Abstract
We discuss the Bayesian approach to the solution of inverse problems and apply the formalism to analyse the closure tests performed by the NNPDF collaboration. Starting from a comparison with the approach that is currently used for the determination of parton distributions (PDFs), we discuss some analytical results that can be obtained for linear problems and use these results as a guidance for more complicated non-linear problems. We show that, in the case of Gaussian distributions, the posterior probability density of the parametrized PDFs is fully determined by the results of the NNPDF fitting procedure. Building on the insight that we obtain from the analytical results, we introduce new estimators to assess the statistical faithfulness of the fit results in closure tests. These estimators are defined in data space, and can be studied analytically using the Bayesian formalism in a linear model in order to clarify their meaning. Finally we present results from a number of closure tests performed with current NNPDF methodologies. These further tests allow us to validate the NNPDF4.0 methodology and provide a quantitative comparison of the NNPDF4.0 and NNPDF3.1 methodologies. As PDFs determinations move into precision territory, the need for a careful validation of the methodology becomes increasingly important: the error bar has become the focal point of contemporary PDFs determinations. In this perspective, theoretical assumptions and other sources of error are best formulated and analysed in the Bayesian framework, which provides an ideal language to address the precision and the accuracy of current fits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Inverse problems are the typical example of inference where a model is sought starting from a finite-dimensional set of experimental observations. These problems are notoriously difficult, and often require trying to guess a continuous function, i.e. an element of an infinite dimensional space, from a finite amount of data. As emphasised by Hadamard a long time ago [1], it is easy to end up in a situation where we deal with ill-posed problems, in the sense that the solution may not exist, may not be unique, or may be unstable under small variations of the input data. The determination of parton distributions from experimental data, or the reconstruction of spectral densities from lattice QCD simulations, are just two examples where these problems arise in particle physics. In all these cases, finding a robust solution becomes very challenging, if not impossible, making these questions all the more urgent, especially at a time when precision studies are the ultimate challenge in order to highlight divergences from the Standard Model.
A Bayesian approach provides an apter tool for addressing inverse problems. Starting from a prior distribution for the model, which encodes our theoretical knowledge or prejudice, basic statistical tools allow us to determine the posterior distribution of the solution, after taking into account a set of experimental observations. The prior and posterior probabilities encode our knowledge about the model before and after incorporating the experimental results. There are multiple advantages to this formulation: the inverse problem is well defined, and the prior distribution ensures that all the assumptions are explicit, while regulating the likelihood distribution.
Note also that probability measures can be defined in infinite-dimensional spaces. In cases where we are looking to determine a continuous function, the Bayesian approach provides, at least in principle, a formulation of the problem directly in terms of the posterior probability measure of the model function. It is often convenient for practical reasons to define a parametrization of the model function in terms of a finite, albeit large, number of parameters and reduce the problem to a finite dimensional one. The price to pay for this simplification is the introduction of some bias, which needs to be understood and possibly quantified. An expressive parametrization clearly helps in this case.
A Bayesian approach to inverse problems has been actively developed by mathematicians for a long time, and this development has dramatically accelerated in the last decade. In this paper we aim at summarising the existing framework and adapt it to analyse the fits of parton distribution functions obtained by the NNPDF collaboration. We review the Bayesian formalism in Sect. 2, where we define the notation that will be used in the rest of the paper. We report some known examples for illustrative purposes. Even though these are well-known results, we find it useful to summarise them in the context of our specific problem. In Sect. 3, we try to connect the Bayesian approach with the NNPDF fits based on a Monte Carlo methodology, where the distribution of the PDFs is encoded in a set of fits to artificially generated data, called replicas. We can anticipate here that, under the hypotheses that the data are Gaussian and the model is linear, the NNPDF procedure does characterise completely the posterior probability density. When the model is non-linear two modifications need to be taken into account. First of all the analytical calculation that we present in Sect. 2.1 is no longer possible, and one needs to rely on the fact that a linearization of the model yields an accurate prediction. Even though linear models are known to provide good approximations, the systematic errors introduced by this approximation are not easy to quantify. There is also a more subtle effect that needs to be taken into account. When working with linear models, the minimization procedure is know to have a unique minimum, which can be computed exactly. Non-linear models can be plagued by multiple minima, and more importantly by inefficiencies of the algorithm in finding them. While it would be foolish to ignore the limitations of the analytical calculation, it is nonetheless very useful to have an explicit result as a template to guide the analysis of our numerical investigations.
In the NNPDF fitting framework, the posterior probability of the parton distribution functions, is encoded in an ensemble of fits to replicas of the data, where the data replicas have been generated in order to reproduce the fluctuations described by the experimental central values and uncertainties. This bootstrap procedure propagates the data fluctuations in the space of fitted PDFs. A successful fit should yield robust confidence intervals for observables, in particular for those that are more relevant for phenomenology.
The idea of closure tests is to test this procedure in a fit to artificial data that have been generated from a known set of input PDFs. In this case the underlying law is known and we can check how the posterior distribution compares to the underlying law. This is the basis of a closure test, which is summarised at the end of Sect. 3. Closure tests have already been used to test to validity of previous iterations of the NNPDF methodology. Here we aim to refine some of the pre-existing closure test estimators and with the help of fast fitting methodology perform a more extensive study of how faithful our uncertainties are. For this purpose we introduce in Sect. 4 new estimators that allow us to quantify the quality of our fits. These estimators are defined in the space of data and need to be understood as stochastic variables that are characterised by a probability density. Where possible, we use the Bayesian formalism in order to compute analytically these probability densities and compare with numerical results. In order to perform analytical calculations we often need to make some simplifying assumptions. While it is incorrect to use the analytical results for making quantitative predictions for the results of the realistic case of a non-linear fit, the analytical results provide a guide to the interpretation of the observed patterns. It is important to stress that what we test in a closure test in terms of faithfulness, despite important and non-trivial, does not fully validate the uncertainties of a global fit: the artificial data used in a closure test are by construction compatible between each other, because generated starting from the same underlying law; this is not always the case when considering global fits, where tensions between different datasets can be present.
The results of our numerical studies are summarised in Sect. 5, where we also compare the NNPDF4.0 methodology introduced in the latest NNPDF fit [2] to the methodology used by the collaboration in previous fits.
The Bayesian formalism, by providing posterior probability distributions, paves the way to explore a number of issues. We highlight in our conclusions some possible questions that we defer to future studies.
2 Inverse problems
The problem of determining PDFs from a set of experimental data falls under the general category of inverse problems, i.e. the problem of finding the input to a given model knowing a set of observations, which are often finite and noisy. In this section we are going to review the Bayesian formulation of inverse problems. It is impossible to do justice to this vast subject here. Instead we try to emphasise the aspects that are relevant for quantifying uncertainties on PDF determinations.
2.1 Statement of the problem
The space of inputs is denoted by \(X\), while R denotes the space of responses. The model is specified by a forward map
which associates a response \(r \in R\) to the input \(u\in X\), where we assume that \(X\) and R are Banach spaces.Footnote 1 As an example we can think of \(u\) as being a PDF, i.e. a function defined on the interval [0, 1], and r a DIS structure function. The structure function is related to the PDF by a factorization formula involving perturbative coefficient functions:
Note that in this example the forward map maps one real function into another real function. In this case the space of models X is the space of continuous functions defined on the interval [0, 1], which satisfy integrability conditions. Even though this is an infinite-dimensional space, it is possible to define a probability measure on such a space and construct a Bayesian solution to the inverse problem. In current determinations of PDFs, the functional form of the PDF is dictated by some kind of parametrization, with different parametrizations being used by different collaborations. In all cases, the space X is a finite-dimensional space, \(\mathbb {R}^{N_{\mathrm{model}}}\), where \(N_{\mathrm{model}}\) is the number of parameters. In the case of the NNPDF fits discussed below, the weights of the neural networks are the parameters that determine the functional form of the PDFs. Alternatively, one could think of a finite-dimensional representation defined by the value of the PDF at selected values of x, i.e. \(u_i=u(x_i)\) for \(i=1,\ldots ,N_{\mathrm{model}}\). Depending on the context we will denote by u either the function u(x), or the vector of real parameters that are used to determine the function u(x). Disambiguation should hopefully be straightforward.
Experiments will not have access to the full function r but only to a subset of \(N_{\mathrm{data}}\) observations. In order to have a formal mathematical expression that takes into account the fact that we have a finite number of measurements, we introduce an observation operator
where \(y\in Y\) is a vector in a finite-dimensional space Y of experimental results, e.g. the value of the structure function for some values of the kinematic variables x and \(Q^2\). In general we will assume that \(y\in \mathbb {R}^{N_{\mathrm{data}}}\), i.e. we have a finite number \(N_{\mathrm{data}}\) of real experimental values. The quantity of interest is the composed operator
which maps the input \(u\) to the set of data. Taking into account the fact that experimental data are subject to noise, we can write
where \(\eta \) is a random variable defined over \(\mathbb {R}^{N_{\mathrm{data}}}\) with probability density \(\rho (\eta )\). We will refer to \(\eta \) as the observational noise. In this setting, the inverse problem becomes finding \(u\) given \(y\). It is often the case that inverse problems are ill-defined in the sense that the solution may not exist, may not be unique, or may be unstable under small variations of the problem.
In solving the inverse problem, we are going to adopt a Bayesian point of view, as summarised e.g. in Ref. [3]: our prior knowledge about \(u\) is encoded in a prior probability measure \(\mu _X^0\), where the suffix X indicates that the measure is defined in the space of models, and the suffix 0 refers to the fact that this is a prior distribution. We use Bayes’ theorem to compute the posterior probability measure of \(u\) given the data \(y\), which we denote as \(\mu _X^\mathcal {G}\). When the probability measure can be described by a probability density, we denote the probability densities associated to \(\mu _X^0\) and \(\mu _X^\mathcal {G}\), by \(\pi _X^0\) and \(\pi _X^\mathcal {G}\) respectively. Then, using Eq. (5), we can write the data likelihood, i.e. the probability density of \(y\) given \(u\),
and Bayes’ theorem yields
Even though the concepts that we have introduced so far should sound familiar, it is worthwhile clarifying some ideas and present an explicit case where all the probability densities are carefully defined. This is best exemplified by considering the case where both the observational noise and the model prior are Gaussian. We assume that we are given a set of central values \(y_0\in \mathbb {R}^{N_{\mathrm{data}}}\) and their covariance matrix \(C_{Y}\). Then the prior probability density of the observable \(y\) is
where, similarly to the convention used above, the suffix Y emphasises the fact that this is a probability density in data space, and the notation explicitly reminds us that this is the probability density given the central values \(y_0\) (and the covariance matrix \(C_{Y}\)). Similarly we can choose a Gaussian distribution for the prior distribution of the input model, characterized by a central value \(u_0\) and a covariance \(C_X\):
Following the convention above, we use a suffix X here to remind the reader that we are looking at a probability density in the space of models. Note that in the expressions above we used the norms in \(X\) and \(\mathbb {R}^{N_{\mathrm{data}}}\) respectively, and introduced the short-hand notation
where a denotes a generic element of \(X\), R or \(\mathbb {R}^{N_{\mathrm{data}}}\). For the case where \(a \in \mathbb {R}^{N_{\mathrm{data}}}\), we use the Euclidean norm and
where the indices i, j run from 1 to \(N_{\mathrm{data}}\), which eventually yields the usual expression for the \(\chi ^2\) of correlated data. Up to this point data and models are completely independent, and the joint distribution is simply the product of \(\pi _{Y}^0\) and \(\pi _{X}^0\).
