Abstract—
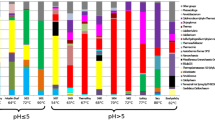
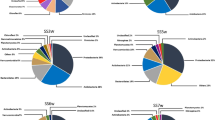
Changes in microbial community composition during formation of an acid mine drainage were studied on a model of two water reservoirs located in the Ozernoye open-cast mine for polymetallic ores in Eastern Siberia. The first reservoir, Bu-18, was filled with groundwater, had a neutral pH and low levels of sulfate and dissolved metal ions. The second reservoir, Bu-16, was an acid mine drainage (pH 2.85) filled with the water from Bu-18, which passed through rocks containing sulfide minerals. The Bu-16 water contained 1405 mg/L of sulfate, 164 mg/L of manganese, 78 mg/L of magnesium, and 26 mg/L of iron. Molecular analysis of the microbial communities of two reservoirs, carried out using high-throughput sequencing of the 16S rRNA gene fragments, showed that formation of the acid mine drainage was accompanied by a decrease in microbial diversity and by selection of several dominant taxonomic and functional groups. Chemolithoautotrophic iron- and sulfur-oxidizing bacteria of the genera Leptospirillum, Acidithiobacillus, Gallionella, Sulfuriferula, and Sulfobacillus constituted most of the prokaryotic community in Bu-16. Heterotrophic bacteria of the genera Ferrimicrobium and Metallibacterium, capable of reducing Fe(III) under anaerobic conditions, were present as minor components. Over 20% of the community were members of the Candidate Phyla Radiation group and of the candidate phylum Dependentiae (TM6), known for their parasitic or symbiotic lifestyle. These groups of bacteria were rarely found in acid mine drainage and only in minor quantities. Potential hosts of the Dependentiae, flagellates of the genus Spumella, were found among eukaryotes in Bu-16.

Similar content being viewed by others
REFERENCES
Anderson, I., Chertkov, O., Chen, A., Saunders, E., Lapidus, A., Nolan, M., Lucas, S., Hammon, N., Deshpande, S., Cheng, J.F., Han, C., Tapia, R., Goodwin, L.A., Pitluck, S., Liolios, K., et al., Complete genome sequence of the moderately thermophilic mineral-sulfide-oxidizing firmicute Sulfobacillus acidophilus type strain (NAL(T)), Stand. Genomic Sci. 2012, vol. 6 , no. 3, pp. 1‒13. https://doi.org/10.4056/sigs.2736042
Baker, B.J. and Banfield, J.F., Microbial communities in acid mine drainage, FEMS Microbiol. Ecol., 2003, vol. 44, pp. 139–152.
Behnke, A., Engel, M., Christen, R., Nebel, M., Klein, R.R., and Stoeck, T., Depicting more accurate pictures of protistan community complexity using pyrosequencing of hypervariable SSU rRNA gene regions, Environ. Microbiol., 2011, vol. 13, pp. 340–349.
Boenigk, J., Pfandl, K., Stadler, P., and Chatzinotas, A., High diversity of the “Spumella-like” flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions, Environ. Microbiol., 2005, vol. 7, pp. 685‒697.
Brown, C.T., Hug, L.A., Thomas, B.C., Sharon, I., Castelle, C.J., Singh, A., Wilkins, M.J., Wrighton, K.C., Williams, K.H., and Banfield, J.F., Unusual biology across a group comprising more than 15% of domain Bacteria, Nature, 2015, vol. 523, pp. 208‒211.
Bruneel, O., Duran, R., Casiot, C., Elbaz-Poulichet, F., and Personne, J.C., Diversity of microorganisms in Fe-As-rich acid mine drainage waters of Carnoules, France, Appl. Environ. Microbiol., 2006, vol. 72, pp. 551–556.
Castelle, C.J., Brown, C.T., Anantharaman, K., Probst, A.J., Huang, R.H., and Banfield, J.F., Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations, Nat. Rev. Microbiol., 2018, vol. 16, pp. 629‒645.
