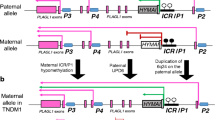
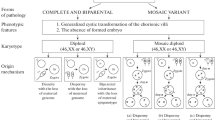
Abstract—
Epigenetic regulation is hereditary and non-hereditary changes in the expression of a particular gene without any corresponding structural changes in its nucleotide sequence. Genomic imprinting is an epigenetic mechanism for regulating the expression of homologous genes depending on parental origin, i.e., they are expressed monoallelically in the mammalian diploid cell. Being genetically imprinted, only the maternal or only the paternal genome is unable to ensure normal embryonic development. The most studied epigenetic modification, which plays one of the main roles in the maintenance of imprinting processes, is the specific methylation of cytosine in CpG-dinucleotides. All known imprinted genes contain differential DNA methylation regions on homologous parent chromosomes, which are necessary for their monoallelic expression. However, it is now known that not only DNA methylation, but chromatin remodeling, histone modifications, and non-coding RNAs also ensure the proper functioning of imprinted genes in the human body. Structural and functional disturbances of epigenetic mechanisms lead to imprinting diseases.
Similar content being viewed by others
REFERENCES
McGrath J., Solter D. 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 37, 179–183.
Surani M.A., Barton S.C., Norris M.L. 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 308, 548–550.
Sazhenova E.A., Lebedev I.N. 2021. Evolutionary aspects of genomic imprinting. Mol. Biol. (Moscow). 55 (1), 1–16.
Monk D., Mackay D.J.G., Eggermann T., Maher E.R., Riccio A. 2019. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 20, 235–248.
Singh P., Wu X., Lee D.-H., Li A.X., Rauch T.A., Pfeifer G.P., Mann J.R., Szabó P.E. 2011. Chromosome-wide analysis of parental allele-specific chromatin and DNA methylation. Mol. Cell. Biol. 31, 1757–1770.
Sanli I., Feil R. 2015. Chromatin mechanisms in the developmental control of imprinted gene expression. Int. J. Biochem. Cell. Biol. 67, 139–147.
Kota S.K., Llères D., Bouschet T., Hirasawa R., Marchand A., Begon-Pescia C., Sanli I., Arnaud P., Journot L., Girardot M., Feil R. 2014. ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1–Dio3 domain. Dev. Cell. 31, 19–33.
Kanduri C. 2016. Long noncoding RNAs: Lessons from genomic imprinting. Biochim. Biophys. Acta. 1859, 102–111.
Abi Habib W., Brioude F., Azzi S., Rossignol S., Linglart A., Sobrier M-L., Giabicani É., Steunou V., Harbison M.D., Le Bouc Y., Netchine I. 2019. Transcriptional profiling at the DLK1/MEG3 domain explains clinical overlap between imprinting disorders. Sci. Adv. 5, eaau9425.
MacDonald W.A., Mann M.R.W. 2020. Long noncoding RNA functionality in imprinted domain regulation. PLoS Genet. 16, e1008930.
Horsthemke B. 2014. In brief: genomic imprinting and imprinting diseases. J. Pathol. 232, 485–487.
Barlow D.P., Bartolomei M.S. 2014. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6 (2), a018382. https://doi.org/10.1101/cshperspect.a018382
Tucci V., Isles A.R., Kelsey G., Ferguson-Smith A.C., Erice Imprinting Group. 2019. Genomic imprinting and physiological processes in mammals. Cell. 176, 952–965.
Patten M.M., Cowley M., Oakey R.J., Feil R. 2016. Regulatory links between imprinted genes: Evolutionary predictions and consequences. Proc. Biol. Sci. 283 (1824), 20152760. https://doi.org/10.1098/rspb.2015.2760
Jadhav B., Monajemi R., Gagalova K.K., Ho D., Draisma H.H.M., van de Wiel M.A., Franke L., Heijmans B.T., van Meurs J., Jansen R., GoNL Consortium, BIOS Consortium, ’t Hoen PAC, Sharp A.J., Kiełbasa S.M. 2019. RNA-Seq in 296 phased trios provides a high-resolution map of genomic imprinting. BMC Biol. 17, 50.
Chaves T.F., Oliveira L.F., Ocampos M., Barbato I.T., de Luca G.R., Barbato Filho J.H., de Camargo Pinto L.L., Bernardi P., Maris A.F. 2019. Long contiguous stretches of homozygosity detected by chromosomal microarrays (CMA) in patients with neurodevelopmental disorders in the South of Brazil. BMC Med. Genomics. 12, 50.
Elbracht M., Mackay D., Begemann M., Kagan K.O., Eggermann T. 2020. Disturbed genomic imprinting and its relevance for human reproduction: Causes and clinical consequences. Hum. Reprod. Update. 26, 197–213.
Cerrato F., Sparago A., Ariani F., Brugnoletti F., Calzari L., Coppedè F., De Luca A., Gervasini C., Giardina E., Gurrieri F., Lo Nigro C., Merla G., Miozzo M., Russo S., Sangiorgi E., et al. 2020. DNA methylation in the diagnosis of monogenic diseases. Genes (Basel). 11 (4), 355. https://doi.org/10.3390/genes11040355
Temple I.K., Mackay D.J. 1993. Diabetes mellitus, 6q24-related transient neonatal. In: GeneReviews. Eds Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Mirzaa G., Amemiya A. Seattle (WA): Univ. of Washington, 1993–2021. https://www.ncbi. nlm.nih.gov/books/NBK1534/
Temple I.K., Gardner R.J., Robinson D.O., Kibirige M.S., Ferguson A.W., Baum J.D., Barber J.C., James R.S., Shield J.P. 1996. Further evidence for an imprinted gene for neonatal diabetes localised to chromosome 6q22–q23. Hum. Mol. Genet. 5, 1117–1121.
Gardner R.J., Mackay D.J., Mungall A.J., Polychronakos C., Siebert R., Shield J.P., Temple I.K., Robinson D.O. 2000. An imprinted locus associated with transient neonatal diabetes mellitus. Hum. Mol. Genet. 9, 589–596.
Su H.-C., Wu S.-C., Yen L.-C., Chiao L.-K., Wang J.-K., Chiu Y.-L., Ho C.-L., Huang S.-M. 2020. Gene expression profiling identifies the role of Zac1 in cervical cancer metastasis. Sci. Rep. 10, 11837.
Hoffmann A., Spengler D. 2015. Role of ZAC1 in transient neonatal diabetes mellitus and glucose metabolism. World J. Biol. Chem. 6, 95–109.
Iglesias-Platas I., Court F., Camprubi C., Sparago A., Guillaumet-Adkins A., Martin-Trujillo A., Riccio A., Moore G.E., Monk D. 2013. Imprinting at the PLAGL1 domain is contained within a 70-kb CTCF/cohesin-mediated non-allelic chromatin loop. Nucleic Acids Res. 41, 2171–2179.
Varrault A., Dantec C., Le Digarcher A., Chotard L., Bilanges B., Parrinello H., Dubois E., Rialle S., Severac D., Bouschet T., Journot L. 2017. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res. 45, 10466–10480.
Mackay D.J.G., Coupe A.-M., Shield J.P.H., Storr J.N.P., Temple I.K., Robinson D.O. 2002. Relaxation of imprinted expression of ZAC and HYMAI in a patient with transient neonatal diabetes mellitus. Hum. Genet. 110, 139–144.
Touati A., Errea-Dorronsoro J., Nouri S., Halleb Y., Pereda A., Mahdhaoui N., Ghith A., Saad A., Perez de Nanclares G., H′mida Ben Brahim D. 2019. Transient neonatal diabetes mellitus and hypomethylation at additional imprinted loci: Novel ZFP57 mutation and review on the literature. Acta Diabetol. 56, 301–307.
Kerr E.R., Stuhlmiller G.M., Maha G.C., Ladd M.A., Mikhail F.M., Koester R.P., Hurst A.C.E. 2018. Maternal uniparental isodisomy for chromosome 6 discovered by paternity testing: a case report. Mol. Cytogenet. 11, 60.
Court F., Camprubi C., Garcia C.V., Guillaumet-Adkins A., Sparago A., Seruggia D., Sandoval J., Esteller M., Martin-Trujillo A., Riccio A., Montoliu L., Monk D. 2014. The PEG13-DMR and brain-specific enhancers dictate imprinted expression within the 8q24 intellectual disability risk locus. Epigenetics Chromatin. 7, 5.