The forward map induces a correlation between the input model and the observables, so we introduce a probability density \(\theta \) that describes these correlations due to the underlying theory,
where the Dirac delta corresponds to the case where there are no theoretical uncertainties. Theoretical uncertainties can be introduced by broadening the distribution of \(y\) away from the exact prediction of the forward map, e.g. using a Gaussian with covariance \(C_T\),
In the context of PDF fitting a similar recipe to take into account theoretical errors has recently been advocated in Refs. [4, 5]. Note that there are no rigorous arguments favouring the assumption that theoretical errors are normally distributed; it is nonetheless a useful working assumption, and a definite improvement compared to ignoring the theoretical errors altogether. The net effect of the theory errors is a redefinition of the covariance of the data, which has no major impact in our discussion, and therefore will be ignored. Taking the correlation \(\theta (y,u|\mathcal {G})\) into account, the joint distribution of \(y\) and \(u\) is
We can now marginalize with respect to y, neglecting theory errors,
We see that we have recovered Eq. 7. The log-likelihood in the Gaussian case is simply the \(\chi ^2\) of the data, \(y_0\), to the theory prediction, \(\mathcal {G}(u)\):
In the notation of Eq. 7
where in this case
The probability density \(\pi ^\mathcal {G}_{X}(u|y_0, C_{Y},u_0,C_X)\) was called \(\pi _{X}^\mathcal {G}(u)\) in Eq. 7, where the suffix \(\mathcal {G}\) is a short-hand to denote the posterior probability in model space, taking into account all the conditional variables. Hence, for the Gaussian case, the result from Bayes’ theorem reduces to
Note that in the argument of the likelihood function we have the central values of the data points \(y_0\) as given by the experiments. Eq. (19) is the Bayesian answer to the inverse problem, our knowledge of the model \(u\) is encoded in the probability measure \(\mu _{X}^\mathcal {G}\), which is fully specified by the density \(\pi _{X}^\mathcal {G}\). There are several ways to characterise a probability distribution, a task that becomes increasingly difficult in high-dimensional spaces. As discussed later in this study, the NNPDF approach is focused on the determination of the Maximum A Posteriori (MAP) estimator, i.e. the element \(u_* \in X\) that maximises \(\pi _{X}^\mathcal {G}(u)\):
For every instance of the data \(y_0\), the MAP estimator is computed by minimising a regulated \(\chi ^2\), where the regularization is determined by the prior that is assumed on the model u. We will refer to this procedure as the classical fit of experimental data to a model. Note that in the Bayesian approach, the regulator appears naturally after having specified carefully all the assumptions that enter in the prior. In this specific example the regulator arises from the Gaussian prior for the model input \(u\), which is normally distributed around a solution \(u_0\). The MAP estimator provides the explicit connection between the Bayesian approach and the classical fit.
2.2 Comparison with classical fitting
Analytical results make the connection between the two approaches more quantitative, and therefore more transparent. We are going to summarise these results here without proofs, referring the reader to the mathematical literature for the missing details. Working in the finite-dimensional case, we assume
and we are going to review in detail two examples from Ref. [3], which illustrate the role of the priors in the Bayesian setting. We report here the results in Ref. [3] because their simplicity provides a striking example of the role of priors, which is sometimes underestimated. It is particularly useful to distinguish the case of an underdetermined system from the case of an overdetermined one.
Underdetermined system The first case that we are going to analyse is the case of a linear system that is underdetermined by the data. The linear model is completely specified by a vector of coefficients \(g\in \mathbb {R}^{N_{\mathrm{model}}}\),
Assuming that we have only one datapoint, i.e. \(N_{\mathrm{data}}=1\),
where \(\eta \sim {\mathcal {N}}(0,\gamma ^2)\) is one Gaussian number, whose probability density is centred at 0 and has variance \(\gamma ^2\).
For simplicity we are going to assume that the prior on \(u\) is also a multi-dimensional Gaussian, centred at 0 with covariance matrix \(C_X\). In this case the posterior distribution can be written as
which is still a Gaussian distribution for \(u\). The mean and covariance are respectively
Because the argument of the exponential is a quadratic form, the mean of the distribution coincides with the MAP estimator. Hence in this case a fit that minimises the \(\chi ^2\) of the data to the theory prediction yields exactly the mean of the posterior distribution. It is instructive to look at these quantities in the limit of infinitely precise data, i.e. in the limit \(\gamma \rightarrow 0\):
These values satisfy
which shows that the mean of the distribution is such that the data point is exactly reproduced by the model, and that the uncertainty in the direction defined by g vanishes. It should be noted that the uncertainty in directions perpendicular to g does not vanish and is determined by a combination of the prior and the model, viz. \(C_X\) and g in our example. This is a particular example of a more general feature: for underdetermined systems the information from the prior still shapes the probability distribution of the solution even in the limit of vanishing statistical noise.
A linear model in a two-dimensional space is constrained by a single data point y. Once the vector g is given, the model is fully specified by the vector u, viz. \(y=g^T u\). All models u along the vertical red line reproduce exactly the data point. In the absence of prior knowledge, every model along that one-dimensional subspace is a legitimate solution of the inverse problem, which is clearly underdetermined
It is interesting to analyse what happens when the prior on the model is removed. For this purpose we can start from a covariance in model space that is proportional to the identity matrix,
and take the limit \(\sigma \rightarrow \infty \). In this limit
while the posterior covariance becomes
This covariance is the projector on the subspace orthogonal to g multiplied by \(\sigma \). In the simple two-dimensional example depicted in Fig. 1, choosing the direction of g as one of the basis vector, we obtain
which shows that the error in the g direction vanishes, while the error in the orthogonal subspace diverges as we remove the prior.
Overdetermined system We are now going to consider an example of an overdetermined system and discuss again the case of small observational noise. We consider \(N_{\mathrm{data}}\ge 2\) and \(N_{\mathrm{model}}=1\), with a linear forward map such that
where \(\eta \) is an \(N_{\mathrm{data}}\)-dimensional Gaussian variable with a diagonal covariance \(\gamma ^2 {\mathbb {1}}\), and \({\mathbb {1}}\) denotes the identity matrix. For simplicity we are going to assume a Gaussian prior with unit variance for \(u\), which yields for the posterior distribution:
The posterior is Gaussian and we can easily compute its mean and variance:
In this case, in the limit of vanishing observational noise, we obtain
The mean is given by the weighted average of the datapoints, which is also the solution of the \(\chi ^2\) minimization
Note that in this case the variance \(\sigma _\star \) vanishes independently of the prior. In the limit of small noise, the distribution tends to a Dirac delta around the value of the MAP estimator.
2.3 Linear problems
Linear problems in finite-dimensional spaces are characterized by a simple forward law,
where \(\mathcal {G}\) is a matrix. In this framework one can readily derive analytical solutions that are useful to understand the main features of the Bayesian approach. Assuming that the priors are Gaussian again, the cost function \(S(u)\) is a quadratic function of \(u\),
where
The case where we have no prior information on the model is recovered by taking the limit \(C_X^{-1} \rightarrow 0\), which yields
The action of \(C_{Y}^{-1}\) on the vector of observed data \(y_0\) is best visualised using a spectral decomposition
where \(P_n\) denotes the projector on the n-th eigenvector of \(C_{Y}\), and \(\sigma _n^2\) is the corresponding eigenvalue. The action of \(C_{Y}^{-1}\) is to perform a weighted average of the components of \(y_0\) in the directions of the eigenvectors of \(C_{Y}\).
An explicit expression for the posterior distribution of data can be obtained from the joint distribution by marginalising over the model input \(u\):
where
Note that this is the naive error propagation from the covariance of the model, \(\tilde{C}_X\), to the space of data.
Posterior distribution of unseen data In real-life cases we are also interested in the posterior distribution of a set of data that have not been included in the fit. Because different datasets are described by the same theory, the knowledge of one dataset will inform our knowledge of the underlying theory – i.e. we will determine a posterior distribution for the model. That new knowledge about the model will then propagate to any other – unseen – set of data, even if the experiments are completely unrelated. In the Bayesian framework that we have developed, this situation can be modeled by having two independent sets of data y and \(y'\), for which we have a prior distribution
Note that the prior distribution is factorised as the product of individual distributions for y and \(y'\) since the datasets are assumed to be independent. Following the derivation above, we can write the joint distribution for the data and the model
Because both sets of data are derived from the same model u, the joint distribution above introduces a correlation between the data sets. The same model u appears in both delta functions in the equation above. We can now marginalise with respect to the dataset y,
where \({\tilde{C}}_{X}\) and \({\tilde{u}}\) are given respectively in Eqs. 44 and 45. By marginalising again, this time with respect to the model, we derive the posterior distribution of the unseen data,
where
and we have introduced the variables \({\tilde{u}}'\) and \({\tilde{C}}'_{X}\),
The variables \({\tilde{C}}_{X}\) and \({\tilde{u}}\) have been defined above. We repeat their definition here in order to have all the necessary equations collected together:
Equations 55–60 yield the central value and the variance of the posterior distribution of unseen data as a function of \(y_0\), \(y_0'\), \(C_Y\), \(C_Y'\), \(u_0\) and \(C_X\).
A comment on non-linear models The linear models that we have discussed so far may look over-simplified at first sight. In practice, it turns out that non-linear models can often be linearised around the central value of the prior distribution,
where
and we have neglected higher-order terms in the expansion of \(\mathcal {G}(u)\).
If these terms are not negligible, another option is to find the MAP estimator, and then expand the forward map around it, which yields equations very similar to the previous ones, with \(u_0\) replaced by \(u_*\). If the posterior distribution of u is sufficiently peaked around the MAP estimator, then the linear approximation can be sufficiently accurate.
2.4 The infinite-dimensional case
In the finite-dimensional case, where the probability measures are specified by their densities with respect to the Lebesgue measure, Eq. (7) can be rephrased by saying that \(\rho \) is the Radon–Nikodym derivative of the probability measure \(\mu ^\mathcal {G}\) with respect to \(\mu _0\), viz.
Using the fact that the density \(\rho \) is a positive function, we can rewrite
and therefore
In finite-dimensional spaces, the three equations above are just definitions that do not add anything to the above discussion in terms of probability densities. Their interest resides in the fact that the last expression, Eq. (65), can be properly defined when \(X\) is infinite-dimensional, allowing a rigorous extension of the Bayesian formulation of inverse problems to the case of infinite-dimensional spaces.
