of Mycobacterium tuberculosis. Eur Respir J. 2018;52:537–45.
Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45:1255–60.
Walker TM, Kohl TA, Omar SV, Hedge J, Del Ojo EC, Bradley P, et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;3099:1–10.
Coll F, Phelan J, Hill-Cawthorne GA, Nair MB, Mallard K, Ali S, et al. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat Genet. 2018;50:307–16.
The CRyPTIC Consortium and the 100,000 Genomes Project. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379:1403–15.
Eldholm V, Norheim G, von der Lippe B, Kinander W, Dahle UR, Caugant DA, et al. Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol. 2014. https://doi.org/10.1186/s13059-014-0490-3.
Trauner A, Liu Q, Via LE, Liu X, Ruan X, Liang L, et al. The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy. Genome Biol. 2017. https://doi.org/10.1186/s13059-017-1196-0.
Nikolayevskyy V, Kranzer K, Niemann S, Drobniewski F. Whole genome sequencing of Mycobacterium tuberculosis for detection of recent transmission and tracing outbreaks: a systematic review. Tuberculosis. (Edinb.). 2016;98:77–85.
Ioerger TR, Feng Y, Chen X, Dobos KM, Victor TC, Streicher EM, et al. The non-clonality of drug resistance in Beijing-genotype isolates of Mycobacterium tuberculosis from the Western Cape of South Africa. BMC Genomics. 2010. https://doi.org/10.1186/1471-2164-11-670.
Shah NS, Auld SC, Brust JCM, Mathema B, Ismail N, Moodley P, et al. Transmission of extensively drug-resistant tuberculosis in South Africa. N Engl J Med. 2017;376:243–53.
Manson AL, Cohen KA, Abeel T, Desjardins CA, Armstrong DT, Barry CE, et al. Genomic analysis of globally diverse Mycobacterium tuberculosis strains provides insights into the emergence and spread of multidrug resistance. Nat Genet. 2017;49:395–402.
Cohen KA, Manson AL, Abeel T, Desjardins CA, Chapman SB, Hoffner S, et al. Extensive global movement of multidrug-resistant M. tuberculosis strains revealed by whole-genome analysis. Thorax. 2019. https://doi.org/10.1136/thoraxjnl-2018-211616.
Nelson KN, Shah NS, Mathema B, Ismail N, Brust JCM, Brown TS, et al. Spatial patterns of extensively drug-resistant tuberculosis transmission in KwaZulu-Natal. South Africa. J Infect Dis. 2018;218:1964–73.
Cohen KA, Abeel T, Manson McGuire A, Desjardins CA, Munsamy V, Shea TP, et al. Evolution of extensively drug-resistant tuberculosis over four decades: whole genome sequencing and dating analysis of Mycobacterium tuberculosis isolates from KwaZulu-Natal. PLoS Med. 2015. https://doi.org/10.1371/journal.pmed.1001880.
Eldholm V, Monteserin J, Rieux A, Lopez B, Sobkowiak B, Ritacco V, et al. Four decades of transmission of a multidrug-resistant Mycobacterium tuberculosis outbreak strain. Nat Commun. 2015. https://doi.org/10.1038/ncomms8119.
Fonseca JD, Knight GM, McHugh TD. The complex evolution of antibiotic resistance in Mycobacterium tuberculosis. Int J Infect Dis. 2015;32:94–100.
Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet. 2012;44:106–10.
Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, Kontsevaya I, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46:279–86.
Merker M, Barbier M, Cox H, Rasigade J-P, Feuerriegel S, Kohl TA, et al. Compensatory evolution drives multidrug-resistant tuberculosis in Central Asia. Elife. 2018. https://doi.org/10.7554/eLife.38200.
Köser CU, Bryant JM, Becq J, Török ME, Ellington MJ, Marti-Renom MA, et al. Whole-genome sequencing for rapid susceptibility testing of M. tuberculosis. N Engl J Med. 2013;369:290–2.
Liu Q, Via LE, Luo T, Liang L, Liu X, Wu S, et al. Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci Rep. 2015. https://doi.org/10.1038/srep17507.
Lieberman TD, Wilson D, Misra R, Xiong LL, Moodley P, Cohen T, et al. Genomic diversity in autopsy samples reveals within-host dissemination of HIV-associated Mycobacterium tuberculosis. Nat Med. 2016;22:1470–4.