Ruf N., Bähring S., Galetzka D., Pliushch G., Luft F.C., Nürnberg P., Haaf T., Kelsey G., Zechner U. 2007. Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum. Mol. Genet. 16, 2591–2599.
Liang Z.S., Cimino I., Yalcin B., Raghupathy N., Vancollie V.E., Ibarra-Soria X., Firth H.V., Rimmington D., Farooqi I.S., Lelliott C.J., Munger S.C., O’Rahilly S., Ferguson-Smith A.C., Coll A.P., Logan D.W. 2020. Trappc9 deficiency causes parent-of-origin dependent microcephaly and obesity. PLoS Genet. 16, e1008916.
Zadeh N., Graham J.M. 1993. KCNK9 imprinting syndrome. In: GeneReviews. Eds. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Mirzaa G., Amemiya A. Seattle (WA): Univ. of Washington, 1993–2021. https://www.ncbi.nlm.nih.gov/ books/NBK425128/
Graham J.M., Zadeh N., Kelley M., Tan E.S., Liew W., Tan V., Deardorff M.A., Wilson G.N., Sagi-Dain L., Shalev S.A. 2016. KCNK9 imprinting syndrome-further delineation of a possible treatable disorder. Am. J. Med. Genet. A. 170, 2632–2637.
Šedivá M., Laššuthová P., Zámečník J., Sedláčková L., Seeman P., Haberlová J. 2020. Novel variant in the KCNK9 gene in a girl with Birk Barel syndrome. Eur. J. Med. Genet. 63, 103619.
Kashevarova A.A., Nikitina T.V., Mikhailik L.I., Belyaeva E.O., Vasilyev S.A., Lopatkina M.E., Fedotov D.A., Fonova E.A., Zarubin A.A., Sivtsev A.A., Skryabin N.A., Nazarenko L.P., Lebedev I.N. 2020. 46,XY,r(8)/45,XY,-8 mosaicism as a possible mechanism of the imprinted Birk–Barel syndrome: A case study. Genes (Basel). 11(12), 1473. https://doi.org/10.3390/genes11121473
Besson A., Dowdy S.F., Roberts J.M. 2008. CDK inhibitors: Cell cycle regulators and beyond. Dev. Cell. 14, 159–169.
Creff J., Besson A. 2020. Functional versatility of the CDK inhibitor p57Kip2. Front. Cell. Dev. Biol. 8, 584590.
Neyroud N., Richard P., Vignier N., Donger C., Denjoy I., Demay L., Shkolnikova M., Pesce R., Chevalier P., Hainque B., Coumel P., Schwartz K., Guicheney P. 1999. Genomic organization of the KCNQ1 K+ channel gene and identification of C-terminal mutations in the long-QT syndrome. Circ. Res. 84, 290–297.
Mitsuya K., Meguro M., Lee M.P., Katoh M., Schulz T.C., Kugoh H., Yoshida M.A., Niikawa N., Feinberg A.P., Oshimura M. 1999. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum. Mol. Genet. 8, 1209–1217.
Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J., Nagano T., Mancini-Dinardo D., Kanduri C. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 32, 232–246.
Kanduri C. 2011. Kcnq1ot1: A chromatin regulatory RNA. Semin. Cell Dev. Biol. 22, 343–350.
Monk D., Sanches R., Arnaud P., Apostolidou S., Hills F.A., Abu-Amero S., Murrell A., Friess H., Reik W., Stanier P., Constância M., Moore G.E. 2006. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum. Mol. Genet. 15, 1259–1269.
Ghafouri-Fard S., Esmaeili M., Taheri M. 2020. H19 lncRNA: Roles in tumorigenesis. Biomed. Pharmacother. 123, 109774.
Jinno Y., Ikeda Y., Yun K., Maw M., Masuzaki H., Fukuda H., Inuzuka K., Fujishita A., Ohtani Y., Okimoto T. 1995. Establishment of functional imprinting of the H19 gene in human developing placentae. Nat. Genet. 10, 318–324.
Higashimoto K., Jozaki K., Kosho T., Matsubara K., Fuke T., Yamada D., Yatsuki H., Maeda T., Ohtsuka Y., Nishioka K., Joh K., Koseki H., Ogata T., Soejima H. 2014. A novel de novo point mutation of the OCT-binding site in the IGF2/H19-imprinting control region in a Beckwith–Wiedemann syndrome patient. Clin. Genet. 86, 539–544.
Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 405, 486–489.
Nativio R., Sparago A., Ito Y., Weksberg R., Riccio A., Murrell A. 2011. Disruption of genomic neighbourhood at the imprinted IGF2-H19 locus in Beckwith–Wiedemann syndrome and Silver–Russell syndrome. Hum. Mol. Genet. 20, 1363–1374.
Lopes S., Lewis A., Hajkova P., Dean W., Oswald J., Forné T., Murrell A., Constância M., Bartolomei M., Walter J., Reik W. 2003. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 12, 295–305.
Lee M.P., DeBaun M.R., Mitsuya K., Galonek H.L., Brandenburg S., Oshimura M., Feinberg A.P. 1999. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith–Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. U. S. A. 96, 5203–5208.
Mancini-Dinardo D., Steele S.J.S., Levorse J.M., Ingram R.S., Tilghman S.M. 2006. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 20, 1268–1282.
Ferrero G.B., Boonen S.E., Cole T., Baker R., Bertoletti M., Cocchi G., Coze C., De Pellegrin M., Hussain K., Ibrahim A., et al. 2018. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith–Wiedemann syndrome: An international consensus statement. Nat. Rev. Endocrinol. 14, 229–249.
Wang K.H., Kupa J., Duffy K.A., Kalish J.M. 2019. Diagnosis and management of Beckwith–Wiedemann syndrome. Front. Pediatr. 7, 562.
Eggermann T., Brück J., Knopp C., Fekete G., Kratz C., Tasic V., Kurth I., Elbracht M., Eggermann K., Begemann M. 2020. Need for a precise molecular diagnosis in Beckwith–Wiedemann and Silver–Russell syndrome: What has to be considered and why it is important. J. Mol. Med. (Berl.). 98, 1447–1455.
Papulino C., Chianese U., Nicoletti M.M., Benedetti R., Altucci L. 2020. Preclinical and clinical epigenetic-based reconsideration of Beckwith–Wiedemann syndrome. Front. Genet. 11, 563718.
Fontana L., Bedeschi M.F., Maitz S., Cereda A., Faré C., Motta S., Seresini A., D′Ursi P., Orro A., Pecile V., Calvello M., Selicorni A., Lalatta F., Milani D., Sirchia S.M., et al. 2018. Characterization of multi-locus imprinting disturbances and underlying genetic defects in patients with chromosome 11p15.5 related imprinting disorders. Epigenetics. 13, 897–909.
Nemtsova M.V., Strel’nikov V.V., Babenko S.V., Zemlyakova V.V., Zaletaev D.V. 2005. Molecular diagnosis of epigenetic disorders in Beckwith–Wiedemann syndrome. Med. Genet. 4, 33–38.
Chang S., Bartolomei M.S. 2020. Modeling human epigenetic disorders in mice: Beckwith–Wiedemann syndrome and Silver–Russell syndrome. Dis. Model. Mech. 13 (5), dmm044123. https://doi.org/10.1242/dmm.044123
Yamaguchi Y., Tayama C., Tomikawa J., Akaishi R., Kamura H., Matsuoka K., Wake N., Minakami H., Kato K., Yamada T., Nakabayashi K., Hata K. 2019. Placenta-specific epimutation at H19-DMR among common pregnancy complications: Its frequency and effect on the expression patterns of H19 and IGF2. Clin. Epigenetics. 11, 113.
Brioude F., Netchine I., Praz F., Le Jule M., Calmel C., Lacombe D., Edery P., Catala M., Odent S., Isidor B., Lyonnet S., Sigaudy S., Leheup B., Audebert-Bellanger S., Burglen L., et al. 2015. Mutations of the imprinted CDKN1C gene as a cause of the overgrowth Beckwith–Wiedemann syndrome: Clinical spectrum and functional characterization. Hum. Mutat. 36, 894–902.
Eggermann T., Begemann M., Pfeiffer L. 2021. Unusual deletion of the maternal 11p15 allele in Beckwith–Wiedemann syndrome with an impact on both imprinting domains. Clin. Epigenetics. 13, 30.