Summarising the details of probability measure in infinite-dimensional spaces, is beyond the scope of this work. Adopting instead a heuristic approach, we can say that a function f is a random function if f(x) is a random variable for all values of x. Since the values of the function at different values of x can be correlated, a random function is fully characterised by specifying the joint probability densities
where \(f_i=f(x_i)\), for all values of n, and all values of \(x_1, \ldots , x_n\). This infinite set of finite-dimensional densities allows the definition of a probability measure.
For a Gaussian random function, these densities only depend on a mean value function m(x) and a covariance \(C(x,x')\). The probability densities for the variables \(f_i\), for any value of n is
The covariance C is such that
which shows that the two-point probability density determines all the other distributions.
Functional linear problems This formalism allows us to formulate a Bayesian solution of the inverse problem
where \(y^i\) is a discrete set of observables and u(x) is a random function, with a Gaussian prior with mean \(u_0(x)\) and covariance \(C_{X}(x,x')\). The vector of observed values is denoted \(y_0\), and we assume that the prior distribution of y is a Gaussian centred at \(y_0\) with covariance \(C_{Y}\).
Similarly to the finite-dimensional case, the Bayesian solution yields a Gaussian random function for the posterior distribution of the solution u(x). In order to characterise the posterior Gaussian distribution we need explicit expressions for its mean and its covariance. Introducing the matrix
and its inverse \(T^{ij}=\left( S^{-1}\right) ^{ij}\), the posterior Gaussian field is centred at
which is the expected generalization of the finite-dimensional example discussed above. Interestingly, this can be rewritten as
where
and the weighted residuals are given by
Defining
the posterior covariance can be written as
It is instructive to compare the Bayesian result summarised above with the method proposed by Backus and Gilbert [6] to solve the same inverse problem. Assuming that there exists an unknown ‘true’ model \(u_\mathrm {true}\), such that the observed data are
we look for an estimate \(u_\mathrm {est}\) of the true solution in the form
so that the problem is now recast as finding the functions \(Q^i(x)\). Using Eq. 77 we obtain
which states that with a finite amount of data we can only resolve a filtered version of the true solution. The kernel R is given by
The coefficient functions \(Q^i(x)\) can be chosen so that the kernel is as close as possible to a delta function,
Approximating the delta function can be achieved by minimising
which yields
where
The interesting observation is that the central value of the Bayesian solution presented above reduces to the Backus–Gilbert \(u_\mathrm {est}\) in the case where \(u_0\) is just white noise and therefore
The connections between the Bayesian treatment and the Backus–Gilbert solution and its regulated variations, deserves further investigations, which we defer to future studies. Note that the Bayesian solution allows a variety of priors to be explicitly declared and compared to the Backus–Gilbert solution.
3 NNPDF Monte Carlo approach to inverse problems
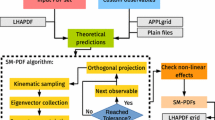
In this section we discuss the NNPDF approach to inverse problems, and make contact explicitly with the formalism laid out in Sect. 2. In the Bayesian formulation, Eq. 19 gives a quantitative description of how the information contained in the experimental data propagates into our knowledge of the space of models. In practice, it should be possible to sample directly from the posterior distribution, or to find a solution using some kind of Backus–Gilbert methodology [6]. These new types of approach are not straightforward and we defer their investigation to further, dedicated studies. Here we focus instead on the standard NNPDF fitting procedure and investigate its relation with the Bayesian result. The NNPDF approach generates an ensemble of fit results, which are supposed to describe the posterior probability distribution for the model (i.e. in the space of PDFs) given the experimental data. In the case of a linear map, we show here that this is exactly the case: the NNPDF replicas are distributed exactly according to the posterior density that was obtained in the previous section.
3.1 Fitting replicas
The approach for generating a sample in model space utilised by NNPDF can broadly be described as fitting model replicas to pseudo-data replicas. As discussed in Eq. 5 the experimental values are subject to observational noise. If we assume this observational noise to be multigaussian then the experimental central values, \(y_0\), are given explicitly by
where \(f\) is the vector of true observable values, and the observational noise is drawn from a Gaussian centred on zero such as in Eq. 18, i.e. \(\eta \sim {\mathcal {N}}(0, C_{Y})\) where \(C_{Y}\) is the experimental covariance matrix. In Eq. 86, each basis vector corresponds to a separate data point, and the vector of shifts \(\eta \) permits correlations between data points according to the covariance matrix provided by the experiments. Given the data, the NNPDF approach is to compute a MAP estimator along the lines discussed in the previous section, i.e. finding the model that minimises the \(\chi ^2\) to the data.Footnote 2 The key difference between the NNPDF approach and the classical MAP estimator is that instead of fitting the observational data given by Eq. 86, an ensemble of model replicas are fitted each to an independently sampled instance of pseudo-data, which is generated by augmenting \(y_0\) with some noise, \(\epsilon ^{(k)}\),
where k is the replica index and each instance of the noise, \(\epsilon \), is drawn independently from the same Gaussian from which the observational noise is drawn from, i.e. \(\epsilon \sim {\mathcal {N}}(0, C_{Y})\). For each replica k, \(\mu ^{(k)}\) is a vector in \(\mathbb {R}^{N_{\mathrm{data}}}\). The distribution of pseudo-data in a simple one-dimensional example is shown in Fig. 2. Note that, if we were to repeat this construction multiple times, the true value f would be within a 1\(\sigma \) interval centred at \(y_0\) with a 68% probability.
Histogram showing the distribution of \(10^4\) replicas generated around an experimental value \(y_0\) with unit variance. The central value \(y_0\), which is represented by the solid dot at the centre of the replica distribution, is drawn from a Gaussian distribution with unit variance centred at the true value f, which is assumed to be the origin in this plot. The central value \(y_0\) is kept fixed for the whole set of replicas
The parameters for each model replica maximise the likelihood evaluated on the corresponding pseudo-data. We can think of this approach as a special case of MAP estimation, as described in Eq. 20, where there is no model prior that regulates the likelihood. Another way of viewing this is to take \(C_X^{-1} \rightarrow 0\) in Eq. 20, as was done to obtain the result in Eq. 46. Either way, there is no prior information about the model. The parameterisation of the model is fixed, so the model space is the space of parameters \(u\in \mathbb {R}^{N_{\mathrm{model}}}\). In \(\mathbb {R}^{N_{\mathrm{model}}}\), we find the parameters which minimise the \(\chi ^2\) between the predictions from the model and the corresponding pseudo-data \(\mu ^{(k)}\)
where, as usual, minimising the \(\chi ^2\) is equivalent to maximising the likelihood, \({\mathcal {L}}\), since \(\chi ^2 \equiv -\log {{\mathcal {L}}}\).
As a final note: since we do not include the model prior, overall normalisations can be omitted in Eq. 88. It is clear however that if we were including a model prior in our MAP, it is important that the relative normalisation between the likelihood function and the model prior is clearly specified.
3.2 Fluctuations of fitted values
Whilst this MC methodology, maximising the likelihood on an ensemble of pseudo-data replicas, makes intuitive sense, we would like to explicitly draw the connection between the distribution of model replicas and the posterior distribution of the parameters given the data as described e.g. in Eq. 19. In order to investigate this issue, we will again consider a model whose predictions are linear in the model parameters, so that the posterior distribution of model parameters can be computed explicitly. DIS observables can be seen as an example of a linear model. Let us assume that the PDF are parametrized by their values at selected values of \(x_i\), so that u is a finite-dimensional vector
The observables are then computed by taking a matrix-vector multiplication of the vector u by a \(N_{\mathrm{data}}\times N_{\mathrm{model}}\) matrix, \(F_{ij}\), which is called FK table in the NNPDF jargon,
where \(i=1,\ldots ,N_{\mathrm{data}}\). In this case the forward map coincides exactly with the FK table, \(\mathcal {G}_{ij} = F_{ij}\) and the sum in Eq. 90 approximates the convolution in Eq. 2. However it should be clear that the discussion below applies to any model whose forward map can be approximated as Eq. 90, like a linear approximation of neural networks [7]. Let us consider for instance the actual NNPDF parametrization, where the value of the PDF is given by a neural network whose state is determined by a set of weights \(\theta \). In this case X is the space of weights, the model will be specified by a vector \(\theta \) in the space of weights, and we are going to continue using u to denote the PDF, so that
If the parameters have a sufficiently narrow distribution around some value \({\bar{\theta }}\), then we can expand the expression for the observables:
and therefore
where
In order to get an exact analytical solution for the linear model, we additionally require \(\mathcal {G}\) to have linearly independent rows, and therefore \(\mathcal {G}C_{Y}\mathcal {G}^T\) is invertible. As discussed in Sect. 2, in the absence of prior information on the model, the posterior distribution of model parameters is a Gaussian with mean and covariance given by Eqs. 46 and 47.
Let us now stick to the parametrization in Eq. 89 and let us deploy the NNPDF Monte Carlo method to fitting model replicas, then in the case under study \(\arg \min _{u^{(k)}} {\chi ^2}^{(k)}\) is found analytically by imposing that the derivative of \({\chi ^2}^{(k)}\) with respect to the model parameters is zero, i.e.
Equation 97 shows that \(u_*\) is a linear combination of the Gaussian variables \(\epsilon \), and therefore is also a Gaussian variable. Its probability density is then completely specified by the mean and covariance, which can be calculated explicitly, given that the probability density for \(\epsilon \) is known:
These two equations show that, under the assumptions specified above, \(u_* \sim {\mathcal {N}}(\tilde{u}, \, \tilde{C}_X)\). In other words, when the model predictions are linear in the model parameters, the NNPDF MC method is shown to produce a sample of models exactly distributed according to the expected posterior distribution of model parameters given the data. When we fit PDFs, parameterised as deep fully connected neural networks, to data which includes hadronic observables, it is clear that the forward map is non-linear, and therefore this proof does not strictly apply. As discussed above, even for non-linear models we can make a linear approximation of the forward map provided that we are expanding around the MAP estimator. This means the NNPDF MC methodology should reproduce the posterior distribution of the model given the data, at least close to \(\tilde{u}\), the central value of the fitted replicas. Furthermore, by fluctuating the data and fitting the replicas, the fluctuations in data space are propagated to model space non-linearly. So even for non-linear problems, the NNPDF MC methodology can be expected to produce a sample of models which are at least approximately distributed according to the posterior model distribution. It remains to be shown, however, that further away from the MAP estimator the approximation holds despite the non-linear dependence of the model replicas on the data uncertainties.
3.3 Closure test
The concept of the closure test, which was first introduced in Ref. [8], is to construct artificial data by using a known pre-existing function to generate the true observable values, \(f\). One way of achieving this is by choosing a model \(w\) and then compute \(f= \mathcal {G}(w)\). Then the experimental central values are artificially generated according to Eq. 86, where the observational noise is pseudo-randomly generated from the assumed distribution. In Ref. [8], \(f\) is referred to as level 0 (L0) data and \(y_0\) is referred to as level 1 (L1) data. Finally, if we use the NNPDF MC method to fit artificially generated closure data, the pseudo-data replicas that are fitted by the model replicas are referred to as level 2 (L2) data.
The outcome of the fit is then compared with the known true value to check the precision and accuracy of the fitting procedure. Finding quantitative estimators that allow us to characterise the quality of the closure fit is one of the main problems that need to be addressed. We will discuss a new class of estimators in the next section.