Dheda K, Lenders L, Magombedze G, Srivastava S, Raj P, Arning E, et al. Drug-penetration gradients associated with acquired drug resistance in patients with tuberculosis. Am J Respir Crit Care Med. 2018;198:1208–19.
Sobkowiak B, Glynn JR, Houben RMGJ, Mallard K, Phelan JE, Guerra-Assunção JA, et al. Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics. 2018. https://doi.org/10.1186/s12864-018-4988-z.
Pankhurst LJ, Del Ojo EC, Votintseva AA, Walker TM, Cole K, Davies J, et al. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med. 2016;4:49–58.
Doughty EL, Sergeant MJ, Adetifa I, Antonio M, Pallen MJ. Culture-independent detection and characterisation of Mycobacterium tuberculosis and M. africanum in sputum samples using shotgun metagenomics on a benchtop sequencer. PeerJ. 2014. doi:https://doi.org/10.7717/peerj.585.
Votintseva AA, Bradley P, Pankhurst L, Del Ojo EC, Loose M, Nilgiriwala K, et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55:1285–98.
Liu X, Gutacker MM, Musser JM, Fu Y-X. Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol. 2006;188:8169–77.
Ramaswamy S, Musser JM. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79:3–29.
Simons SO, van Ingen J, van der Laan T, Mulder A, Dekhuijzen PNR, Boeree MJ, et al. Validation of pncA gene sequencing in combination with the mycobacterial growth indicator tube method to test susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol. 2012;50:428–34.
Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–37.
Chen J, Zhang S, Cui P, Shi W, Zhang W, Zhang Y. Identification of novel mutations associated with cycloserine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2017;72:3272–6.
Zampieri M, Szappanos B, Buchieri MV, Trauner A, Piazza I, Picotti P, et al. High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds. Sci Transl Med. 2018. https://doi.org/10.1126/scitranslmed.aal3973.
Gagneux S. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–6.
Bergval IL, Schuitema ARJ, Klatser PR, Anthony RM. Resistant mutants of Mycobacterium tuberculosis selected in vitro do not reflect the in vivo mechanism of isoniazid resistance. J Antimicrob Chemother. 2009;64:515–23.
Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, et al. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–90.
Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, et al. Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet. 2011;43:482–6.
Papaventsis D, Casali N, Kontsevaya I, Drobniewski F, Cirillo DM, Nikolayevskyy V. Whole genome sequencing of Mycobacterium tuberculosis for detection of drug resistance: a systematic review. Clin Microbiol Infect. 2017;23:61–8.
Gygli SM, Keller PM, Ballif M, Blöchliger N, Hömke R, Reinhard M, et al. Whole genome sequencing for drug resistance profile prediction in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2019. https://doi.org/10.1128/AAC.02175-18.
Cohen KA, El-Hay T, Wyres KL, Weissbrod O, Munsamy V, Yanover C, et al. Paradoxical hypersusceptibility of drug-resistant Mycobacterium tuberculosis to β-lactam antibiotics. EBioMedicine. 2016;9:170–9.
World Health Organization. Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Geneva: WHO; 2014. https://apps.who.int/iris/bitstream/handle/10665/130918/9789241548809_eng.pdf?sequence=1. Accessed 9 July 2019
Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet. 2017;18:41–50.
Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ, et al. Genome wide association with quantitative resistance phenotypes in Mycobacterium tuberculosis reveals novel resistance genes and regulatory regions. Nat Commun. 2019. https://doi.org/10.1038/s41467-019-10110-6.
Earle SG, Wu C-H, Charlesworth J, Stoesser N, Gordon NC, Walker TM, et al. Identifying lineage effects when controlling for population structure improves power in bacterial association studies. Nat Microbiol. 2016. https://doi.org/10.1038/nmicrobiol.2016.41.
Jaillard M, Lima L, Tournoud M, Mahé P, van Belkum A, Lacroix V, et al. A fast and agnostic method for bacterial genome-wide association studies: bridging the gap between k-mers and genetic events. PLoS Genet. 2018. https://doi.org/10.1371/journal.pgen.1007758.
Lees JA, Vehkala M, Välimäki N, Harris SR, Chewapreecha C, Croucher NJ, et al. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat Commun. 2016. https://doi.org/10.1038/ncomms12797.
Alam MT, Petit RA, Crispell EK, Thornton TA, Conneely KN, Jiang Y, et al. Dissecting vancomycin-intermediate resistance in staphylococcus aureus using genome-wide association. Genome Biol Evol. 2014;6:1174–85.