Sun F.L., Dean W.L., Kelsey G., Allen N.D., Reik W. 1997. Transactivation of Igf2 in a mouse model of Beckwith–Wiedemann syndrome. Nature. 389, 809–815.
Wakeling E.L., Brioude F., Lokulo-Sodipe O., O’Connell S.M., Salem J., Bliek J., Canton A.P.M., Chrzanowska K.H., Davies J.H., Dias R.P., Dubern B., Elbracht M., Giabicani E., Grimberg A., Grønskov K., et al. 2017. Diagnosis and management of Silver–Russell syndrome: First international consensus statement. Nat. Rev. Endocrinol. 13, 105–124.
Lokulo-Sodipe O., Ballard L., Child J., Inskip H.M., Byrne C.D., Ishida M., Moore G.E., Wakeling E.L., Fenwick A., Mackay D.J.G., Davies J.H., Temple I.K. 2020. Phenotype of genetically confirmed Silver–Russell syndrome beyond childhood. J. Med. Genet. 57, 683–691.
Tümer Z., López-Hernández J.A., Netchine I., Elbracht M., Grønskov K., Gede L.B., Sachwitz J., den Dunnen J.T., Eggermann T. 2018. Structural and sequence variants in patients with Silver–Russell syndrome or similar features – curation of a disease database. Hum. Mutat. 39, 345–364.
Forbes B.E., Blyth A.J., Wit J.M. 2020. Disorders of IGFs and IGF-1R signaling pathways. Mol. Cell. Endocrinol. 518, 111035.
Dörr S., Midro A.T., Färber C., Giannakudis J., Hansmann I. 2001. Construction of a detailed physical and transcript map of the candidate region for Russell–Silver syndrome on chromosome 17q23–q24. Genomics. 71, 174–181.
Chiesa N., De Crescenzo A., Mishra K., Perone L., Carella M., Palumbo O., Mussa A., Sparago A., Cerrato F., Russo S., Lapi E., Cubellis M.V., Kanduri C., Cirillo Silengo M., Riccio A., Ferrero G.B. 2012. The KCNQ1OT1 imprinting control region and non-coding RNA: New properties derived from the study of Beckwith–Wiedemann syndrome and Silver–Russell syndrome cases. Hum. Mol. Genet. 21, 10–25.
Cytrynbaum C., Chong K., Hannig V., Choufani S., Shuman C., Steele L., Morgan T., Scherer S.W., Stavropoulos D.J., Basran R.K., Weksberg R. 2016. Genomic imbalance in the centromeric 11p15 imprinting center in three families: Further evidence of a role for IC2 as a cause of Russell–Silver syndrome. Am. J. Med. Genet A. 170, 2731–2739.
Gicquel C., Rossignol S., Cabrol S., Houang M., Steunou V., Barbu V., Danton F., Thibaud N., Le Merrer M., Burglen L., Bertrand A.-M., Netchine I., Le Bouc Y. 2005. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver–Russell syndrome. Nat. Genet. 37, 1003–1007.
Inoue T., Nakamura A., Iwahashi-Odano M., Tanase-Nakao K., Matsubara K., Nishioka J., Maruo Y., Hasegawa Y., Suzumura H., Sato S., Kobayashi Y., Murakami N., Nakabayashi K., Yamazawa K., Fuke T., et al. 2020. Contribution of gene mutations to Silver–Russell syndrome phenotype: Multigene sequencing analysis in 92 etiology-unknown patients. Clin. Epigenet. 12, 86.
Saal H.M., Harbison M.D., Netchine I. 1993. Silver–Russell syndrome. In: GeneReviews. Eds Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J., Mirzaa G., Amemiya A. Seattle (WA): Univ. of Washington, 1993–2021. https://www.ncbi.nlm.nih.gov/ books/NBK1324/
Hannula-Jouppi K., Muurinen M., Lipsanen-Nyman M., Reinius L.E., Ezer S., Greco D., Kere J. 2014. Differentially methylated regions in maternal and paternal uniparental disomy for chromosome 7. Epigenetics. 9, 351–365.
Hitchins M.P., Monk D., Bell G.M., Ali Z., Preece M.A., Stanier P., Moore G.E. 2001. Maternal repression of the human GRB10 gene in the developing central nervous system; evaluation of the role for GRB10 in Silver–Russell syndrome. Eur. J. Hum. Genet. 9, 82–90.
Schöherr N., Jäger S., Ranke M.B., Wollmann H.A., Binder G., Eggermann T. 2008. No evidence for isolated imprinting mutations in the PEG1/MEST locus in Silver–Russell patients. Eur. J. Med. Genet. 51, 322–324.
Su J., Wang J., Fan X., Fu C., Zhang S., Zhang Y., Qin Z., Li H., Luo J., Li C., Jiang T., Shen Y. 2017. Mosaic UPD(7q)mat in a patient with Silver–Russell syndrome. Mol. Cytogenet. 10, 36.
Brioude F., Oliver-Petit I., Blaise A., Praz F., Rossignol S., Jule M.L., Thibaud N., Faussat A.-M., Tauber M., Bouc Y.L., Netchine I. 2013. CDKN1C mutation affecting the PCNA-binding domain as a cause of familial Russell–Silver syndrome. J. Med. Genet. 50, 823–830.
Sabir A.H., Ryan G., Mohammed Z., Kirk J., Kiely N., Thyagarajan M., Cole T. 2019. Familial Russell–Silver syndrome like phenotype in the PCNA domain of the CDKN1C gene, a further case. Case Rep. Genet. 2019, 1398250. https://doi.org/10.1155/2019/1398250
Binder G., Ziegler J., Schweizer R., Habhab W., Haack T.B., Heinrich T., Eggermann T. 2020. Novel mutation points to a hot spot in CDKN1C causing Silver–Russell syndrome. Clin. Epigenetics. 12, 152.
Rockstroh D., Pfäffle H., Le Duc D., Rößler F., Schlensog-Schuster F., Heiker J.T., Kratzsch J., Kiess W., Lemke J.R., Abou Jamra R., Pfäffle R. 2019. A new p.(Ile66Serfs*93) IGF2 variant is associated with pre- and postnatal growth retardation. Eur. J. Endocrinol. 180, K1–13.
Masunaga Y., Inoue T., Yamoto K., Fujisawa Y., Sato Y., Kawashima-Sonoyama Y., Morisada N., Iijima K., Ohata Y., Namba N., Suzumura H., Kuribayashi R., Yamaguchi Y., Yoshihashi H., Fukami M., et al. 2020. IGF2 mutations. J. Clin. Endocrinol. Metabolism. 105, 116–125.
Hübner C.T., Meyer R., Kenawy A., Ambrozaityte L., Matuleviciene A., Kraft F., Begemann M., Elbracht M., Eggermann T. 2020. HMGA2 variants in Silver–Russell syndrome: Homozygous and heterozygous occurrence. J. Clin. Endocrinol. Metab. 105, 2401–2407.
Vado Y., Pereda A., Llano-Rivas I., Gorria-Redondo N., Díez I., Perez de Nanclares G. 2020. Novel variant in PLAG1 in a familial case with Silver–Russell syndrome suspicion. Genes. 11, 1461.
Akhtar M., Holmgren C., Göndör A., Vesterlund M., Kanduri C., Larsson C., Ekström T.J. 2012. Cell type and context-specific function of PLAG1 for IGF2 P3 promoter activity. Int. J. Oncol. 41, 1959–1966.
Hara-Isono K., Matsubara K., Fuke T., Yamazawa K., Satou K., Murakami N., Saitoh S., Nakabayashi K., Hata K., Ogata T., Fukami M., Kagami M. 2020. Genome-wide methylation analysis in Silver–Russell syndrome, Temple syndrome, and Prader–Willi syndrome. Clin. Epigenetics. 12, 159.
Vilain E., Le Merrer M., Lecointre C., Desangles F., Kay M.A., Maroteaux P., McCabe E.R. 1999. IMAGe, a new clinical association of intrauterine growth retardation, metaphyseal dysplasia, adrenal hypoplasia congenita, and genital anomalies. J. Clin. Endocrinol. Metab. 84, 4335–4340.
Borges K.S., Arboleda V.A., Vilain E. 2015. Mutations in the PCNA-binding site of CDKN1C inhibit cell proliferation by impairing the entry into S phase. Cell Div. 10, 2.