Note that in a closure test, the assumed prior of the data is fully consistent with the particular instance of observed central values, \(y_0\): by construction, there are no inconsistent data sets. In the original closure test of Ref. [8] there was also no modelisation uncertainty, the true observable values were assumed to be obtained by applying the forward map \(\mathcal {G}\) to a vector in model space \(w\). It is worth noting that the assumption of zero modelisation uncertainties is quite strong and likely unjustified in many areas of physics. In the context of fitting parton distribution functions there are potentially missing higher order uncertainties (MHOUs) from using fixed order perturbative calculations as part of the forward map. MHOUs have been included in parton distribution fits [4, 5] and in the future these should be included in the closure test, however this is beyond the scope of the study presented here, since MHOUs are still not included in the NNPDF methodology by default. In the results presented in the rest of this paper we do include nuclear and deuteron uncertainties, as presented in [9,10,11], since they are to be included in NNPDF fits by default. Extensive details for including theoretical uncertainties, modelled as theoretical covariance matrices can be found in those references. For the purpose of this study the modelisation uncertainty is absorbed into the prior of the data, since
where \(\delta \sim {\mathcal {N}}(0, C^\mathrm{theory})\). As long as the modelisation uncertainty is independent of the data uncertainty, we can absorb \(\delta \) into \(\eta \) by modifying the data prior: \(\eta \sim {\mathcal {N}}(0, C + C^\mathrm{theory})\). In doing that, we must also update the likelihood of the data given the model to use the total covariance \((C + C^\mathrm{theory})\). From now onwards we will omit \(C^\mathrm{theory}\) because it is implicit that we always sample and fit data using the total covariance matrix which includes any modelisation uncertainty we currently take into account as part of our methodology.
Mapping the closure test procedure to the quantities used in the Bayesian treatment presented in the previous section will allow us to derive a number of analytical results in Sect. 4.4.
4 Data space estimators
In order to perform a quantitative analysis of the results obtained in the closure tests, we discuss several estimators, which are computed from the outcome of the closure test fits. These results depend on the pseudo-data that have been generated and therefore are stochastic variables which can fluctuate. The values of the estimators on a single replica will not tell anything about the quality of our fits: we need to understand their probability distributions in order to validate our fitting procedure. We begin this section by defining estimators in data space, i.e. estimators that are computed from the model predictions for a set of experimental data points. Having defined the estimators, we define criteria to characterise faithful uncertainties. We conclude this section with a discussion of the predictions that can be obtained for these estimators in the case of a linear model, where analytical calculations can be performed. As we already saw in the previous section, the analytical results cannot be applied directly to the NNPDF fits, but they are useful examples that illustrate the expected behaviour of these quantities.
4.1 Deriving the data space estimators
For a given model \(u_*^{(k)}\), obtained from fitting the k-th replica, we start by defining the model error as the \(\chi ^2\) between the model predictions and some data central values \( {{y_0}^{\prime }} \), normalised by the number of data points
and we purposely denoted the data which the model error is evaluated on as \( {{y_0}^{\prime }} \), as opposed to the training data \(y_0\), which is used to determine the model parameters. The corresponding covariance is denoted \( {{C_{Y}}^{\prime }} \). Note that in Eq. 101, \( {{y_0}^{\prime }} \) is a stochastic variable, but also \(u_*^{(k)}\) is a stochastic variable, with its pattern of fluctuations, since the fitted model depends on the data \(\mu ^{(k)}\) that enter the fit. We define the model error \(\mathcal {E}^{\mathrm{out}}\) on the set of data \( {{y_0}^{\prime }} \) by taking the average over the models,
where we defined the expectation value over the ensemble of model replicas as
We could of course set \( {{y_0}^{\prime }} = y_0\) and evaluate the model performance on the fitted data however, as is common in machine learning literature, we intend to use a separate set of test data. Ideally we would choose \( {{y_0}^{\prime }} \) such that \( {{y_0}^{\prime }} \) and \(y_0\) are statistically independent, as in Eq. 52. This is achieved by choosing the split such that the experimental covariance matrix is block diagonal:
It is useful to perform a decomposition of Eq. 102, following usual manipulations of the likelihood function associated with least-squares regression in [12]. Least-squares regression is a special case of minimum likelihood estimation, where the uncertainty on each data point is equal in magnitude and uncorrelated. Here we review the decomposition in the more general framework of data whose uncertainty is multigaussian. Starting with Eq. 102 (evaluated on the ideal test data), we can complete the square
Let us now discuss these terms one by one, starting from the last two. The second term is the shift associated with evaluating the model error on noisy test data and the final term is a cross term which we will deal with later. We therefore focus next on further decomposing the first term,
where we have used the fact that the second term is constant across replicas and the cross term that arises in this decomposition is zero when the expectation value across replicas is taken. The first term in this expression we call the variance and the second term is the bias.
As previously mentioned \(\mathcal {E}^{\mathrm{out}}\) should be considered a stochastic estimator, in theory we could take the expectation value across training data \(y_0\) and test data \( {{y_0}^{\prime }} \). In other words, \(\mathcal {E}^{\mathrm{out}}\) is a function of the Gaussian parameters \(\eta \) and \(\eta '\), and we can take the average over them to compute the average over training and test data. Denoting as \(\rho \) the Gaussian describing the distribution of \(\eta \) and \(\eta '\), we can define
The average over the test data cancels the cross term in Eq. 105 and the final result would be
We are not interested in the observational noise term, since it is independent of the model and in the limit of infinite test data \({\mathbf {E}}_{ {{y_0}^{\prime }} }[\mathrm{noise}] \rightarrow 1\). The two estimators of interest are independent of the parameter \(\eta '\) associated with the test data, however they do depend implicitly of \(\eta \), entering the definition of the training data used to determine \(u_*^{(k)}\). Therefore we only need to take the expectation value over the parameter \(\eta \), associated with the training data. In practical terms, this can be achieved by running multiple closure fits, each with a different observational noise vector \(\eta \), and taking the average i.e.
Clearly this is resource intensive, and requires us to perform many fits. In NNPDF3.0 [8], single replica proxy fits, namely fits run over different training sets and made by a single replica, were used to perform a study of the uncertainties. Here we have expanded the data-space estimators used in the closure fits and also will be using multiple full replica fits to calculate various expectation values - made possible by our next generation fitting code.
4.2 Geometric interpretation
It is possible to interpret the relevant data space estimators geometrically, by considering a coordinate system where each basis vector corresponds to an eigenvector of the experimental covariance matrix normalised by the square root of the corresponding eigenvalue. An example of this is given in Fig. 3, where for simplicity we have considered a system with just two data points, i.e. a two-dimensional data space, with a diagonal covariance and we take these for both the training and the test set. The origin of the coordinate system is the true value of the observable. The observational noise in these coordinates corresponds to a unit circle centred in the origin as shown in Fig. 3. If the experimental covariance is faithful, there is a 68% probability that the experimental value \(y_0\) is within this unit circle. Fig. 3 shows one possible instance of \(y_0\). Repeating the entire fit procedure multiple times requires generating new sets of experimental data \(y_0\). The average over \(y_0\) mentioned above, is precisely the average over multiple fits, restarting the procedure each time from a new instance of \(y_0\).
For a given \(y_0\) the replicas are generated as a set of points Gaussianly distributed around it and therefore, in the limit of a large number of replicas, 68% of them will fall within a unit circle centred in \(y_0\). This is the dashed circle in the figure. Clearly there is also a 68% probability that the true value (i.e. the origin in our plot) is inside this second circle. The model predictions, one for each replica, are then a set of points, whose mean is \(E_\epsilon [g]\). The mean squared radius of those points is what we call the variance. The bias is the l2-norm of the vector between the origin and the mean of the model predictions.
A faithful representation of the errors requires that the true value, i.e. the origin of the coordinate system in our figure, has 68% probability of being within 1\(\sigma \) from the central value of the fit, which is given by \(E_\epsilon [g]\). Looking at the figure again, the probability for the origin to be inside the shaded circle must be 68%. We will discuss faithful errors in more detail in the next subsection.
Example of geometric interpretation of closure test estimators. The origin is the true observable values for each data point. The level one data (or experimental central values) are shifted away from this by \(\eta \). In this example the covariance matrix is diagonal, so the eigenvectors correspond to the two data points, the square root of the eigenvalues are simply the standard deviation of those points. This is without loss of generality because any multivariate distribution can be rotated into a basis which diagonalises the covariance matrix. The 1-sigma observational noise confidence interval is a unit circle centered on the origin. Some closure estimators can be understood as l2-norms of the vectors connecting points, i.e the bias is the l2-norm of the vector from the origin to the central value of the predictions
4.3 Faithful uncertainties in data space
The two closure estimators of interest, bias and variance, can be used to understand faithful uncertainties in a practical sense. If we return to Eq. 108 we can examine both estimators in a more detail.
Variance The variance in the above decomposition refers to the variance of the model predictions in units of the covariance
which can be interpreted as the model uncertainty in the space of the test data. It is instructive to rephrase Eq. 110 as
where
is the covariance matrix of the predictions from the model replicas. Note that we can rotate to a basis where \( {{C_{Y}}^{\prime }} \) is diagonal,
then we can rewrite Eq. 111 as
The numerator in the right-hand side of the equation above is the variance of the theoretical prediction obtained from the fitted replicas, while the denominator is the experimental variance, the average is now taken over eigenvectors of the experimental covariance matrix. Note that this ratio does not need to be equal to one, the model error on a given point can be smaller than the experimental one, since all theoretical estimates are correlated by the underlying theoretical law.
Bias Similarly, the bias is defined as the difference between the expectation value of the model predictions and the true observable values in units of the covariance, i.e.
The smaller the bias, the closer the central value of the predictions is to the underlying law. In Eq. 108, the expectation value is taken across the prior distribution of the training data, which yields
where we have introduced \( {{C}^{\prime }} ^{({\mathrm{central}})}\) as the covariance of the expectation value of the model predictions,
The point is that the bias on the test data is a stochastic variable which depends on the central value of the training data through \(u_*^{(k)}\). The matrix \( {{C}^{\prime }} ^{({\mathrm{central}})}\) describes the fluctuations of the central value of the model prediction around the true observable values as we scan different realisations of the training data.
It is important to stress the difference between variance and bias. In the case of the variance, we are looking at the fluctuations of the replicas around their central value for fixed \(y_0\). This is related to the ensemble of model replicas we provide as the end product of a fit and can be calculated when we have one instance of \(y_0\), provided by the experiments. In the case of the bias we consider the fluctuations of the central value over replicas around the true theoretical prediction as the values of \(y_0\) fluctuate around \(f\). This latter procedure is only possible in a closure test, where the underlying true observable is known. The bias as defined here yields an estimate of the fluctuations of the MAP estimator if we could do multiple independent instances of their measurements from each experiment.