Collins C, Didelot X. A phylogenetic method to perform genome-wide association studies in microbes that accounts for population structure and recombination. PLoS Comput Biol. 2018. https://doi.org/10.1371/journal.pcbi.1005958.
Barker D, Meade A, Pagel M. Constrained models of evolution lead to improved prediction of functional linkage from correlated gain and loss of genes. Bioinformatics. 2007;23:14–20.
Hicks ND, Yang J, Zhang X, Zhao B, Grad YH, Liu L, et al. Clinically prevalent mutations in Mycobacterium tuberculosis alter propionate metabolism and mediate multidrug tolerance. Nat Microbiol. 2018;3:1032–42.
Merker M, Kohl TA, Roetzer A, Truebe L, Richter E, Rüsch-Gerdes S, et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS One. 2013. https://doi.org/10.1371/journal.pone.0082551.
Stucki D, Gagneux S. Single nucleotide polymorphisms in Mycobacterium tuberculosis and the need for a curated database. Tuberculosis (Edinb). 2013;93:30–9.
Reddy TBK, Riley R, Wymore F, Montgomery P, DeCaprio D, Engels R, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37:D499–508.
Gillespie JJ, Wattam AR, Cammer SA, Gabbard JL, Shukla MP, Dalay O, et al. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human pathogenic species. Infect Immun. 2011;79:4286–98.
Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009. https://doi.org/10.1371/journal.pmed.1000002.
Iwai H, Kato-Miyazawa M, Kirikae T, Miyoshi-Akiyama T. CASTB (the comprehensive analysis server for the Mycobacterium tuberculosis complex): a publicly accessible web server for epidemiological analyses, drug-resistance prediction and phylogenetic comparison of clinical isolates. Tuberculosis (Edinb). 2015;95:843–4.
Steiner A, Stucki D, Coscolla M, Borrell S, Gagneux S. KvarQ. Targeted and direct variant calling from fastq reads of bacterial genomes. BMC Genomics. 2014. doi:https://doi.org/10.1186/1471-2164-15-881.
Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, et al. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun. 2015. https://doi.org/10.1038/ncomms10063.
Feuerriegel S, Schleusener V, Beckert P, Kohl TA, Miotto P, Cirillo DM, et al. PhyResSE: a web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol. 2015;53:1908–14.
Coll F, McNerney R, Preston MD, Guerra-Assunção JA, Warry A, Hill-Cawthorne G, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med. 2015. https://doi.org/10.1186/s13073-015-0164-0.
Sekizuka T, Yamashita A, Murase Y, Iwamoto T, Mitarai S, Kato S, et al. TGS-TB: total genotyping solution for Mycobacterium tuberculosis using short-read whole-genome sequencing. PLoS One. 2015. https://doi.org/10.1371/journal.pone.0142951.
Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S. Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools. Sci Rep. 2017. https://doi.org/10.1038/srep46327.
van Beek J, Haanperä M, Smit PW, Mentula S, Soini H. Evaluation of whole genome sequencing and software tools for drug susceptibility testing of Mycobacterium tuberculosis. Clin Microbiol Infect. 2019;25:82–6.
Ngo T-M, Teo Y-Y. Genomic prediction of tuberculosis drug-resistance: benchmarking existing databases and prediction algorithms. BMC Bioinformatics. 2019. https://doi.org/10.1186/s12859-019-2658-z.
ReSeqTB. Relational sequencing TB data platform. https://platform.reseqtb.org. Accessed 9 July 2019.
Starks AM, Avilés E, Cirillo DM, Denkinger CM, Dolinger DL, Emerson C, et al. Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform. Clin Infect Dis. 2015;61:S141–6.
CRyPTIC. Comprehensive Resistance Prediction for Tuberculosis: an International Consortium. http://www.crypticproject.org. Accessed 9 July 2019.
Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J. 2017. https://doi.org/10.1183/13993003.01354-2017.
Falush D, Bowden R. Genome-wide association mapping in bacteria? Trends Microbiol. 2006;14:353–5.
Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, et al. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–46.
Guerra-Assunção J, Crampin A, Houben R, Mzembe T, Mallard K, Coll F, et al. Large scale population-based whole genome sequencing of Mycobacterium tuberculosis provides insights into transmission in a high prevalence area. Elife. 2015. https://doi.org/10.7554/eLife.05166.