Suntharalingham J.P., Ishida M., Buonocore F., Del Valle I., Solanky N., Demetriou C., Regan L., Moore G.E., Achermann J.C. 2019. Analysis of CDKN1C in fetal growth restriction and pregnancy loss. F1000Res. 8, 90.
Eggermann T., Binder G., Brioude F., Maher E.R., Lapunzina P., Cubellis M.V., Bergadá I., Prawitt D., Begemann M. 2014. CDKN1C mutations: Two sides of the same coin. Trends Mol Med. 20, 614–622.
Babenko O.V., Zemlyakova V.V., Saakyan S.V., Brovkina A.F., Strelnikov V.V., Zaletaev D.V., Nemtsova M.V. 2002. RB1 and CDKN2A functional defects resulting in retinoblastoma. Mol. Biol. (Moscow) 36 (5), 625–630.
Gelli E., Pinto A.M., Somma S., Imperatore V., Cannone M.G., Hadjistilianou T., De Francesco S., Galimberti D., Currò A., Bruttini M., Mari F., Renieri A., Ariani F. 2019. Evidence of predisposing epimutation in retinoblastoma. Hum. Mutat. 40, 201–206.
Kanber D., Berulava T., Ammerpohl O., Mitter D., Richter J., Siebert R., Horsthemke B., Lohmann D., Buiting K. 2009. The human retinoblastoma gene is imprinted. PLoS Genet. 5 (12), e1000790. https://doi.org/10.1371/journal.pgen.1000790
Buiting K., Kanber D., Horsthemke B., Lohmann D. 2010. Imprinting of RB1 (the new kid on the block). Brief Funct. Genomics. 9, 347–353.
Taylor M., Dehainault C., Desjardins L., Doz F., Levy C., Sastre X., Couturier J., Stoppa-Lyonnet D., Houdayer C., Gauthier-Villars M. 2007. Genotype–phenotype correlations in hereditary familial retinoblastoma. Hum. Mutat. 28, 284–293.
Eloy P., Dehainault C., Sefta M., Aerts I., Doz F., Cassoux N., Lumbroso le Rouic L., Stoppa-Lyonnet D., Radvanyi F., Millot G.A., Gauthier-Villars M., Houdayer C. 2016. A parent-of-origin effect impacts the phenotype in low penetrance retinoblastoma families segregating the c.1981C>T/p.Arg661Trp mutation of RB1. PLoS Genet. 12, e1005888.
Alekseeva E.A., Babenko O.V., Kozlova V.M., Ushakova T.L., Kazubskaya T.P., Saakyan S.V., Tanas A.S., Zaletaev D.V., Strelnikov V.V. 2019. The effect of parental origin of RB1 mutations in hereditary retinoblastoma with low penetrance. Med. Genet. 18 (8), 21–28.
Kagami M., Sekita Y., Nishimura G., Irie M., Kato F., Okada M., Yamamori S., Kishimoto H., Nakayama M., Tanaka Y., Matsuoka K., Takahashi T., Noguchi M., Tanaka Y., Masumoto K., et al. 2008. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 40, 237–242.
Falix F.A., Aronson D.C., Lamers W.H., Gaemers I.C. 2012. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim. Biophys. Acta – Mol. Basis Disease. 1822, 988–995.
Gomes L.G., Cunha-Silva M., Crespo R.P., Ramos C.O., Montenegro L.R., Canton A., Lees M., Spoudeas H., Dauber A., Macedo D.B., Bessa D.S., Maciel G.A., Baracat E.C., Jorge A.A.L., Mendonca B.B., et al. 2019. DLK1 is a novel link between reproduction and metabolism. J. Clin. Endocrinol. Metab. 104, 2112–2120.
Kitazawa M., Sutani A., Kaneko-Ishino T., Ishino F. 2021. The role of eutherian-specific RTL1 in the nervous system and its implications for the Kagami–Ogata and Temple syndromes. Genes Cells. 26, 165–179.
Martinez M.E., Cox D.F., Youth B.P., Hernandez A. 2016. Genomic imprinting of DIO3, a candidate gene for the syndrome associated with human uniparental disomy of chromosome 14. Eur. J. Hum. Genet. 24, 1617–1621.
Hamilton S., de Cabo R., Bernier M. 2020. Maternally expressed gene 3 in metabolic programming. Biochim. Biophys. Acta – Gene Regul. Mech. 1863, 194396.
Kagami M., O′Sullivan M.J., Green A.J., Watabe Y., Arisaka O., Masawa N., Matsuoka K., Fukami M., Matsubara K., Kato F., Ferguson-Smith A.C., Ogata T. 2010. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: Hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 6, e1000992.
da Rocha S.T., Edwards C.A., Ito M., Ogata T., Ferguson-Smith A.C. 2008. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306–316.
Ioannides Y., Lokulo-Sodipe K., Mackay D.J.G., Davies J.H., Temple I.K. 2014. Temple syndrome: Improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: An analysis of 51 published cases. J. Med. Genet. 51, 495–501.
Kagami M., Nagasaki K., Kosaki R., Horikawa R., Naiki Y., Saitoh S., Tajima T., Yorifuji T., Numakura C., Mizuno S., Nakamura A., Matsubara K., Fukami M., Ogata T. 2017. Temple syndrome: Comprehensive molecular and clinical findings in 32 Japanese patients. Genet. Med. 19, 1356–1366.
Ogata T., Kagami M. 2016. Kagami–Ogata syndrome: A clinically recognizable upd(14)pat and related disorder affecting the chromosome 14q32.2 imprinted region. J. Hum. Genet. 61, 87–94.
Soellner L., Begemann M., Mackay D.J.G., Grønskov K., Tümer Z., Maher E.R., Temple I.K., Monk D., Riccio A., Linglart A., Netchine I., Eggermann T. 2017. Recent advances in imprinting disorders. Clin. Genet. 91, 3–13.
van der Werf I.M., Buiting K., Czeschik C., Reyniers E., Vandeweyer G., Vanhaesebrouck P., Lüdecke H.-J., Wieczorek D., Horsthemke B., Mortier G., Leroy J.G., Kooy R.F. 2016. Novel microdeletions on chromosome 14q32.2 suggest a potential role for non-coding RNAs in Kagami–Ogata syndrome. Eur. J. Hum. Genet. 24, 1724–1729.
Stelzer Y., Sagi I., Yanuka O., Eiges R., Benvenisty N. 2014. The noncoding RNA IPW regulates the imprinted DLK1–DIO3 locus in an induced pluripotent stem cell model of Prader–Willi syndrome. Nat. Genet. 46, 551–557.
Cavaillé J. 2017. Box C/D small nucleolar RNA genes and the Prader–Willi syndrome: A complex interplay. Wiley Interdisc. Rev. RNA. 8 (4). https://doi.org/10.1002/wrna.1417
Wang T.-S., Tsai W.-H., Tsai L.-P., Wong S.-B. 2020. Clinical characteristics and epilepsy in genomic imprinting disorders: Angelman syndrome and Prader–-Willi syndrome. Ci Ji Yi Xue Za Zhi. 32, 137–144.
Mendiola A.J.P., LaSalle J.M. 2021. Epigenetics in Prader–Willi syndrome. Front. Genet. 12, 624581.
Christian S. 1999. Large genomic duplicons map to sites of instability in the Prader–Willi/Angelman syndrome chromosome region (15q11–q13). Hum. Mol. Genet. 8, 1025–1037.
Nicholls R.D., Knepper J.L. 2001. Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2, 153–175.
Kim S.-J., Miller J.L., Kuipers P.J., German J.R., Beaudet A.L., Sahoo T., Driscoll D.J. 2012. Unique and atypical deletions in Prader–Willi syndrome reveal distinct phenotypes. Eur. J. Hum. Genet. 20, 283–290.
Chung M.S., Langouët M., Chamberlain S.J., Carmichael G.G. 2020. Prader–Willi syndrome: Reflections on seminal studies and future therapies. Open Biol. 10, 200195.
Butler M.G. 2020. Imprinting disorders in humans: A review. Curr. Opin. Pediatr. 32, 719–729.
Cheon C.K. 2016. Genetics of Prader–Willi syndrome and Prader–Will-like syndrome. Ann. Pediatr. Endocrinol. Metab. 21, 126–135.