Bias-variance ratio Finally, the bias-variance ratio is defined as
where we have taken the square root, since bias and variance are both mean squared quantities. The value of \(\mathcal {R}_{bv}\) yields a measurement of how much uncertainties are over or under estimated. If the uncertainties are completely faithful, then \(\mathcal {R}_{bv}= 1\). We note that \(\mathcal {R}_{bv}\) is not completely general: it is not a measure defined in model space and depends on the choice of test data. Therefore it only gives local information on the model uncertainties. If the distribution of the expectation value of model predictions is gaussian centered on the true observable values, with covariance \( {{C}^{\prime }} ^{({\mathrm{central}})}\) and the distribution of the model replicas is also gaussian, with covariance \( {{C}^{\prime }} ^{({\mathrm{replica}})}\) then model uncertainties are faithful if
The difficulty with calculating Eq. 119 comes from the fact that \( {{C}^{\prime }} ^{({\mathrm{replica}})}\) is likely to have large correlations which would lead it to be singular or ill-conditioned. As a result, any error estimating \( {{C}^{\prime }} ^{({\mathrm{replica}})}\) from finite number of replicas could lead to unstable results. \(\mathcal {R}_{bv}\) overcomes this instability by taking the ratio of the average across test data of these matrices, in units of the experimental covariance matrix. There may still be large relative errors for smaller eigenvalues of \( {{C}^{\prime }} ^{({\mathrm{replica}})}\), but these should not lead to instabilities in \(\mathcal {R}_{bv}\) unless they correspond to directions with very low experimental uncertainty. As an extra precaution, we shall estimate an uncertainty on \(\mathcal {R}_{bv}\) by performing a bootstrap sample on fits and replicas.
Quantile statistics When the closure test was first presented in [8], there was an estimator introduced in the space of PDFs which also aimed to estimate faithfulness of PDF uncertainties using the combined assumption of Gaussian PDF uncertainties and quantile statistics, called \(\xi _{1\sigma }\). Here we can define an analogous expression in the space of data,
where \( {{\sigma }^{\prime }} _i^{(l)} = \sqrt{ {{C}^{\prime }} ^{({\mathrm{replica}})}_{ii}}\) is the standard deviation of the theory predictions estimated from the replicas of fit l and \(I_{[a, b]}(x)\) is the indicator function, which is one when \(a \le x \le b\) and zero otherwise. In other words, \(\xi ^{({\mathrm{data}})}_{n {{\sigma }^{\prime }} }\) is counting how often the difference between the prediction from the MAP estimator and the true observable value is within the \(n\sigma \)-confidence interval of the replicas, assuming they’re Gaussian. Since \( {{C}^{\prime }} ^{({\mathrm{replica}})}\) is primarily driven by the replica fluctuations, we assume that it is roughly constant across fits, or independent upon the specific instance of observational noise. This allows us to write \(\xi ^{({\mathrm{data}})}_{n {{\sigma }^{\prime }} }\) for a specific data point in the limit of infinite fits, each to a different instance of the data as
where \( {\mathbf {E}}_{\{ u_* \}} \left[ \mathcal {G}_i \right] ^{(l)}\) has implicit conditional dependence on \(\eta \). If the distribution of
is Gaussian, centered on zero, we can define \({ {{ \hat{\sigma } }^{\prime }} }_i = \sqrt{ {{C}^{\prime }} ^{({\mathrm{central}})}_{ii}}\). In this case
which is simply the standard result of integrating a gaussian over some finite symmetric interval.
The analogy between \(\mathcal {R}_{bv}\) and \(\xi ^{({\mathrm{data}})}_{n {{\sigma }^{\prime }} }\) is clear, the ratios of uncertainties are both attempts to quantify Eq. 119 whilst keeping effects due to using finite statistics under control. Whilst with \(\mathcal {R}_{bv}\) we take the average over test data before taking the ratio, \(\xi ^{({\mathrm{data}})}_{n {{\sigma }^{\prime }} }\) instead takes the ratio of the diagonal elements - ignoring correlations. Since the predictions from the model will be compared with experimental central values, taking into account experimental error, we find it more natural to calculate \(\xi ^{({\mathrm{data}})}_{n {{\sigma }^{\prime }} }\) in the basis which diagonalises the experimental covariance of the test data as in Eq. 113. If we assume that in this new basis, that both \(\frac{ {{C}^{\prime }} ^{({\mathrm{replica}})}_{ii}}{\left( \sigma '_i\right) ^2}\) and \(\frac{ {{C}^{\prime }} ^{({\mathrm{central}})}_{ii}}{\left( \sigma '_i\right) ^2}\) are approximately constant for all eigenvectors of the experimental covariance matrix, then we recover the approximation
Whilst it is clear that Eq. 123 is reliant on a fair few assumptions which may not hold, we will use the comparison of \(\xi ^{({\mathrm{data}})}_{n {{\sigma }^{\prime }} }\) with \(\mathcal {R}_{bv}\) to consider how valid these assumptions may be.
4.4 Closure estimators: linear problems
Once again we return to the linear model framework set out in Sect. 3.2. We can perform an analytical closure test in this framework, and check our understanding of the closure estimators. Consider the true observable values for the test data are given by
where \(w\in X\), and we assume that the number of (non-zero) parameters in the underlying law is less than or equal to the number of parameters in the model, \(N_{\mathrm{law}}\le N_{\mathrm{model}}\). Using the previous results from Sect. 3.2, we can write down the difference between the true observables and the predictions from the MAP estimator (or the expectation of the model predictions across model replicas - in the linear model these are the same)
where we recall that \(\mathcal {G}\) is the forward map to the training observables. Because the training observables, previously denoted as \(y_0\), do not necessarily coincide with the data used to compute the estimators, denoted as \(y_0'\), we have two different maps \(\mathcal {G}\) and \(\mathcal {G}'\). Calculating the covariance across training data of \( {\mathbf {E}}_{\{ u_* \}} \left[ {{\mathcal {G}}^{\prime }} \left( u_*^{(k)}\right) \right] - {{f}^{\prime }} \) gives
so the full expression for \({\mathbf {E}}_{y_0}[\mathrm{bias}]\) is given by
We note that if the test data is identical the data the model was fitted on, we recover an intuitive result \({\mathbf {E}}_{y_0}[\mathrm{bias}] = \frac{N_{\mathrm{model}}}{N_{\mathrm{data}}}\). In the case of a polynomial model, where the parameter of the model are the coefficients of the polynomial function, the maximum value which \(N_{\mathrm{model}}\) can take whilst \(\mathcal {G}\) still has linearly independent rows is \(N_{\mathrm{data}}\) and in this case the \({\mathbf {E}}_{y_0}[\mathrm{bias}]\) takes its maximum value of 1. The central predictions from the model exactly pass through each data point.
We can perform a similar exercise on the model replica predictions. The difference between the predictions from model replica \((k)\) and the expectation value of the model predictions is
Since \(\epsilon \) and \(\eta \) follow the same distribution, it is clear that
which simply means that
We recall that when the map is linear, the NNPDF MC methodology generates replicas which are sampled from the posterior distribution of the model given the data. We have shown here that provided the underlying law belongs to the model space, the posterior distribution of the model predictions satisfy the requirement that \(\mathcal {R}_{bv}= 1\).
We note that due to the invariance of the trace under cyclic permutations, we can rearrange Eq. 127 as
where the term \( {{\mathcal {G}}^{\prime }} ^T {{C_{Y}}^{\prime }} ^{-1} {{\mathcal {G}}^{\prime }} \) can be understood as the covariance matrix of the posterior distribution in model space given the test data, with zero prior knowledge of the model, which we denote as \(\tilde{C}_X'\):
where we emphasise that the covariance matrices \(\tilde{C}_X\) and \( {{\tilde{C}_X}^{\prime }} \) are obtained from completely independent Bayesian inferences with no prior information on the model parameters, unlike in Eq. 57 where a sequential marginalisation causes \( {{\tilde{C}_X}^{\prime }} \) to depend on \(\tilde{C}_X\).
Alternatively, if we perform a sequential marginalisation of the data, and use the result in Eq. 57, but then take \( {{C_{Y}}^{\prime }} ^{-1} \rightarrow 0\), i.e. there is no information on the observables in the test set, then the covariance of the posterior model distribution is
which is identical to the posterior model distribution given just the training data - as one would expect. Now we can express bias (or variance) as
where \( {{\tilde{C}_Y}^{\prime }} \) is the covariance of the posterior distribution of \( {{y}^{\prime }} \) with no prior information on that data. This might seem peculiar because in determining \( {{\tilde{C}_Y}^{\prime }} \) we took the limit \( {{C_{Y}}^{\prime }} ^{-1} \rightarrow 0\), which encodes the fact that we had no prior information on the unseen data, however in Eq. 134 we require \( {{C_{Y}}^{\prime }} ^{-1}\) to be finite. We rationalise Eq. 134 as a comparison between the posterior distribution in the space of data of some unseen observables to an independently determined prior from performing the relevant experiment which measures the same observables. The two distributions can be compared when the independently measured experimental data is published.
Underparameterised model Note that if we were to choose the number of model parameters such that \(N_{\mathrm{law}}> N_{\mathrm{model}}\), then the variance would be unaffected, since the underlying law parameters cancel. However, the bias would now contain an extra term from the extra parameters in the underlying law, schematically:
which would mean that \(\mathcal {R}_{bv}\ne 1\). This demonstrates that requiring \(\mathcal {R}_{bv}= 1\) demands that the model space is suitably flexible, if the underlying law is parameterised then this can be summarised as requiring \(w\in X\). Note that in the underparameterised regime the model replicas are still drawn from the posterior distribution, however because \(w\notin X\) we have somehow invalidated the assumptions that go into the relation between model predictions and the data-space prior.
Although \(\mathcal {R}_{bv}\) was largely chosen on practical grounds, we see that it is still a stringent test that our assumptions are correct and that the distribution our model replicas are drawn from is meaningful, this is what we mean when we say faithful uncertainties.
An unfortunate trade-off when using \(\mathcal {R}_{bv}\) is that it cannot be used as a diagnostic tool, and is instead used simply for validation. For example, if \(\mathcal {R}_{bv}> 1\), then we cannot know whether there was a problem with the fitting methodology used to generate the model replicas or a deeper issue such as an inflexible model.
5 Numerical setup and results
In this section we first introduce the experimental setup used to run the closure tests, and then discuss the actual results. Following the study performed in Ref. [8], we first analyse the relative size of the different components of PDFs uncertainty, comparing the changes between methodologies used to produce the NNPDF3.1 [13] and NNPDF4.0 [2] sets of PDFs respectively. We then move to the data space estimators \(\mathcal {R}_{bv}\) and \(\xi ^{({\mathrm{data}})}_{1 {{\sigma }^{\prime }} }\), which have been computed only for NNPDF4.0.Footnote 3 The results here act both as a proof of principle of new estimators presented in this paper but also as part of a suite of methodological validation tools, see also the “future tests” [15], used to understand the PDF uncertainties of the recent NNPDF4.0 set of PDFs. For the purpose of understanding how the results here were produced, we will briefly describe the key features of the NNPDF4.0 methodology, but refer the reader to Ref. [2] for a full discussion on how these methodological choices were made.