Bryant JM, Schürch AC, van Deutekom H, Harris SR, de Beer JL, de Jager V, et al. Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect Dis. 2013. https://doi.org/10.1186/1471-2334-13-110.
Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med. 2013. https://doi.org/10.1371/journal.pmed.1001387.
World Health Organization. Policy guidance on drug-susceptibility testing (DST) of second-line antituberculosis drugs. Geneva: WHO; 2008. https://apps.who.int/iris/bitstream/handle/10665/70500/WHO_HTM_TB_2008.392_eng.pdf?sequence=1. Accessed 9 July 2019
Colangeli R, Jedrey H, Kim S, Connell R, Ma S, Chippada Venkata UD, et al. Bacterial factors that predict relapse after tuberculosis therapy. N Engl J Med. 2018;379:823–33.
Rinder H. Hetero-resistance: an under-recognised confounder in diagnosis and therapy? J Med Microbiol. 2001;50:1018–20.
Rinder H, Mieskes KT, Löscher T. Heteroresistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2001;5:339–45.
Folkvardsen DB, Thomsen VØ, Rigouts L, Rasmussen EM, Bang D, Bernaerts G, et al. Rifampin heteroresistance in Mycobacterium tuberculosis cultures as detected by phenotypic and genotypic drug susceptibility test methods. J Clin Microbiol. 2013;51:4220–2.
Liang B, Tan Y, Li Z, Tian X, Du C, Li H, et al. Highly sensitive detection of isoniazid heteroresistance in Mycobacterium tuberculosis by DeepMelt assay. J Clin Microbiol. 2018. https://doi.org/10.1128/JCM.01239-17.
Zhang D, Gomez JE, Chien J-Y, Haseley N, Desjardins CA, Earl AM, et al. Genomic analysis of the evolution of fluoroquinolone resistance in Mycobacterium tuberculosis prior to tuberculosis diagnosis. Antimicrob Agents Chemother. 2016;60:6600–8.
Phelan JE, Coll F, Bergval I, Anthony RM, Warren R, Sampson SL, et al. Recombination in pe/ppe genes contributes to genetic variation in Mycobacterium tuberculosis lineages. BMC Genomics. 2016. https://doi.org/10.1186/s12864-016-2467-y.
Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: Perhaps more? Biochimie. 2012;94:110–6.
Farhat MR, Shapiro B, Sheppard SK, Colijn C, Murray M. A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens. Genome Med. 2014. https://doi.org/10.1186/s13073-014-0101-7.
Casali N, Broda A, Harris SR, Parkhill J, Brown T, Drobniewski F. Whole genome sequence analysis of a large isoniazid-resistant tuberculosis outbreak in London: a retrospective observational study. PLoS Med. 2016. https://doi.org/10.1371/journal.pmed.1002137.
van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9.
Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14.
Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003.
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–510.
Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–31.
Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, et al. Assessment of an optimized mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol. 2007;45:691–7.
Chihota VN, Müller B, Mlambo CK, Pillay M, Tait M, Streicher EM, et al. Population structure of multi- and extensively drug-resistant Mycobacterium tuberculosis strains in South Africa. J Clin Microbiol. 2012;50:995–1002.
Gandhi NR, Brust JCM, Moodley P, Weissman D, Heo M, Ning Y, et al. Minimal diversity of drug-resistant Mycobacterium tuberculosis strains. South Africa. Emerg Infect Dis. 2014;20:394–401.
Bifani PJ, Plikaytis BB, Kapur V, Stockbauer K, Pan X, Lutfey ML, et al. Origin and interstate spread of a New York City multidrug-resistant Mycobacterium tuberculosis clone family. JAMA. 1996;275:452–7.
Lutfey M, Della-Latta P, Kapur V, Palumbo LA, Gurner D, Stotzky G, et al. Independent origin of mono-rifampin-resistant Mycobacterium tuberculosis in patients with AIDS. Am J Respir Crit Care Med. 1996;153:837–40.
Post FA, Willcox PA, Mathema B, Steyn LM, Shean K, Ramaswamy SV, et al. Genetic polymorphism in Mycobacterium tuberculosis isolates from patients with chronic multidrug-resistant tuberculosis. J Infect Dis. 2004;190:99–106.
Roof I, Jajou R, Kamst M, Mulder A, de Neeling A, van Hunen R, et al. Prevalence and characterization of heterogeneous variable-number tandem-repeat clusters comprising drug-susceptible and/or variable resistant Mycobacterium tuberculosis complex isolates in the Netherlands from 2004 to 2016. J Clin Microbiol. 2018. https://doi.org/10.1128/JCM.00887-18.