Runte M., Hüttenhofer A., Gross S., Kiefmann M., Horsthemke B., Buiting K. 2001. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 10, 2687–2700.
Galiveti C.R., Raabe C.A., Konthur Z., Rozhdestvensky T.S. 2014. Differential regulation of non-protein coding RNAs from Prader–Willi syndrome locus. Sci. Rep. 4, 6445.
Maina E.N., Webb T., Soni S., Whittington J., Boer H., Clarke D., Holland A. 2007. Analysis of candidate imprinted genes in PWS subjects with atypical genetics: A possible inactivating mutation in the SNURF/SNRPN minimal promoter. J. Hum. Genet. 52, 297–307.
Green Finberg Y., Kantor B., Hershko A.Y., Razin A. 2003. Characterization of the human SNRPN minimal promoter and cis elements within it. Gene. 304, 201–206.
Cassidy S.B., Schwartz S., Miller J.L., Driscoll D.J. 2012. Prader–Willi syndrome. Genet. Med. 14, 10–26.
Geuns E., De Rycke M., Van Steirteghem A., Liebaers I. 2003. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum. Mol. Genet. 12, 2873–2879.
Meng L., Person R.E., Huang W., Zhu P.J., Costa-Mattioli M., Beaudet A.L. 2013. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 9, e1004039.
Smith E.Y., Futtner C.R., Chamberlain S.J., Johnstone K.A., Resnick J.L. 2011. Transcription Is required to establish maternal imprinting at the Prader–Willi syndrome and Angelman syndrome locus. PLoS Genet. 7 (12), e1002422. https://doi.org/10.1371/journal.pgen.1002422
Buiting K., Lich C., Cottrell S., Barnicoat A., Horsthemke B. 1999. A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum. Genet. 105, 665–666.
Lewis M.W., Brant J.O., Kramer J.M., Moss J.I., Yang T.P., Hansen P.J., Williams R.S., Resnick J.L. 2015. Angelman syndrome imprinting center encodes a transcriptional promoter. Proc. Natl. Acad. Sci. U. S. A. 112, 6871–6875.
Lewis M.W., Vargas-Franco D., Morse D.A., Resnick J.L. 2019. A mouse model of Angelman syndrome imprinting defects. Hum. Mol. Genet. 28, 220–229.
Wu M.-Y., Tsai T.-F., Beaudet A.L. 2006. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 20, 2859–2870.
Buiting K., Gross S., Lich C., Gillessen-Kaesbach G., el-Maarri O., Horsthemke B. 2003. Epimutations in Prader–Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am. J. Hum. Genet. 72, 571–577.
Ohta T., Gray T.A., Rogan P.K., Buiting K., Gabriel J.M., Saitoh S., Muralidhar B., Bilienska B., Krajewska-Walasek M., Driscoll D.J., Horsthemke B., Butler M.G., Nicholls R.D. 1999. Imprinting-mutation mechanisms in Prader–Willi syndrome. Am. J. Hum. Genet. 64, 397–413.
Saitoh S., Buiting K., Cassidy S.B., Conroy J.M., Driscoll D.J., Gabriel J.M., Gillessen-Kaesbach G., Glenn C.C., Greenswag L.R., Horsthemke B., Kondo I., Kuwajima K., Niikawa N., Rogan P.K., Schwartz S., et al. 1997. Clinical spectrum and molecular diagnosis of Angelman and Prader–Willi syndrome patients with an imprinting mutation. Am. J. Med. Genet. 68, 195–206.
Bielinska B., Blaydes S.M., Buiting K., Yang T., Krajewska-Walasek M., Horsthemke B., Brannan C.I. 2000. De novo deletions of SNRPN exon 1 in early human and mouse embryos result in a paternal to maternal imprint switch. Nat. Genet. 25, 74–78.
Gray T.A., Saitoh S., Nicholls R.D. 1999. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc. Natl. Acad. Sci. U. S. A. 96, 5616–5621.
Glenn C.C., Saitoh S., Jong M.T., Filbrandt M.M., Surti U., Driscoll D.J., Nicholls R.D. 1996. Gene structure, DNA methylation, and imprinted expression of the human SNRPN gene. Am. J. Hum. Genet. 58, 335–346.
de los Santos T., Schweizer J., Rees C.A., Francke U. 2000. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader–Willi deletion region, which Is highly expressed in brain. Am. J. Hum. Genet. 67, 1067–1082.
Gallagher R.C., Pils B., Albalwi M., Francke U. 2002. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader–Willi syndrome. Am. J. Hum. Genet. 71, 669–678.
Bortolin-Cavaille M.-L., Cavaille J. 2012. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader–Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Res. 40, 6800–6807.
Bratkovič T., Božič J., Rogelj B. 2020. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 48, 1627–1651.
Raabe C.A., Voss R., Kummerfeld D-M., Brosius J., Galiveti C.R., Wolters A., Seggewiss J., Huge A., Skryabin B.V., Rozhdestvensky T.S. 2019. Ectopic expression of Snord115 in choroid plexus interferes with editing but not splicing of 5-Ht2c receptor pre-mRNA in mice. Sci. Rep. 9, 4300.
Leung K.N., Vallero R.O., DuBose A.J., Resnick J.L., LaSalle J.M. 2009. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum. Mol. Genet. 18, 4227–4238.
Bieth E., Eddiry S., Gaston V., Lorenzini F., Buffet A., Conte Auriol F., Molinas C., Cailley D., Rooryck C., Arveiler B., Cavaillé J., Salles J.P., Tauber M. 2015. Highly restricted deletion of the SNORD116 region is implicated in Prader–Willi syndrome. Eur. J. Hum Genet. 23, 252–255.
Powell W.T., Coulson R.L., Crary F.K., Wong S.S., Ach R.A., Tsang P., Alice Yamada N., Yasui D.H., Lasalle J.M. 2013. A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet. 22, 4318–4328.
Coulson R.L., Yasui D.H., Dunaway K.W., Laufer B.I., Vogel Ciernia A., Zhu Y., Mordaunt C.E., Totah T.S., LaSalle J.M. 2018. Snord116-dependent diurnal rhythm of DNA methylation in mouse cortex. Nat. Commun. 9, 1616.
Wu H., Yin Q.-F., Luo Z., Yao R.-W., Zheng C.-C., Zhang J., Xiang J.-F., Yang L., Chen L.-L. 2016. Unusual processing generates SPA lncRNAs that sequester multiple RNA binding proteins. Mol. Cell. 64, 534–548.
Yin Q.-F., Yang L., Zhang Y., Xiang J.-F., Wu Y.-W., Carmichael G.G., Chen L.-L. 2012. Long noncoding RNAs with snoRNA ends. Mol. Cell. 48, 219–230.
Wevrick R., Kerns J.A., Francke U. 1994. Identification of a novel paternally expressed gene in the Prader–Willi syndrome region. Hum. Mol. Genet. 3, 1877–1882.
Cavaillé J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Hüttenhofer A. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. U. S. A. 97, 14311–14316.
Castle J.C., Armour C.D., Löwer M., Haynor D., Biery M., Bouzek H., Chen R., Jackson S., Johnson J.M., Rohl C.A., Raymond C.K. 2010. Digital genome-wide ncRNA expression, including snoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 5, e11779.
Chamberlain S.J., Chen P.-F., Ng K.Y., Bourgois-Rocha F., Lemtiri-Chlieh F., Levine E.S., Lalande M. 2010. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader–Willi syndromes. Proc. Natl. Acad. Sci. U. S. A. 107, 17668–17673.
Hsiao J.S., Germain N.D., Wilderman A., Stoddard C., Wojenski L.A., Villafano G.J., Core L., Cotney J., Chamberlain S.J. 2019. A bipartite boundary element restricts UBE3A imprinting to mature neurons. Proc. Natl. Acad. Sci. U. S. A. 116, 2181–2186.
Wijesuriya T.M., De Ceuninck L., Masschaele D., Sanderson M.R., Carias K.V., Tavernier J., Wevrick R. 2017. The Prader–Willi syndrome proteins MAGEL2 and necdin regulate leptin receptor cell surface abundance through ubiquitination pathways. Hum. Mol. Genet. 26, 4215–4230.
Tacer K.F., Potts P.R. 2017. Cellular and disease functions of the Prader–Willi syndrome gene MAGEL2. Biochem. J. 474, 2177–2190.