5.1 Closure test setup
Using neural networks to fit PDFs has been discussed many times in previous NNPDF publications, see for example [8, 13]. A new feature of NNPDF4.0 is that, for the default fit performed in the evolution basis, a single neural network parameterises all 8 PDF flavours
at the initial scale. The PDF for a single flavour j, at the initial scale \(Q_0 = 1.65~\mathrm{GeV}\) is given by
where \(\alpha \) and \(\beta \) are the preprocessing exponents, which control the PDF behaviour at \(x \rightarrow 0\) and \(x \rightarrow 1\) respectively and \(NN(x, \ln x | u)_j\) is the \(j^\mathrm{th}\) output of the neural network, which takes x and \(\ln x\) as input. As discussed in Sect. 3.1, an ensemble of models is fitted, each one is a MAP estimator of the corresponding pseudo-data it is fitted on. An optimization algorithm is used to try and find the parameters which maximise the likelihood. There are clearly many choices with respect to hyperparameters, the discussion of how these choices have been made is beyond the scope of this paper and left to the full NNPDF4.0 release [2]. A summary of the hyperparameters used to produce results presented in this paper are provided in Table 3.
As input to the closure test, a single replica was drawn randomly from a previous NNPDF fit to experimental data. This has generally more structure than the final central PDF and it is therefore a more general choice than any final central fit. We refer to this as the underlying law and the corresponding predictions as the true observable values. An example of the gluon input is provided in Fig. 4. In principle any function could be used as underlying law, however it makes sense to use a realistic input.
The observables used in the fits are a subset of the full NNPDF4.0 dataset. For convenience, we chose to fit the PDFs on a variant of the NNPDF3.1 dataset used in Ref. [16], which is described in detail in a study of the determination of the strange PDF [17]. The datasets used in the calculation of statistical estimators are the new datasets which have been included in NNPDF4.0 and discussed in detail in the main release [2]. For a full summary of the observables used in the test data and a visual representation of the kinematic region of both the training and testing data, see App. B.
The partitioning of the available data into fitting and testing should not affect the interpretation of the closure test results. However, one could consider splitting the data into fitting and testing in a way which was physically motivated e.g. the partitioning could have been stratified such that the kinematic coverage of the data of each partition was approximately equal. Alternatively, since the data is generated from the theory predictions produced by the input underlying law, one could even produce completely artificial data using a different set of FK tables.
In order to compute the expectation value over training data defined in Eq. 109, we generate 30 different sets of experimental central values (or L1 data), as discussed in Sect. 3.3, for the fitted 3.1-like dataset. Each set of experimental central values was then fitted following NNPDF4.0 methodology [2], producing 40 pseudo-data replicas.
5.2 Different components of the PDF uncertainty
As already discussed in Ref. [8], fitting to L0, L1 and L2 pseudo-data allows us to validate different aspects of the fitting procedure. In an L0 fit, we fit multiple time the exact same set of data, which corresponds to the theory prediction from the chosen model. The fitted pseudo-data is the result obtained by applying the forward map to the model. It is clear that in this case the quality of the fit can be improved at will, provided that the parametrization is flexible enough and the minimization algorithm is efficient. There are indeed multiple solutions that reproduce exactly the data set, while interpolating between the data points. In an L1 fit the data have been fluctuated around the theoretical prediction – mimicking thereby the central values of experimental measurements. The true model no longer reproduces the data; instead it will yield a \(\chi ^2\) of order one. The pseudo-data are held fixed, fluctuations from one replica to the next are due to the existence of multiple solutions that hold a similar value for the residual \(\chi ^2\) at the end of the minimization process. Finally, in the L2 fits, the fluctuations of the data are reproduced by the replicas, and propagated to the model function when fitting the data for each replica. Since the NNPDF fitting methodology has changed in the latest release, adopting the procedure described in Ref. [18], it is important to compare the uncertainties at L0, L1 and L2 that are obtained with the NNPDF4.0 methodology with the corresponding ones obtained with the NNPDF3.1 framework.
An example of these relative errors on fitted PDFs is shown in the upper panel of Fig. 5 where results from L0, L1 and L2 closure tests are displayed on the same plot in the case of the gluon distribution. Each fit is normalized to the corresponding central value. We note how the L0 and L1 uncertainties tend to increase in the x regions where less experimental data are available, namely at small and large-x, where the model is left unconstrained and has more freedom to fluctuate, while they are considerably reduced in the data region where the contribution of the L2 error, induced by the actual experimental data, becomes more important. The fact that the data uncertainty is not always the dominant component of the PDF uncertainty was already stressed in Ref. [8]. Improved methodologies should therefore aim to reduce the L0 and L1 error in the data region. Where less experimental data are available, we should still expect the dominant component of PDFs error to come from the L0 and L1 uncertainties, which however should be driven by the lack of experimental information rather than by the unefficiency of the methodology.
In this respect, it is interesting to compare these results with what we find with the NNPDF3.1 methodology. The corresponding plots are shown in the lower panel of Fig. 5: unlike the NNPDF4.0 case, here the PDF uncertainty is always dominated by the L0 uncertainty, even in those kinematic regions where experimental data are present. We can conclude that moving to the NNPDF4.0 methodology, thanks to the optimized hyperparameters listed in Table 3, we observe a marked reduction of the L0 uncertainty, see Fig. 6, where the latter is plotted on the same panel for both NNPDF4.0 and NNPDF3.1. The better efficiency of the NNPDF4.0 methodology can be also appreciated by looking at Fig. 7, where we plot the distribution across replicas of the L0 closure test \(\chi ^2\) as a function of the training epoch and genetic algorithm generation for the NNPDF4.0 and NNPDF3.1 methodology respectively. As mentioned before, in a L0 closure test the quality of the fit can in principle be improved at will, assuming the use of an efficient optimizer. For the final \(\chi ^2\) of the central value of each fit, plotted as a black dashed line, we find 0.002 and 0.012 in the NNPDF4.0 and NNPDF3.1 fitting code respectively, showing the better efficiency of the new methodology.
It should be noted how Fig. 5 only provide a qualitative assessment of the relative size of the different components of PDFs uncertainty. Despite being useful to assess how the methodology has improved with respect to the previous one, they do not provide any quantitative estimation of the faithfulness of PDF uncertainty. This is best achieved in data space, using the new estimators introduced in the previous sections. How to validate PDFs uncertainty in the extrapolation regions remains an open problem, and will be addressed in a future work.
\(\chi ^2\) distribution across replicas as a function of the training epoch and genetic algorithm generation for the NNPDF4.0 (left) and NNPDF3.1 (right) methodology. The black dotted line in each plot represents the final \(\chi ^2\) of the central value of the corresponding fit, equal to 0.002 and 0.012 respectively
5.3 Data space estimators
Results for \(\mathcal {R}_{bv}\) and the corresponding uncertainty calculated on the test data are shown in the first column of Table 1. The uncertainties, which take into account the finite size of the replicas and fits samples, have been computed by performing bootstrap [19], where we randomly sample from both fits and replicas and re-calculate \(\mathcal {R}_{bv}\). The value and error presented in the table is then the mean and standard deviation across bootstrap samples. We checked that the distribution of the estimator across bootstrap samples is indeed Gaussian. We also checked that increasing the number of fits and replicas reduced the bootstrap error but the central values were the same within the estimated bootstrap uncertainties. We see that overall \(\mathcal {R}_{bv}\) is consistent with 1, which gives a good indication that, at least for the unseen data used in this study, the uncertainties are faithful
In Fig. 8 we compare qualitatively the distribution of bias across fits, to the distribution of the difference between replica predictions and expectation values of predictions (in units of the covariance) across different fits and replicas. The square root ratio of the mean of these two distributions is precisely \(\mathcal {R}_{bv}\).
The green histogram is the distribution of the total bias across fits, the orange histogram is the distribution of the difference between the replica and central predictions squared, in units of the covariance across all fits and replicas. In both of these cases, the x-axis represents the total distance in units of the covariance summed across all unseen data. We expect shape of the distributions to be equal, and qualitatively we see that this is approximately the case. We attribute difference in shape between the distributions to low statistics, in particular for the bias distribution across fits. For a qualitative comparison we compare the square root of the mean of each distribution shown here, getting the quoted value for \(\mathcal {R}_{bv}\) in Table 1, where we observe the first moment of the distributions are equal within uncertainties
As discussed in Sect. 4.3, one can define an analogous estimator in data space, based upon \(\xi _{n\sigma }\), which was defined on a grid of points in x and \(Q^2\) in PDF space in [8]. There is not a one-to-one correspondence between this and \(\mathcal {R}_{bv}\), but a loose approximation using Eq. 122. In the second and third columns of Table 1 we compare the estimated \(\xi _{1\sigma }\) from substituting \(\mathcal {R}_{bv}\) into Eq. 122 and to the measured value. Despite the assumptions entering each of the two estimators differing, we see good agreement between the \(\xi _{1\sigma }\) estimated from \(\mathcal {R}_{bv}\) and that measured directly. We find this result reassuring, since it indicates not only that the total uncertainty averaged across all data is faithful, but also that the uncertainty on each data point seems faithful. If the results differed it would indicate some kind of imbalance, where some components of the uncertainty are correctly represented by the replicas but other directions are not. Finally we note how not only are the measured value and estimated value from \(\mathcal {R}_{bv}\) self consistent, but they are also consistent with 0.68, which further supports the argument that the model uncertainties are faithful.
As mentioned before, the improved efficiency of the new NNPDF4.0 code makes it possible to easily compute the data space estimators for the new methodology, but not for the NNPDF3.1 one, for which a much higher computational cost would be required to perform the same exercise. For this reason, here we choose to present numbers for NNPDF4.0 only. Given the fact that uncertainties in NNPDF4.0 are much smaller than those in NNPDF3.1, one might still wonder to what extend such estimators can or cannot distinguish between the two methodologies. In this respect, we notice that the faithfulness of uncertainties as defined here is not directly related to the corresponding size, and that the estimators presented here cannot distinguish between two different faithful methodologies. Looking at the toy example of Fig. 3, if we had bigger uncertainty, like in the NNPDF3.1 case, we would expect to get a bigger blue circle, with the true value of the observable still within its larger \(68\%\). The data space estimators for any faithful methodology should therefore have similar values to those obtained for NNPDF4.0, independently of the relative size of the uncertainty.
6 Summary
We have presented a theoretical framework for treating inverse problems from a Bayesian perspective. In particular, the framework provides a more formal description of what it means when we talk about propagating experimental uncertainties into the space of models by focusing on the posterior probability measure in model space given the available data. Strictly speaking, there is no fitting required to obtain the expression for the posterior distributions in the space of the data or the model, instead these are obtained by marginalising the joint distribution. We note that sampling from the posterior distribution of the model is, in general, highly non-trivial; however we can show that at least for linear problems the NNPDF MC methodology produces a sample of models which are distributed according to the posterior model distribution predicted from the Bayesian formalism. Furthermore, we provide evidence that even for non-linear models this result at least holds as a good approximation close to the MAP estimator.