Witney AA, Bateson AL, Jindani A, Phillips PP, Coleman D, Stoker NG, et al. Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med. 2017. https://doi.org/10.1186/s12916-017-0834-4.
Van Rie A, Victor TC, Richardson M, Johnson R, Van Der Spuy GD, Murray EJ, et al. Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med. 2005;172:636–42.
Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, et al. Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–4.
Hatherell H-A, Colijn C, Stagg HR, Jackson C, Winter JR, Abubakar I. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 2016. https://doi.org/10.1186/s12916-016-0566-x.
Sun G, Luo T, Yang C, Dong X, Li J, Zhu Y, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J Infect Dis. 2012;206:1724–33.
Mariam SH, Werngren J, Aronsson J, Hoffner S, Andersson DI. Dynamics of antibiotic resistant mycobacterium tuberculosis during long-term infection and antibiotic treatment. PLoS One. 2011. https://doi.org/10.1371/journal.pone.0021147.
Manson AL, Abeel T, Galagan JE, Sundaramurthi JC, Salazar A, Gehrmann T, et al. Mycobacterium tuberculosis whole genome sequences from Southern India suggest novel resistance mechanisms and the need for region-specific diagnostics. Clin Infect Dis. 2017;64:1494–501.
Wollenberg KR, Desjardins CA, Zalutskaya A, Slodovnikova V, Oler AJ, Quiñones M, et al. Whole-genome sequencing of Mycobacterium tuberculosis provides insight into the evolution and genetic composition of drug-resistant tuberculosis in Belarus. J Clin Microbiol. 2017;55:457–69.
Koch A, Wilkinson RJ. The road to drug resistance in Mycobacterium tuberculosis. Genome Biol. 2014. https://doi.org/10.1186/s13059-014-0520-1.
Walker TM, Lalor MK, Broda A, Saldana Ortega L, Morgan M, Parker L, et al. Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–12, with whole pathogen genome sequences: an observational study. Lancet Respir Med. 2014;2:285–92.
Gandhi NR, Moll A, Sturm AW, Pawinski R, Govender T, Lalloo U, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–80.
Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, et al. Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res. 2012;22:735–45.
Guerra-Assunção JA, Houben RMGJ, Crampin AC, Mzembe T, Mallard K, Coll F, et al. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J Infect Dis. 2015;211:1154–63.
Perdigão J, Silva H, Machado D, Macedo R, Maltez F, Silva C, et al. Unraveling Mycobacterium tuberculosis genomic diversity and evolution in Lisbon, Portugal, a highly drug resistant setting. BMC Genomics. 2014. https://doi.org/10.1186/1471-2164-15-991.
Lalor MK, Casali N, Walker TM, Anderson LF, Davidson JA, Ratna N, et al. The use of whole-genome sequencing in cluster investigation of a multidrug-resistant tuberculosis outbreak. Eur Respir J. 2018. https://doi.org/10.1183/13993003.02313-2017.
Bjorn-Mortensen K, Soborg B, Koch A, Ladefoged K, Merker M, Lillebaek T, et al. Tracing Mycobacterium tuberculosis transmission by whole genome sequencing in a high incidence setting: a retrospective population-based study in East Greenland. Sci Rep. 2016. https://doi.org/10.1038/srep33180.
Packer S, Green C, Brooks-Pollock E, Chaintarli K, Harrison S, Beck CR. Social network analysis and whole genome sequencing in a cohort study to investigate TB transmission in an educational setting. BMC Infect Dis. 2019. https://doi.org/10.1186/s12879-019-3734-8.
Auld SC, Shah NS, Mathema B, Brown TS, Ismail N, Omar SV, et al. Extensively drug-resistant tuberculosis in South Africa: genomic evidence supporting transmission in communities. Eur Respir J. 2018. https://doi.org/10.1183/13993003.00246-2018.
Ford C, Yusim K, Ioerger T, Feng S, Chase M, Greene M, et al. Mycobacterium tuberculosis—heterogeneity revealed through whole genome sequencing. Tuberculosis (Edinb). 2012;92:194–201.
Farmer P, Bayona J, Becerra M, Furin J, Henry C, Hiatt H, et al. The dilemma of MDR-TB in the global era. Int J Tuberc Lung Dis. 1998;2:869–76.