Pagliardini S., Ren J., Wevrick R., Greer J.J. 2005. Developmental abnormalities of neuronal structure and function in prenatal mice lacking the Prader–Willi syndrome gene necdin. Am. J. Pathol. 167, 175–191.
Wawrzik M., Spiess A.-N., Herrmann R., Buiting K., Horsthemke B. 2009. Expression of SNURF-SNRPN upstream transcripts and epigenetic regulatory genes during human spermatogenesis. Eur. J. Hum. Genet. 17, 1463–1470.
Buiting K., Nazlican H., Galetzka D., Wawrzik M., Groß S., Horsthemke B. 2007. C15orf2 and a novel noncoding transcript from the Prader–Willi/Angelman syndrome region show monoallelic expression in fetal brain. Genomics. 89, 588–595.
Neumann L.C., Markaki Y., Mladenov E., Hoffmann D., Buiting K., Horsthemke B. 2012. The imprinted NPAP1/C15orf2 gene in the Prader–Willi syndrome region encodes a nuclear pore complex associated protein. Hum. Mol. Genet. 21, 4038–4048.
Rougeulle C., Cardoso C., Fontés M., Colleaux L., Lalande M. 1998. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 19, 15–16.
Kishino T., Wagstaff J. 1998. Genomic organization of the UBE3A/E6-AP gene and related pseudogenes. Genomics. 47, 101–107.
Rougeulle C., Glatt H., Lalande M. 1997. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat. Genet. 17, 14–15.
Dindot S.V., Antalffy B.A., Bhattacharjee M.B., Beaudet A.L. 2008. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum. Mol. Genet. 17, 111–118.
DuBose A.J., Johnstone K.A., Smith E.Y., Hallett R.A.E., Resnick J.L. 2010. Atp10a, a gene adjacent to the PWS/AS gene cluster, is not imprinted in mouse and is insensitive to the PWS-IC. Neurogenetics. 11, 145–151.
Mohamad F.H., Has A.T.C. 2019. The α5-containing GABAA receptors-a. Brief summary. J. Mol. Neurosci. 67, 343–351.
DeLorey T.M., Sahbaie P., Hashemi E., Homanics G.E., Clark J.D. 2008. Gabrb3 gene deficient mice exhibit impaired social and exploratory behaviors, deficits in non-selective attention and hypoplasia of cerebellar vermal lobules: a potential model of autism spectrum disorder. Behav. Brain Res. 187, 207–220.
Delahanty R.J., Zhang Y., Bichell T.J., Shen W., Verdier K., Macdonald R.L., Xu L., Boyd K., Williams J., Kang J.-Q. 2016. Beyond epilepsy and autism: Disruption of GABRB3 causes ocular hypopigmentation. Cell Repts. 17, 3115–3124.
Buiting K., Williams C., Horsthemke B. 2016. Angelman syndrome: Insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 12, 584–593.
Zemlyakova V.V., Ermakova M.A., Zaletaev D.V., Nemtsova M.V. 2009. Molecular diagnosis of Prader–Willi and Angelman syndromes. Med. Genet. 8, 16–20.
Bird L.M. 2014. Angelman syndrome: Review of clinical and molecular aspects. Appl. Clin. Genet. 7, 93–104.
Butler M.G., Miller J.L., Forster J.L. 2019. Prader–Willi syndrome: Clinical genetics, diagnosis and treatment approaches: an update. Curr. Pediatr. Rev. 15, 207–244.
Anderlid B.-M., Lundin J., Malmgren H., Lehtihet M., Nordgren A. 2014. Small mosaic deletion encompassing the snoRNAs and SNURF-SNRPN results in an atypical Prader–Willi syndrome phenotype. Am. J. Med. Genet. A. 164A, 425–431.
de Smith A.J., Purmann C., Walters R.G., Ellis R.J., Holder S.E., Van Haelst M.M., Brady A.F., Fairbrother U.L., Dattani M., Keogh J.M., Henning E., Yeo G.S.H., O’Rahilly S., Froguel P., Farooqi I.S., Blakemore A.I.F. 2009. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 18, 3257–3265.
Fridman C., Koiffmann C.P. 2000. Origin of uniparental disomy 15 in patients with Prader–Willi or Angelman syndrome. Am. J. Med. Genet. 94, 249–253.
Robinson W.P., Christian S.L., Kuchinka B.D., Peñaherrera M.S., Das S., Schuffenhauer S., Malcolm S., Schinzel A.A., Hassold T.J., Ledbetter D.H. 2000. Somatic segregation errors predominantly contribute to the gain or loss of a paternal chromosome leading to uniparental disomy for chromosome 15. Clin. Genet. 57, 349–358.
Beygo J., Buiting K., Ramsden S.C., Ellis R., Clayton-Smith J., Kanber D. 2019. Update of the EMQN/ACGS best practice guidelines for molecular analysis of Prader–Willi and Angelman syndromes. Eur. J. Hum. Genet. 27, 1326–1340.
Beygo J., Grosser C., Kaya S., Mertel C., Buiting K., Horsthemke B. 2020. Common genetic variation in the Angelman syndrome imprinting centre affects the imprinting of chromosome 15. Eur. J. Hum. Genet. 28, 835–839.
Le Fevre A., Beygo J., Silveira C., Kamien B., Clayton-Smith J., Colley A., Buiting K., Dudding-Byth T. 2017. Atypical Angelman syndrome due to a mosaic imprinting defect: case reports and review of the literature. Am. J. Med. Genet. A. 173, 753–757.
Ermakova M.A., Babenko O.V., Zaletaev D.V., Nemtsova M.V. 2010. Analysis of UBE3A gene mutations in patients with Angelman syndrome. Med. Genet. 9 (5), 18–23.
Eggermann T., Perez de Nanclares G., Maher E.R., Temple I.K., Tümer Z., Monk D., Mackay D.J.G., Grønskov K., Riccio A., Linglart A., Netchine I. 2015. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin. Epigenetics. 7, 123.
Buiting K., Clayton-Smith J., Driscoll D.J., Gillessen-Kaesbach G., Kanber D., Schwinger E., Williams C., Horsthemke B. 2015. Clinical utility gene card for Angelman syndrome. Eur. J. Hum. Genet. 23 (2). https://doi.org/10.1038/ejhg.2014.93
Beasley S.A., Kellum C.E., Orlomoski R.J., Idrizi F., Spratt D.E. 2020. An Angelman syndrome substitution in the HECT E3 ubiquitin ligase C-terminal lobe of E6AP affects protein stability and activity. PLoS One. 15, e0235925.
Aguilera C., Viñas-Jornet M., Baena N., Gabau E., Fernández C., Capdevila N., Cirkovic S., Sarajlija A., Miskovic M., Radivojevic D., Ruiz A., Guitart M. 2017. Novel intragenic deletions within the UBE3A gene in two unrelated patients with Angelman syndrome: Case report and review of the literature. BMC Med. Genet. 18, 137.
Bossuyt S.N.V., Punt A.M., de Graaf I.J., van den Burg J., Williams M.G., Heussler H., Elgersma Y., Distel B. 2021. Loss of nuclear UBE3A activity is the predominant cause of Angelman syndrome in individuals carrying UBE3A missense mutations. Hum. Mol. Genet. 30 (6), 430–442. https://doi.org/10.1093/hmg/ddab050
McCarthy J.M., McCann-Crosby B.M., Rech M.E., Yin J., Chen C.-A., Ali M.A., Nguyen H.N., Miller J.L., Schaaf C.P. 2018. Hormonal, metabolic and skeletal phenotype of Schaaf–Yang syndrome: A comparison to Prader–Willi syndrome. J. Med. Genet. 55, 307–315.
Negishi Y., Ieda D., Hori I., Nozaki Y., Yamagata T., Komaki H., Tohyama J., Nagasaki K., Tada H., Saitoh S. 2019. Schaaf–Yang syndrome shows a Prader–Willi syndrome-like phenotype during infancy. Orphanet. J. Rare Dis. 14, 277.
Chen X., Ma X., Zou C. 2020. Phenotypic spectrum and genetic analysis in the fatal cases of Schaaf–Yang syndrome: Two case reports and literature review. Medicine. 99, e20574.
