Abstract
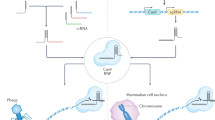
In the last few decades, genomic manipulation has made significant progress as a result of the development of recombinant DNA technologies; however, more often than not, these techniques have been costly and labor intensive. In contrast, recently developed next-generation sequencing (NGS) technologies have provided a cheaper, faster, and easier process to study genomics. In particular, an NGS technique emerged from bacterial CRISPR-associated protein-9 nuclease (Cas9) as a revolutionary method to modify, regulate, or mark specific genomic sequences on virtually any organism. A later adaptation of this bacterial defense mechanism that successfully and permanently edits dysfunctional genes and corrects missing proteins has resulted in a new era for disease genetic engineering. Clinical trials using this technique are already being performed, and the applicability of CRISPR-Cas9 techniques is actively being investigated using in vivo studies. However, the concept of genome correction poses great concerns from a regulatory perspective, especially in terms of security, so principles for the regulation of these methodologies are being established. We delved into CRISPR-Cas9 from its natural and ortholog origins to its engineered variants and behaviors to present its notable and diverse applications in the fields of biotechnology and human therapeutics.
Similar content being viewed by others
References
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278.
Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299.
Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Mol Microbiol. 2000;36:244–246.
Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496.
Guy CP, Majerník AI, Chong JPJ, Bolt EL. A novel nuclease-ATPase (Nar71) from archaea is part of a proposed thermophilic DNA repair system. Nucleic Acids Res. 2004;32:6176–6186.
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561.
Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663.
Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mole Microbiol. 2002;43:1565–1575.
Morange M. What history tells us XXXVII. CRISPR-Cas: the discovery of an immune system in prokaryotes. J Biosci. 40:221–223.
Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477.
Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7.
Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821.
Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586.
Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182
Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400.
Horvath P, Romero DA, Coûté-Monvoisin AC, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412.
Shah SA, Erdmann S, Mojica FJM, Garrett RA. Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biol. 2013;10:891–899.
Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607.
Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050.
Sun CL, Barrangou R, Thomas BC, Horvath P, Fremaux C, Banfield JF. Phage mutations in response to CRISPR diversification in a bacterial population. Environ Microbiol. 2013;15:463–70.
Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60.
Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712.
Garneau JE, Dupuis MÈ, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468;67–71.
Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282.
Qi LS, Larson MH, Gilbert LA, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183.
Zhao Y, Dai Z, Liang Y, et al. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci Rep. 2014;4:3943.
Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451.
Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437.
Fonfara I, Le Rhun A, Chylinski K, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590.
Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013;10:841–851.
Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121.
Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823.
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc-finger nucleases. Nat Rev Genet. 2010;11:636–646.
Hsu PD, Zhang F. Dissecting neural function using targeted genome engineering technologies. ACS Chem Neurosci. 2012;3:603–610.
Perez EE, Wang J, Miller JC, et al. Establishment of HIV-1 resistance in CD4+T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816.
Chen F, Pruett-Miller SM, Huang Y, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755.
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67.
Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728.
Chen C, Liu Y, Rappaport AR, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell. 2014;25:652–665.
Wu Y, Liang D, Wang Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662.
U.S. researchers call for greater oversight of powerful genetic technology. Science/AAAS News. News.sciencemag.org. Accessed July 18, 2014.
Science News Staff, December 17, 2015). And Science’s Breakthrough of the Year is…. news.sciencemag.org. Accessed December 21, 2015.
Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843.
Katz G, Pitts PJ. Implications of CRISPR-based germline engineering for cancer survivors. Therapeutic Innovation & Regulatory Science. 2017;51:672–682.
Oude Blenke E, Evers MJW, Mastrobattista E, van der Oost J. CRISPR-Cas9 gene editing: delivery aspects and therapeutic potential. J Control Release. 2016;244(pt B):139–148.
Shim G, Kim D, Park GT, Jin H, Suh SK, Oh YK. Therapeutic gene editing: delivery and regulatory perspectives. Acta Pharmacol Sin. 2017;38:738–753.
Schaefer KA, Wu WH, Colgan DF, Tsang SH, Bassuk AG, Mahajan VB. Unexpected mutations after CRISPR-Cas9 editing in vivo. Nat Methods. 2017;14:547–548.
National Academies of Science, Engineering, and Medicine. Preparing for future products of biotechnology. National Academies Press. https://doi.org/10.17226/24605
Regulation (EC) 1394/2007 on advanced therapy medicinal products and amending Directive 2001/83/EC and Regulation No 726/2004.
Sanzenbacher R, Dwenger A, Schuessler-Lenz M, Cichutek K, Flory E. European regulation tackles tissue-engineering. Nat Biotechnol. 2007;25:1089–1091.
Bellver Capella V. La revolución de la edición genética Mediante crispr-cAS9 y los desafíos éticos y regulatorios que comporta crispr-cAS9 [in Spanish]. Cuad Bioet. 2016;27:223–240. doi:https://doi.org/10.1038/nmeth.1852.2
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Simón, J.E., Rodríguez, Á.S. & Santiago Vispo, N. CRISPR-Cas9: A Precise Approach to Genome Engineering. Ther Innov Regul Sci 52, 701–707 (2018). https://doi.org/10.1177/2168479018762798
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1177/2168479018762798