Xie YL, Chakravorty S, Armstrong DT, Hall SL, Via LE, Song T, et al. Evaluation of a rapid molecular drug-susceptibility test for tuberculosis. N Engl J Med. 2017;377:1043–54.
Lee RS, Behr MA. The implications of whole-genome sequencing in the control of tuberculosis. Ther Adv Infect Dis. 2016;3:47–62.
Jeanes C, O’Grady J. Diagnosing tuberculosis in the 21st century—Dawn of a genomics revolution? Int J Mycobacteriol. 2016;5:384–91.
Lee RS, Pai M. Real-time sequencing of Mycobacterium tuberculosis: are we there yet? J Clin Microbiol. 2017;55:1249–54.
Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, Chan JZM, et al. Rapid whole-genome sequencing of mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol. 2015;53:2230–7.
Doyle RM, Burgess C, Williams R, Gorton R, Booth H, Brown J, et al. Direct whole-genome sequencing of sputum accurately identifies drug-resistant Mycobacterium tuberculosis faster than MGIT culture sequencing. J Clin Microbiol. 2018. https://doi.org/10.1128/JCM.00666-18.
Lowenthal P, Lin S-YG, Desmond E, Shah N, Flood J, Barry PM. Evaluation of the impact of a sequencing assay for detection of drug resistance on the clinical management of tuberculosis. Clin Infect Dis. 2018. https://doi.org/10.1093/cid/ciy937.
Judge K, Harris SR, Reuter S, Parkhill J, Peacock SJ. Early insights into the potential of the Oxford Nanopore MinION for the detection of antimicrobial resistance genes. J Antimicrob Chemother. 2015;70:2775–8.
Schmidt K, Mwaigwisya S, Crossman LC, Doumith M, Munroe D, Pires C, et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J Antimicrob Chemother. 2017;72:104–14.
Cohen T, van Helden PD, Wilson D, Colijn C, Mclaughlin MM, Abubakar I, et al. Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin Microbiol Rev. 2012;25:708–19.
World Health Organization. The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide. Geneva: WHO; 2018. https://apps.who.int/iris/bitstream/handle/10665/274443/WHO-CDS-TB-2018.19-eng.pdf?sequence=1&isAllowed=y. Accessed 9 July 2019
Walker TM, Cruz ALG, Peto TE, Smith EG, Esmail H, Crook DW. Tuberculosis is changing. Lancet Infect Dis. 2017;17:359–61.
Colman RE, Anderson J, Lemmer D, Lehmkuhl E, Georghiou SB, Heaton H, et al. Rapid drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates directly from clinical samples by use of amplicon sequencing: a proof-of-concept study. J Clin Microbiol. 2016;54:2058–67.
Acknowledgements
The authors thank Arlin Keo for her assistance with the creation of Fig. 1.
Funding
The authors’ work has been funded by the National Institute of Allergy and Infectious Diseases (grant number U19AI110818 to AME) and by The National Heart Lung and Blood Institute (K08 HL139994 to KAC) and the Burroughs Wellcome Fund CAMS (to KAC). The funders had no role in preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
KAC, ALM, CAD, TA, and AME wrote the manuscript. All authors read and approved the final manuscript.
Authors’ information
Keira A. Cohen, MD is a pulmonary and critical care medicine specialist at Johns Hopkins University School of Medicine. Abigail L. Manson, PhD is a senior computational biologist and computational group leader for Bacterial Genomics Group in the Infectious Disease and Microbiome Program at the Broad Institute of MIT and Harvard. Christopher Desjardins, PhD is a senior computational biologist working in the Infectious Disease and Microbiome Program at the Broad Institute of MIT and Harvard. Thomas Abeel, PhD is an assistant professor of bioinformatics at Delft University of Technology and a visiting scientist at the Broad Institute of MIT and Harvard. Ashlee M. Earl, PhD is a research scientist and senior group leader for the Bacterial Genomics Group in the Infectious Disease and Microbiome Program at the Broad Institute of MIT and Harvard.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cohen, K.A., Manson, A.L., Desjardins, C.A. et al. Deciphering drug resistance in Mycobacterium tuberculosis using whole-genome sequencing: progress, promise, and challenges. Genome Med 11, 45 (2019). https://doi.org/10.1186/s13073-019-0660-8
Published:
DOI: https://doi.org/10.1186/s13073-019-0660-8