Schaaf C.P., Gonzalez-Garay M.L., Xia F., Potocki L., Gripp K.W., Zhang B., Peters B.A., McElwain M.A., Drmanac R., Beaudet A.L., Caskey C.T., Yang Y. 2013. Truncating mutations of MAGEL2 cause Prader–Willi phenotypes and autism. Nat. Genet. 45, 1405–1408.
Carias K.V., Zoeteman M., Seewald A., Sanderson M.R., Bischof J.M., Wevrick R. 2020. A MAGEL2-deubiquitinase complex modulates the ubiquitination of circadian rhythm protein CRY1. PLoS One. 15, e0230874.
Patak J., Gilfert J., Byler M., Neerukonda V., Thiffault I., Cross L., Amudhavalli S., Pacio-Miguez M., Palomares-Bralo M., Garcia-Minaur S., Santos-Simarro F., Powis Z., Alcaraz W., Tang S., Jurgens J., et al. 2019. MAGEL2-related disorders: A study and case series. Clin. Genet. 96, 493–505.
Ahn H., Seo G.H., Oh A., Lee Y., Keum C., Heo S.H., Kim T., Choi J., Kim G.-H., Ko T.-S., Yum M.-S., Lee B.H., Choi I.H. 2020. Diagnosis of Schaaf–Yang syndrome in Korean children with developmental delay and hypotonia. Medicine (Baltimore). 99, e23864.
Roberts S.A., Kaiser U.B. 2020. Genetics in endocrinology: Genetic etiologies of central precocious puberty and the role of imprinted genes. Eur. J. Endocrinol. 183, R107–R117.
Seraphim C.E., Canton A.P.M., Montenegro L., Piovesan M.R., Macedo D.B., Cunha M., Guimaraes A., Ramos C.O., Benedetti A.F.F., de Castro Leal A., Gagliardi P.C., Antonini S.R., Gryngarten M., Arcari A.J., Abreu A.P., et al. 2021. Genotype–phenotype correlations in central precocious puberty caused by MKRN3 mutations. J. Clin. Endocrinol. Metab. 106, 1041–1050.
Jong M.T.C., Gray T.A., Ji Y., Glenn C.C., Saitoh S., Driscoll D.J., Nicholls R.D. 1999. A novel imprinted gene, encoding a ring zinc-finger protein, and overlapping antisense transcript in the Prader–Willi syndrome critical region. Hum. Mol. Genet. 8, 783–793.
Latronico A.C., Brito V.N., Carel J.-C. 2016. Causes, diagnosis, and treatment of central precocious puberty. Lancet Diabetes Endocrinol. 4, 265–274.
Valadares L.P., Meireles C.G., De Toledo I.P., Santarem de Oliveira R., Gonçalves de Castro L.C., Abreu A.P., Carroll R.S., Latronico A.C., Kaiser U.B., Guerra E.N.S., Lofrano-Porto A. 2019. MKRN3 mutations in central precocious puberty: A systematic review and meta-analysis. J. Endocrine Soc. 3, 979–995.
Maione L., Naulé L., Kaiser U.B. 2020. Makorin RING finger protein 3 and central precocious puberty. Curr. Opin. Endocrine Metab. Res. 14, 152–159.
Fanis P., Skordis N., Toumba M., Papaioannou N., Makris A., Kyriakou A., Neocleous V., Phylactou L.A. 2019. Central precocious puberty caused by novel mutations in the promoter and 5′-UTR region of the imprinted MKRN3 gene. Front. Endocrinol. 10, 677.
Abreu A.P., Macedo D.B., Brito V.N., Kaiser U.B., Latronico A.C. 2015. A new pathway in the control of the initiation of puberty: the MKRN3 gene. J. Mol. Endocrinol. 54, R131–139.
Perry J., Day F., Elks C., et Collaborators. 2014. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 514, 92–97. https://doi.org/10.1038/nature13545
Li H., Du J., Li W., Cheng D., He W., Yi D., Xiong B., Yuan S., Tu C., Meng L., Luo A., Lin G., Lu G., Tan Y.-Q. 2018. Rare partial octosomy and hexasomy of 15q11–q13 associated with intellectual impairment and development delay: report of two cases and review of literature. Mol. Cytogenet. 11, 15.
Lu Y., Liang Y., Ning S., Deng G., Xie Y., Song J., Zuo N., Feng C., Qin Y. 2020. Rare partial trisomy and tetrasomy of 15q11–q13 associated with developmental delay and autism spectrum disorder. Mol. Cytogenet. 13, 21.
Yang L., Zhan G.D., Ding J.J., Wang H.J., Ma D., Huang G.Y., Zhou W.H. 2013. Psychiatric illness and intellectual disability in the Prader–Willi syndrome with different molecular defects – a meta analysis. PLoS One. 8 (8), e72640. https://doi.org/10.1371/journal.pone.0072640
Dykens E.M., Roof E., Hunt-Hawkins H., Dankner N., Lee E.B., Shivers C.M., Daniell C., Kim S.-J. 2017. Diagnoses and characteristics of autism spectrum disorders in children with Prader–Willi syndrome. J. Neurodev. Disord. 9, 18.
Linglart A., Maupetit-Méhouas S., Silve C. 2013. GNAS-related loss-of-function disorders and the role of imprinting. Horm. Res. Paediatr. 79, 119–129.
Turan S., Bastepe M. 2013. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm. Res. Paediatr. 80, 229–241.
Lemos M.C., Thakker R.V. 2015. GNAS mutations in pseudohypoparathyroidism type 1a and related disorders. Hum. Mutat. 36, 11–19.
Mantovani G., Elli F.M. 2019. Inactivating PTH/PTHrP signaling disorders. Front. Horm. Res. 51, 147–159.
Turan S., Bastepe M. 2013. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm. Res. Paediatr. 80, 229–241.
Mantovani G., Bastepe M., Monk D., de Sanctis L., Thiele S., Usardi A., Ahmed S.F., Bufo R., Choplin T., De Filippo G., Devernois G., Eggermann T., Elli F.M., Freson K., García Ramirez A., et al. 2018. Diagnosis and management of pseudohypoparathyroidism and related disorders: First International Consensus Statement. Nat. Rev. Endocrinol. 14, 476–500.
Yang Y., Chu X., Nie M., Song A., Jiang Y., Li M., Xia W., Xing X., Wang O. 2020. A novel long-range deletion spanning STX16 and NPEPL1 causing imprinting defects of the GNAS locus discovered in a patient with autosomal-dominant pseudohypoparathyroidism type 1B. Endocrine. 69, 212–219.
Kiuchi Z., Reyes M., Jüppner H. 2020. Preferential maternal transmission of STX16-GNAS mutations responsible for autosomal dominant pseudohypoparathyroidism type Ib (PHP1B): another example of transmission ratio distortion. J. Bone Miner. Res. 36(4), 696–703. https://doi.org/10.1002/jbmr.4221
Rezwan F.I., Poole R.L., Prescott T., Walker J.M., Karen Temple I., Mackay D.J. 2015. Very small deletions within the NESP55 gene in pseudohypoparathyroidism type 1b. Eur. J. Hum. Genet. 23, 494–499.
Takatani R., Molinaro A., Grigelioniene G., Tafaj O., Watanabe T., Reyes M., Sharma A., Singhal V., Raymond F.L., Linglart A., Jüppner H. 2016. Analysis of multiple families with single individuals affected by pseudohypoparathyroidism type Ib (PHP1B) reveals only one novel maternally inherited GNAS deletion: Only one novel maternally inherited GNAS deletion among multiple PHP1B patients. J. Bone Miner. Res. 31, 796–805.
Swieringa F., Solari F.A., Pagel O., Beck F., Huang J., Feijge M.A.H., Jurk K., Körver-Keularts I.M.L.W., Mattheij N.J.A., Faber J., Pohlenz J., Russo A., Stumpel C.T.R.M., Schrander D.E., Zieger B., et al. 2020. Impaired iloprost-induced platelet inhibition and phosphoproteome changes in patients with confirmed pseudohypoparathyroidism type Ia, linked to genetic mutations in GNAS. Sci. Rep. 10, 11389.
Jüppner H. 2021. Molecular definition of pseudohypoparathyroidism variants. J. Clin. Endocrinol. Metab. 106 (6), 1541–1552. https://doi.org/10.1210/clinem/dgab060
Bastepe M. 2018. GNAS mutations and heterotopic ossification. Bone. 109, 80–85.