We then use this formal framework to think about some of the estimators which we use as part of the NNPDF closure test. In particular we derive bias and variance from decomposing an out of sample error function, which is understood from a classic fitting point of view. The estimators are then related back to the posterior distributions in the Bayesian framework. We note that the estimators themselves are not perfect and suffer from only testing the model uncertainties locally (in regions where the test data probes). Furthermore, the estimators only give an approximate overall picture, and cannot be used to diagnose where the problem arises if we find evidence that the model uncertainties are not faithful.
Given the framework set out here, future work should be undertaken to generalise the closure estimators to model space. This would likely involve a combination of the closure estimators presented here and the extension of the Bayesian framework to infinite-dimensional spaces.
We give some closure results, as a proof of principle. The results presented here are examined in more detail alongside the full NNPDF4.0 release [2] but serve as an example of how the data space estimators can be practically included even in a rather complex setting. The NNPDF4.0 methodology passes the closure test according to the estimators that we analysed, providing evidence that for unseen data the current NNPDF methodology appears to provide faithful uncertainties. The estimators are not limited to this specific application, and the results presented demonstrate how the data space estimators can be incorporated into any inverse problem. As previously mentioned, a more general set of estimators in model space would be the gold-standard and give us confidence in our uncertainties for future observables which probe regions which are not covered by either the training or testing data.
We stress that, if the ultimate goal of closure tests is to validate whether the uncertainty of a given methodology is faithful in the context of a global fit, this is not fully achieved by the estimators presented here: in the closure test framework described in this study, the prior on the data is fully consistent with the generation of the observable central values from the true observables and uncertainties by construction, which is often not the case in real world fits. Something which has to be investigated is how we can guarantee faithful uncertainties when this is no longer the case. For example one could consider a closure test where the generated data was inconsistent with the prior. The closure tests presented here should therefore be seen as the first necessary step in this direction, and future study will aim to complete the picture.
Some of the observed inconsistency between the prior and the data central values might be due to missing theoretical uncertainties, but once all sources of theoretical uncertainties have been accounted for there could still be tension in the data. The advantage of viewing inverse problems from a Bayesian perspective is access to methodologies which deal with inconsistent data (or unknown systematics) in a Bayesian framework, for example in these cosmological studies [20, 21]. These methods potentially offer a more formal approach to dealing with inconsistent data, rather than ad hoc procedures.
The inclusion of MHOUs in the likelihood was justified from a Bayesian perspective [5], but here we have drawn a connection between the model replicas and the posterior distribution in model space, which explains why theoretical uncertainties must be included in both the sampling of the pseudodata replicas and the likelihood. Therefore we emphasise that the framework set out here is not only useful for understanding model uncertainties with the current methodology, but also for motivating future methodological development from a Bayesian perspective. We defer more detailed investigations of the potential of this approach to future work.
Data Availability
This manuscript has no associated data or the data will not be deposited. [Authors’ comment: The results presented in this work do not come with a new data delivery, so no new data are made available.]
Notes
Banach spaces are complete normed vector spaces. We do not need to get into a more detailed discussion here, but it is important to note that working in Banach spaces allows us to generalise the results to infinite-dimensional spaces of functions.
In order to avoid overfitting, the NNPDF fitting procedure does not aim for the absolute minimum of the \(\chi ^2\) defined from the training sample. Instead a conventional split between training and validation is used. The details of this procedure are not relevant for the current discussion and will be neglected here.
References
J. Hadamard, Lectures on Cauchy’s Problem in Linear Partial Differential Equations (Yale University Press, New Haven, 1923)
R.D. Ball et al., The path to proton structure at one-percent accuracy. arXiv:2109.02653
A. Stuart, Inverse problems: a Bayesian perspective. Acta Numer. 19, 451–559 (2010)
NNPDF Collaboration, R. Abdul Khalek et al., A first determination of parton distributions with theoretical uncertainties. Eur. Phys. J. C 79, 838 (2019). arXiv:1905.04311
NNPDF Collaboration, R. Abdul Khalek et al., Parton distributions with theory uncertainties: general formalism and first phenomenological studies. Eur. Phys. J. C 79(11), 931 (2019). arXiv:1906.10698
G. Backus, F. Gilbert, The resolving power of gross earth data. Geophys. J. R. Astron. Soc. 16, 169–205 (1968)
M.S. Advani, A.M. Saxe, H. Sompolinsky, High-dimensional dynamics of generalization error in neural networks. Neural Netw. 132, 428–446 (2020). arXiv:1710.03667
NNPDF Collaboration, R.D. Ball et al., Parton distributions for the LHC Run II. JHEP 04, 040 (2015). arXiv:1410.8849
NNPDF Collaboration, R.D. Ball, E.R. Nocera, R.L. Pearson, Nuclear uncertainties in the determination of proton PDFs. Eur. Phys. J. C 79(3), 282 (2019). arXiv:1812.09074
R.D. Ball, E.R. Nocera, R.L. Pearson, Deuteron uncertainties in the determination of proton PDFs. Eur. Phys. J. C 81(1), 37 (2021). arXiv:2011.00009
R. Abdul Khalek, J.J. Ethier, J. Rojo, G. van Weelden, nNNPDF2.0: quark flavor separation in nuclei from LHC data. JHEP 09, 183 (2020). arXiv:2006.14629
P. Mehta, M. Bukov, C.-H. Wang, A.G. Day, C. Richardson, C.K. Fisher, D.J. Schwab, A high-bias, low-variance introduction to machine learning for physicists. Phys. Rep. 810, 1–124 (2019). arXiv:1803.08823
R.D. Ball, V. Bertone, S. Carrazza, L.D. Debbio, S. Forte, P. Groth-Merrild, A. Guffanti, N.P. Hartland, Z. Kassabov, J.I. Latorre et al., Parton distributions from high-precision collider data. Eur. Phys. J. C 77 (10), 663 (2017). https://doi.org/10.1140/epjc/s10052-017-5199-5. arXiv:1706.00428
R.D. Ball et al., NNPDF Collaboration, An open-source machine learning framework for global analyses of parton distributions. Eur. Phys. J. C 81(10), 958 (2021). https://doi.org/10.1140/epjc/s10052-021-09747-9. arXiv:2109.02671
J. Cruz-Martinez, S. Forte, E. Nocera, Future tests of parton distributions. Acta Phys. Pol. B 52(3), 243 (2021). arXiv:2103.08606
R.D. Ball, S. Carrazza, L.D. Debbio, S. Forte, Z. Kassabov, J. Rojo, E. Slade, M. Ubiali, Precision determination of the strong coupling constant within a global pdf analysis. Eur. Phys. J. C 78 (5), 408 (2018). https://doi.org/10.1140/epjc/s10052-018-5897-7. arXiv:1802.03398
F. Faura, S. Iranipour, E.R. Nocera, J. Rojo, M. Ubiali, The strangest proton? Eur. Phys. J. C 80 (12), 1168 (2020). https://doi.org/10.1140/epjc/s10052-020-08749-3arXiv:2009.00014
S. Carrazza, J. Cruz-Martinez, Towards a new generation of parton densities with deep learning models. Eur. Phys. J. C 79(8), 676 (2019). arXiv:1907.05075
B. Efron, R.J. Tibshirani, An Introduction to the Bootstrap (CRC Press, Boca Raton, 1994)
J. Luis Bernal, J.A. Peacock, Conservative cosmology: combining data with allowance for unknown systematics. J. Cosmol. Astropart. Physi. 2018, 002 (2018). arXiv:1803.04470
M.P. Hobson, S.L. Bridle, O. Lahav, Combining cosmological data sets: hyperparameters and Bayesian evidence. Mon. Not. R. Astron. Soc. 335, 377–388 (2002). arXiv:astro-ph/0203259
J. Dove, B. Kerns, R.E. McClellan, S. Miyasaka, D.H. Morton, K. Nagai, S. Prasad, F. Sanftl, M.B.C. Scott, A.S. Tadepalli et al., The asymmetry of antimatter in the proton. Nature 590, 561–565 (2021). arXiv:2103.04024
ATLAS Collaboration, M. Aaboud et al., Precision measurement and interpretation of inclusive \(W^+\) , \(W^-\) and \(Z/\gamma ^*\) production cross sections with the ATLAS detector. Eur. Phys. J. C 77(6), 367 (2017). arXiv:1612.03016
ATLAS Collaboration, M. Aaboud et al., Measurement of the Drell–Yan triple-differential cross section in \(pp\) collisions at \(\sqrt{s} = 8\) TeV. JHEP 12, 059 (2017). arXiv:1710.05167
ATLAS Collaboration, G. Aad et al., Measurement of the double-differential high-mass Drell–Yan cross section in pp collisions at \( \sqrt{s}=8 \) TeV with the ATLAS detector. JHEP 08, 009 (2016). arXiv:1606.01736
ATLAS Collaboration, G. Aad et al., Measurement of \(W^{\pm }\) and \(Z\)-boson production cross sections in \(pp\) collisions at \(\sqrt{s}=13\) TeV with the ATLAS detector. Phys. Lett. B 759, 601–621 (2016). arXiv:1603.09222
ATLAS Collaboration, M. Aaboud et al., Measurement of differential cross sections and \(W^+/W^-\) cross-section ratios for \(W\) boson production in association with jets at \(\sqrt{s}=8\) TeV with the ATLAS detector. JHEP 05, 077 (2018). arXiv:1711.03296. [Erratum: JHEP 10, 048 (2020)]
ATLAS Collaboration, G. Aad et al., Measurement of the \(t{\bar{t}}\) production cross-section in the lepton+jets channel at \(\sqrt{s}=13\) TeV with the ATLAS experiment. Phys. Lett. B 810, 135797 (2020). arXiv:2006.13076
ATLAS Collaboration, G. Aad et al., Measurements of top-quark pair differential cross-sections in the lepton+jets channel in \(pp\) collisions at \(\sqrt{s}=8\) TeV using the ATLAS detector. Eur. Phys. J. C 76(10), 538 (2016). arXiv:1511.04716
ATLAS Collaboration, M. Aaboud et al., Measurement of top quark pair differential cross-sections in the dilepton channel in \(pp\) collisions at \(\sqrt{s}\) = 7 and 8 TeV with ATLAS. Phys. Rev. D 94(9), 092003 (2016). arXiv:1607.07281. [Addendum: Phys. Rev. D 101, 119901 (2020)]
ATLAS Collaboration, M. Aaboud et al., Measurement of the inclusive jet cross-sections in proton–proton collisions at \( \sqrt{s}=8\) TeV with the ATLAS detector. JHEP 09, 020 (2017). arXiv:1706.03192
ATLAS Collaboration, G. Aad et al., Measurement of dijet cross sections in \(pp\) collisions at 7 TeV centre-of-mass energy using the ATLAS detector. JHEP 1405, 059 (2014). arXiv:1312.3524
M. Aaboud, G. Aad, B. Abbott, J. Abdallah, O. Abdinov, B. Abeloos, S. Abidi, O. AbouZeid, N. Abraham, H. Abramowicz et al., Measurement of the cross section for inclusive isolated-photon production in pp collisions at s=13 TeV using the Atlas detector. Phys. Lett. B 770, 473–493 (2017). arXiv:1701.06882
ATLAS Collaboration, G. Aad et al., Comprehensive measurements of \(t\)-channel single top-quark production cross sections at \(\sqrt{s} = 7\) TeV with the ATLAS detector. Phys. Rev. D 90(11), 112006 (2014). arXiv:1406.7844