Chen, L.X., Hu, M., Huang, L.N., Hua, Z.S., Kuang, J.L., Li, S.J., and Shu, W.S., Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage, ISME J., 2015, vol. 9, pp. 1579‒1592.
Edgar, R.C., Search and clustering orders of magnitude faster than BLAST, Bioinformatics, 2010, vol. 26, pp. 2460–2461.
Fabisch, M., Beulig, F., Akob, D.M., and Kusel, K., Surprising abundance of Gallionella-related iron oxidizers in creek sediments at pH 4.4 or at high heavy metal concentrations, Front. Microbiol., 2013, vol. 4, p. 390.
García-Moyano, A., Austnes, A.E., Lanzén, A., González-Toril, E., Aguilera, Á., and Øvreås, L., Novel and unexpected microbial diversity in acid mine drainage in Svalbard (78° N), revealed by culture-independent approaches, Microorganisms, 2015, vol. 3, pp. 667‒694.
Hallberg, K.B., González-Toril, E., and Johnson, D.B., Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments, Extremophiles, 2010, vol. 14, pp. 9‒19.
He, Z., Xiao, S., Xie, X., Zhong, H., Hu, Y., Li, Q., Gao, F., Li, G., Liu, J., and Qiu, G., Molecular diversity of microbial community in acid mine drainages of Yunfu sulfide mine, Extremophiles, 2007, vol. 11, pp. 305–314.
Ishii, K., Fujitani, H., Soh, K., Nakagawa, T., Takahashi, R., and Tsuneda, S., Enrichment and physiological characterization of a cold-adapted nitrite-oxidizing Nitrotoga sp. from an eelgrass sediment, Appl. Environ. Microbiol., 2017, vol. 83. pii: e00549-17.
Johnson, B.D. and Hallberg, K.B. The microbiology of acidic mine waters, Res. Microbiol., 2003, vol. 154, pp. 466‒473.
Johnson, D.B., Bacelar-Nicolau, P., Okibe, N., Thomas, A., and Hallberg, K.B., Ferrimicrobium acidiphilum gen. nov., sp. nov. and Ferrithrix thermotolerans gen. nov., sp. nov.: heterotrophic, iron-oxidizing, extremely acidophilic actinobacteria, Int. J. Syst. Evol. Microbiol., 2009, vol. 59, pp. 1082‒1089.
Kadnikov, V.V., Ivasenko, D.A., Beletsky, A.V., Mardanov, A.V., Danilova, E.V., Pimenov, N.V., Karnachuk, O.V., and Ravin, N.V., Effect of metal concentration on the microbial community in acid mine drainage of a polysulfide ore deposit, Microbiology (Moscow), 2016a, vol. 85, pp. 745–751. https://doi.org/10.1134/S0026261716060126
Kadnikov, V.V., Ivasenko, D.A., Beletsky, A.V., Mardanov, A.V., Danilova, E.V., Pimenov, N.V., Karnachuk, O.V., and Ravin, N.V., A novel uncultured bacterium of the family Gallionellaceae: description and genome reconstruction based on metagenomic analysis of microbial community in acid mine drainage, Microbiology (Moscow), 2016b, vol. 85, no. 4, pp. 449–461. https://doi.org/10.1134/S002626171604010X
Kimura, S., Bryan, C.G., Hallberg, K.B., and Johnson, D.B., Biodiversity and geochemistry of an extremely acidic, low-temperature subterranean environment sustained by chemolithotrophy, Environ. Microbiol., 2011, vol. 13, pp. 2092–2104.
Kupka, D., Rzhepishevska, O.I., Dopson, M., Lindström, E.B., Karnachuk, O.V., and Tuovinen, O.H., Bacterial oxidation of ferrous sulfate at low temperatures, Biotechnol. Bioeng., 2007, vol. 97, pp. 1470‒1478.
Liljeqvist, M., Valdes, J., Holmes, D.S., and Dopson, M., Draft genome of the psychrotolerant acidophile Acidithiobacillus ferrivorans SS3, J. Bacteriol., 2011, vol. 193, pp. 4304–4305.