Turan S., Bastepe M. 2018. GNAS complex locus. In: Encyclopedia of Signaling Molecules. Ed. Choi S. Cham: Springer, 2173–2185. doi.org/https://doi.org/10.1007/978-3-319-67199-4_101631
Colson C., Decamp M., Gruchy N., Coudray N., Ballandonne C., Bracquemart C., Molin A., Mittre H., Takatani R., Jüppner H., Kottler M-L., Richard N. 2019. High frequency of paternal iso or heterodisomy at chromosome 20 associated with sporadic pseudohypoparathyroidism 1B. Bone. 123, 145–152.
Mulchandani S., Bhoj E.J., Luo M., Powell-Ha-milton N., Jenny K., Gripp K.W., Elbracht M., Eggermann T., Turner C.L.S., Temple I.K., Mackay D.J.G., Dubbs H., Stevenson D.A., Slattery L., Zackai E.H., et al. 2016. Maternal uniparental disomy of chromosome 20: a novel imprinting disorder of growth failure. Genet. Med. 18, 309–315.
Eggermann T., Oehl-Jaschkowitz B., Dicks S., Thomas W., Kanber D., Albrecht B., Begemann M., Kurth I., Beygo J., Buiting K. 2017. The maternal uniparental disomy of chromosome 6 (upd(6)mat) “phenotype”: Result of placental trisomy 6 mosaicism? Mol. Genet. Genom. Med. 5, 668–677.
Mackay D.J.G., Boonen S.E., Clayton-Smith J., Goodship J., Hahnemann J.M.D., Kant S.G., Njølstad P.R., Robin N.H., Robinson D.O., Siebert R., Shield J.P.H., White H.E., Temple I.K. 2006. A maternal hypomethylation syndrome presenting as transient neonatal diabetes mellitus. Hum. Genet. 120, 262–269.
Boonen S.E., Pörksen S., Mackay D.J., Oestergaard E., Olsen B., Brondum-Nielsen K., Temple I.K., Hahne-mann J.M. 2008. Clinical characterisation of the multiple maternal hypomethylation syndrome in siblings. Eur. J. Hum. Genet. 16, 453–461.
Mackay D.J.G., Callaway J.L.A., Marks S.M., White H.E., Acerini C.L., Boonen S.E., Dayanikli P., Firth H.V., Goodship J.A., Haemers A.P., Hahnemann J.M.D., Kordonouri O., Masoud A.F., Oestergaard E., Storr J., et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 40, 949–951.
Quenneville S., Verde G., Corsinotti A., Kapopoulou A., Jakobsson J., Offner S., Baglivo I., Pedone P.V., Grimaldi G., Riccio A., Trono D. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 44, 361–372.
Ecco G., Imbeault M., Trono D. 2017. KRAB zinc finger proteins. Development. 144, 2719–2729.
Farhadova S., Gomez-Velazquez M., Feil R. 2019. Stability and lability of parental methylation imprints in development and disease. Genes (Basel). 10 (12), 999. https://doi.org/10.3390/genes10120999
Baglivo I., Esposito S., De Cesare L., Sparago A., Anvar Z., Riso V., Cammisa M., Fattorusso R., Grimaldi G., Riccio A., Pedone P.V. 2013. Genetic and epigenetic mutations affect the DNA binding capability of human ZFP57 in transient neonatal diabetes type 1. FEBS Lett. 587, 1474–1481.
Monteagudo-Sánchez A., Hernandez Mora J.R., Simon C., Burton A., Tenorio J., Lapunzina P., Clark S., Esteller M., Kelsey G., López-Siguero J.P., de Nanclares G.P., Torres-Padilla M-E., Monk D. 2020. The role of ZFP57 and additional KRAB-zinc finger proteins in the maintenance of human imprinted methylation and multi-locus imprinting disturbances. Nucleic Acids Res. 48, 11394–11407.
Takahashi N., Coluccio A., Thorball C.W., Planet E., Shi H., Offner S., Turelli P., Imbeault M., Ferguson-Smith A.C., Trono D. 2019. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 33, 49–54.
Kim J.D., Kim H., Ekram M.B., Yu S., Faulk C., Kim J. 2011. Rex1/Zfp42 as an epigenetic regulator for genomic imprinting. Hum. Mol. Genet. 20, 1353–1362.
Monk D., Sanchez-Delgado M., Fisher R. 2017. NLRPs, the subcortical maternal complex and genomic imprinting. Reproduction. 154, R161–170.
Begemann M., Rezwan F.I., Beygo J., Docherty L.E., Kolarova J., Schroeder C., Buiting K., Chokkalingam K., Degenhardt F., Wakeling E.L., Kleinle S., González Fassrainer D., Oehl-Jaschkowitz B., Turner C.L.S., Patalan M., et al. 2018. Maternal variants in NLRP and other maternal effect proteins are associated with multilocus imprinting disturbance in offspring. J. Med. Genet. 55, 497–504.
Eggermann T., Kadgien G., Begemann M., Elbracht M. 2020. Biallelic PADI6 variants cause multilocus imprinting disturbances and miscarriages in the same family. Eur. J. Hum. Genet. 29 (4), 575–580. https://doi.org/10.1038/s41431-020-00762-0
Mackay D.J.G., Eggermann T., Buiting K., Garin I., Netchine I., Linglart A., de Nanclares G.P. 2015. Multilocus methylation defects in imprinting disorders. Biomol. Concepts. 6, 47–57.
Nakka P., Pattillo Smith S., O’Donnell-Luria A.H., McManus K.F., Mountain J.L., Ramachandran S., Sathirapongsasuti J.F., Agee M., Auton A., Bell R.K., Bryc K., Elson S.L., Fontanillas P., Furlotte N.A., Hicks B., et al. 2019. Characterization of prevalence and health consequences of uniparental disomy in four million individuals from the general population. Am. J. Hum. Genet. 105, 921–932.
Yauy K., de Leeuw N., Yntema H.G., Pfundt R., Gilissen C. 2020. Accurate detection of clinically relevant uniparental disomy from exome sequencing data. Genet. Med. 22, 803–808.
Scuffins J., Keller-Ramey J., Dyer L., Douglas G., Torene R., Gainullin V., Juusola J., Meck J., Retterer K. 2021. Uniparental disomy in a population of 32 067 clinical exome trios. Genet Med. 23 (6), 1101–1107. https://doi.org/10.1038/s41436-020-01092-8
Gigante S., Gouil Q., Lucattini A., Keniry A., Beck T., Tinning M., Gordon L., Woodruff C., Speed T.P., Blewitt M.E., Ritchie M.E. 2019. Using long-read sequencing to detect imprinted DNA methylation. Nucleic Acids Res. 47, e46–e46.
Klobučar T., Kreibich E., Krueger F., Arez M., Pólvora-Brandão D., von Meyenn F., da Rocha S.T., Eckersley-Maslin M. 2020. IMPLICON: An ultra-deep sequencing method to uncover DNA methylation at imprinted regions. Nucleic Acids Res. 48, e92–e92.
Santoni F.A., Stamoulis G., Garieri M., Falconnet E., Ribaux P., Borel C., Antonarakis S.E. 2017. Detection of imprinted genes by single-cell allele-specific gene expression. Am. J. Hum. Genet. 100, 444–453.
ACKNOWLEDGMENTS
The authors are grateful to I.V. Bure for assistance in preparation of this paper.
Funding
This study was supported by the Russian Foundation for Basic Research, project no. 20-14-50138.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Сonflict of interest. The authors declare that they have no conflict of interest.
Statement on the welfare of humans or animals. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Translated by M. Novikova
Abbreviations: lncRNA, Long Non-Coding RNA; UPD, Uniparental Disomy; AS-IC, Angelman Syndrome Imprinting Center; BP, Break Point; DMR, Differentially Methylated Regions; GOM, Gain of Methylation; IC, Imprinting Centre; LOM, Loss of Methylation; MLID, Multi-Locus Imprinting Disturbances; PWS-IC, Prader-Willi Syndrome Imprinting Center; snoRNA, small nucleolar RNA; miRNA, microRNA.
Rights and permissions
About this article
Cite this article
Zaletaev, D.V., Nemtsova, M.V. & Strelnikov, V.V. Epigenetic Regulation Disturbances on Gene Expression in Imprinting Diseases. Mol Biol 56, 1–28 (2022). https://doi.org/10.1134/S0026893321050149
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893321050149