ATLAS Collaboration, M. Aaboud et al., Measurement of the inclusive cross-sections of single top-quark and top-antiquark \(t\)-channel production in \(pp\) collisions at \(\sqrt{s}\) = 13 TeV with the ATLAS detector. JHEP 04, 086 (2017). arXiv:1609.03920
ATLAS Collaboration, M. Aaboud et al., Fiducial, total and differential cross-section measurements of \(t\)-channel single top-quark production in \(pp\) collisions at 8 TeV using data collected by the ATLAS detector. Eur. Phys. J. C 77(8), 531 (2017). arXiv:1702.02859
CMS Collaboration, S. Chatrchyan et al., Measurements of differential jet cross sections in proton–proton collisions at \(\sqrt{s}=7\) TeV with the CMS detector. Phys. Rev. D 87, 112002 (2013). arXiv:1212.6660
CMS Collaboration, A.M. Sirunyan et al., Measurement of the triple-differential dijet cross section in proton–proton collisions at \(\sqrt{s}=8\,\text{TeV} \) and constraints on parton distribution functions. Eur. Phys. J. C 77(11), 746 (2017). arXiv:1705.02628
CMS Collaboration, A.M. Sirunyan et al., Measurement of the inclusive \( {{\rm t}}\overline{{\rm t}}\) cross section in pp collisions at \( \sqrt{s}=5.02 \) TeV using final states with at least one charged lepton. JHEP 03, 115 (2018). arXiv:1711.03143
CMS Collaboration, A.M. Sirunyan et al., Measurement of double-differential cross sections for top quark pair production in pp collisions at \(\sqrt{s} = 8\) TeV and impact on parton distribution functions. Eur. Phys. J. C 77(7), 459 (2017). arXiv:1703.01630
CMS Collaboration, A.M. Sirunyan et al., Measurement of differential cross sections for the production of top quark pairs and of additional jets in lepton+jets events from pp collisions at \(\sqrt{s} = 13\) TeV. Phys. Rev. D 97(11), 112003 (2018). arXiv:1803.08856
CMS Collaboration, A.M. Sirunyan et al., Measurements of \({\rm t}{\overline{t}}\) differential cross sections in proton–proton collisions at \(\sqrt{s}= 13\) TeV using events containing two leptons. JHEP 02, 149 (2019). arXiv:1811.06625
CMS Collaboration, S. Chatrchyan et al., Measurement of the single-top-quark \(t\)-channel cross section in \(pp\) collisions at \(\sqrt{s}=7\) TeV. JHEP 12, 035 (2012). arXiv:1209.4533
CMS Collaboration, V. Khachatryan et al., Measurement of the t-channel single-top-quark production cross section and of the \(\mid V_{tb} \mid \) CKM matrix element in pp collisions at \(\sqrt{s}= 8\) TeV. JHEP 06, 090 (2014). arXiv:1403.7366
CMS Collaboration, A.M. Sirunyan et al., Cross section measurement of \(t\)-channel single top quark production in pp collisions at \(\sqrt{s} = 13\) TeV. Phys. Lett. B 772, 752–776 (2017). arXiv:1610.00678
LHCb Collaboration, R. Aaij et al., Measurement of the forward Z boson production cross-section in pp collisions at \(\sqrt{s} = 13\) TeV, JHEP 09, 136 (2016). arXiv:1607.06495
Acknowledgements
We thank the members of the NNPDF collaboration for discussions and comments on the present work. L. D. D. is supported by the U.K. Science and Technology Facility Council (STFC) grant ST/P000630/1, T. G. is supported by NWO via a ENW-KLEIN-2 project and M. W. by the STFC grant ST/R504737/1.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A: Gaussian integrals
Theory errors can be included in this framework by allowing the distribution of observables around the theory prediction to have a finite width, e.g. by replacing the Dirac delta
in Eq. 12 with a Gaussian
For the purposes of this study, we do not want to provide a realistic estimate of theory errors. Instead we will be assuming that the errors are uncorrelated and identical for all data points
and we will be interested in the limit where \(\sigma ^2\rightarrow 0\).
1.1 A.1 Integrating out the data
Marginalizing with respect to y in this case yields
The argument of the exponential in the integrand is a quadratic form in y,
The integral can be easily evaluated by completing the square,
Comparing Eqs. 141 and 142 at order \(y^2\) and y, yields
and therefore
Note that the four terms in the equation above are ordered in increasing powers of \(\sigma ^2\) and ultimately we will be interested in the limit \(\sigma ^2\rightarrow 0\), which reproduces the Dirac delta in \(\theta (y,u)\). Plugging Eq. 145 in Eq. 142 and again comparing to Eq. 141, we find
Expanding for small values of \(\sigma ^2\) the terms of order \(1/\sigma ^2\) cancel; keeping only finite terms in the limit \(\sigma ^2 \rightarrow 0\) we finally obtain
This is exactly the result that we obtained earlier when
It should not come as a surprise since in the limit where \(\sigma ^2 \rightarrow 0\) the Gaussian distribution that we chose to describe the fluctuations of the data around the theory predictions reduces indeed to a Dirac delta. The posterior for the model is exactly the one we computed in Sect. 2. We do not learn anything new from this exercise, but it is a useful warm-up for the next example. The integral over y can now be performed easily, since it is yet again a Gaussian integral.
1.2 A.2 Integrating out the model
Using the same approach as above, we now want to marginalise with respect to the model in order to obtain the posterior distribution of the data:
We follow exactly the same procedure outlined above, starting from the argument of the exponential
we complete the square and rewrite it in the form
It can be readily checked that in this case
In order to evaluate \(R_M\), we need
Noting that
we have, in the limit where \(\sigma ^2 \rightarrow 0\)
Collecting all terms we find
Performing the Gaussian integral over u in Eq. 148, we obtain the posterior distribution of the data
As in the case above, we note that this is a Gaussian distribution,
where the mean and the covariance are given in Eqs. 50 and 51. We can rewrite those expressions as
In order to simplify the notation we introduce
and then
Appendix B: Closure test setup details
1.1 B.1 Data
The full list of datasets included in the test set are shown in Table 2. The central values are not actually used in the closure test, however we use the experimental uncertainties in the calculation of both \(\mathcal {R}_{bv}\) and \(\xi ^{({\mathrm{data}})}_{1 {{\sigma }^{\prime }} }\). The corresponding predictions generated from the underlying law are used as the true observable values. Neither of the data-space closure estimators rely on the central values of the test datasets.
For completeness, in Fig. 9 the kinematic coverage of the training datasets, which as mentioned is the NNPDF3.1-like dataset used in [17], and the test datasets shown in Table 2 is plotted.
The kinematic coverage of the training and test data used to train the models and produce results presented in this paper. The kinematics of the two sets of data with this particular split overlaps but there are also kinematic regions which the test dataset probes, for which there are no training data
1.2 B.2 Models
A summary of the hyperparameters for the neural networks used during the closure fits is given in Table 3. These hyperparameters were chosen as part of an extensive hyperparameters scan, which will be explained in detail in NNPDF4.0, for this study we simply provide the values of the hyperparameters as a point of reference.
Appendix C: Understanding NNPDF3.0 data estimators
In the closure test presented in NNPDF3.0 [8] there was a data-space estimator which aimed to measure the level of over or under fitting, \(\varDelta _{\chi ^2}\). Here we discuss how \(\varDelta _{\chi ^2}\) can emerge from the bias-variance decomposition and then use the linear model to try and understand it in the context of viewing the ensemble of model replicas as a sample from the posterior distribution of the model given the data.
Despite the link between the estimators emerging from the decomposition of \(\mathcal {E}^{\mathrm{out}}\) and the posterior distribution for data which is not used to inform the model parameters, if we perform the same decomposition as in Sect. 4.1 but set \( {{y_0}^{\prime }} =y_0\) then we find that the cross term in the final line of Eq. 105 does not go to zero when the expectation across data is taken because there is a dependence on \(y_0\) in both the model predictions and the noisy data. As a result we have to modify Eq. 108 to be
where we refer now to the right hand side of Eq. 163 as \(\mathcal {E}^{\mathrm{in}}\) because it’s evaluated on the data used to inform the model replicas.
Now we examine the definition of \(\varDelta _{\chi ^2}\) introduced in [8], defined as the difference between the \(\chi ^2\) between the expectation value of the model predictions and the level one data, and the \(\chi ^2\) between the underlying observable values and the level one data. In [8] the denominator was also set to be the second term in the numerator, however here we slightly re-define \(\varDelta _{\chi ^2}\) to instead simply be normalised by the number of data points:
where in the second line we show how \(\varDelta _{\chi ^2}\) itself can be decomposed to be equal to two of the terms in Eq. 163.
Constant values of \(\varDelta _{\chi ^2}\) define elliptical contours in data space centered on the level one data. \(\varDelta _{\chi ^2}= 0\), in particular, defines a contour which is centered on the level one data and passes through the underlying law. When viewing \(\varDelta _{\chi ^2}\) from a classical fitting perspective, if \(\varDelta _{\chi ^2}< 0\) then the expectation value of the model predictions fit the level one data better than the underlying observables - which indicates an overfitting of the shift, \(\varvec{\eta }\). Similarly, \(\varDelta _{\chi ^2}> 0\) indicates some underfitting of the level one data.
If we return to the linear model we can write the analytic value of \(\varDelta _{\chi ^2}\). Firstly, since \( {{y_0}^{\prime }} =y_0\) we can simplify Eq. 127
because \(\tilde{C}_X\tilde{C}_X^{-1}\) is an \(N_{\mathrm{model}}\times N_{\mathrm{model}}\) identity matrix. Similarly we can write down the cross term
which leaves us with
The point is that the linear model has already been shown to be a sample from posterior distribution of the model given the data. But from the classical fitting point of view we would say this model has overfitted.
As such, we do not report any results with \(\varDelta _{\chi ^2}\) here, because when \(\mathcal {R}_{bv}= 1\), it doesn’t add much to the discussion. It may still be useful as a diagnostic tool when \(\mathcal {R}_{bv}\ne 1\), which as discussed could be for a variety of reasons - including fitting inefficiency. It also may be used as a performance indicator for deciding between two fitting methodologies: if both fits are shown to have \(\mathcal {R}_{bv}= 1\), the methodology with smaller magnitude of \(\varDelta _{\chi ^2}\) could be preferential. The same could be said for bias and variance however, bias in particular is clearly closely related to \(\varDelta _{\chi ^2}\).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Funded by SCOAP3
About this article
Cite this article
Del Debbio, L., Giani, T. & Wilson, M. Bayesian approach to inverse problems: an application to NNPDF closure testing. Eur. Phys. J. C 82, 330 (2022). https://doi.org/10.1140/epjc/s10052-022-10297-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1140/epjc/s10052-022-10297-x