Magoč, T. and Salzberg, S.L., FLASH: fast length adjustment of short reads to improve genome assemblies, Bioinformatics, 2011, vol. 27, pp. 2957‒2963.
Méndez-García, C., Peláez, A.I., Mesa, V., Sánchez, J., Golyshina, O.V., and Ferrer, M., Microbial diversity and metabolic networks in acid mine drainage habitats, Front. Microbiol., 2015, vol. 29, no. 6, p. 475.
Pruesse, E., Peplies, J., and Glöckner, F.O., SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes, Bioinformatics, 2012, vol. 28, pp. 1823–1829.
Ram, R.J., Verberkmoes, N.C., Thelen, M.P., Tyson, G.W., Baker, B.J., Blake, R.C. 2nd, Shah, M., Hettich, R.L., and Banfield, J.F., Community proteomics of a natural microbial biofilm, Science, 2005, vol. 308, pp. 1915‒1920.
Rohwerder, T., Gehrke, T., Kinzler, K., and Sand, W., Bioleaching review part A: progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation, Appl. Microbiol. Biotechnol., 2003, vol. 63, pp. 239‒248.
Schramm, A., De Beer, D., Wagner, M., and Amann, R., Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor, Appl. Environ. Microbiol., 1998, vol. 64, pp. 3480‒3485.
Schloss, P.D., Westcott, S.L., Ryabin, T., Hall, J.R., Hartmann, M., Hollister, E.B., Lesniewski, R.A., Oakley, B.B., Parks, D.H., Robinson, C.J., Sahl, J.W., Stres, B., Thallinger, G.G., Van Horn, D.J., and Weber, C.F., Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 2009, vol. 75, pp. 7537–7541.
Tyson, G.W., Chapman, J., Hugenholtz, P., Allen, E.E., Ram, R.J., Richardson, P.M., Solovyev, V.V., Rubin, E.M., Rokhsar, D.S., and Banfield, J.F., Community structure and metabolism through reconstruction of microbial genomes from the environment, Nature, 2004, vol. 428, no. 6978, pp. 37–43.
Watanabe, T., Kojima, H., and Fukui, M., Sulfuriferula multivorans gen. nov., sp. nov., isolated from a freshwater lake, reclassification of “Thiobacillus plumbophilus” as Sulfuriferula plumbophilus sp. nov., and description of Sulfuricellaceae fam. nov. and Sulfuricellales ord. nov., Int. J. Syst. Evol. Microbiol., 2015, vol. 65, pp. 1504‒1508.
Yeoh, Y.K., Sekiguchi, Y., Parks, D.H., and Hugenholtz, P., Comparative genomics of candidate phylum TM6 suggests that parasitism is widespread and ancestral in this lineage, Mol. Biol. Evol., 2016, vol. 33, pp. 915‒927.
Ziegler, S., Waidner, B., Itoh, T., Schumann, P., Spring, S., and Gescher, J., Metallibacterium scheffleri gen. nov., sp. nov., an alkalinizing gammaproteobacterium isolated from an acidic biofilm, Int. J. Syst. Evol. Microbiol., 2013, vol. 63, pp. 1499‒1504.
ACKNOWLEDGMENTS
The work was performed using the scientific equipment of the Core Research Facility “Bioengineering.”
Funding
The work was supported by the Russian Science Foundation (project no. 14-14-01016, analysis of prokaryotes) and by the Russian Foundation for Basic Research (project no. 18-34-00356, analysis of eukaryotes). Sampling and physicochemical analysis of acid mine drainage samples were supported by the Russian Foundation for Basic Research, project no. 16-54-150011.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Statement of the welfare of animals. This article does not contain any research using animals as objects.
Conflict of interest. The authors declare that there is no conflict of interest.
Additional information
Translated by A. Bulaev
Rights and permissions
About this article
Cite this article
Kadnikov, V.V., Gruzdev, E.V., Ivasenko, D.A. et al. Selection of a Microbial Community in the Course of Formation of Acid Mine Drainage. Microbiology 88, 292–299 (2019). https://doi.org/10.1134/S0026261719030056
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026261719030056