Abstract
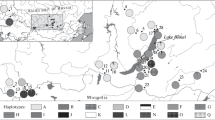
The genetic diversity and population structure of Taxus cuspidata Sieb. et Zucc. ex Endl. on the Russian part of its range was studied for the first time on the basis of the nucleotide polymorphism of intergenic spacers psbA–trnH, trnL–trnF, and trnS–trnfM of chloroplast DNA. A high level of gene (h = 0.807) and nucleotide (π = 0.0227) diversity was revealed. The data of AMOVA showed that the interpopulation component accounted for 12% of genetic variability (F ST = 0.12044, P = 0.0000). We revealed 15 haplotypes, four of which were shown to be unique. The presence of common haplotypes in the majority of populations, the absence of phylogenetic structure, and low values or even the absence of nucleotide divergence show that the T. cuspidata populations studied are fragments of the once common ancestor population. Geographical isolation, which resulted from climatic changes in the Pleistocene–Holocene, as well as from human activities, did not produce a significant effect on the genetic structure of the species.
Similar content being viewed by others
References
Krasnaya Kniga Rossiiskoi Federatsii (rasteniya i griby) (The Red Book of the Russian Federation (Plants and Fungi)), Moscow: Ministerstvo Prirodnykh Resursov i Ekologii Rossiiskoi Federatsii, Rosprirodnadzor, 2008.
Krasnaya kniga Primorskogo kraya: rasteniya. Redkie i nakhodyashchiesya pod ugrozoi ischeznoveniya vidy rastenii i gribov (Red Book of Primorsky Krai: Plants. Rare and Threatened Species of Plants and Fungi), Vladivostok: Apel’sin, 2008.
Katsuki, T. and Luscombe, D., Taxus cuspidata, The IUCN Red List of Threatened Species, 2013. e.T42549A2987373. http://dx.doi.org/. doi 10.2305/IUCN.UK.2013-1.RLTS.T42549A2987373.en
Checklist of CITES Species. http://checklist. cites.org/#/en.
Koropachinskii, I.Yu., Division of Gymnosperms— Pinophyta, in Sosudistye rasteniya sovetskogo Dal’nego Vostoka (Vascular Plants of the Soviet Far East) Leningrad: Nauka, 1989, vol. 4, pp. 9–25.
Kurentsova, G.E., Reliktovye rasteniya Primor’ya (Relict Plants of Primorye), Leningrad: Nauka, 1968.
Yamazaki, T., Taxaceae, in Flora of Japan, vol. 1: Pteridophyta and Gymnospermae, Tokyo: Kodansha, 1995, pp. 286–287.
Nathorst, A.G., Palaeobotanische Mitteillungen: 7. Über Palyssya, Stachyotaxus und Paleotaxus, K. Sven. Vetensk. Akad. Handl., 1908, vol. 8, pp. 1–16.
Florin, R., The distribution of conifers and taxad genera in time and space, Acta Horti. Berg., 1963, vol. 20, pp. 121–312.
Baikovskaya, T.N., Verkhnemiotsenovaya flora Yuzhnogo Primor’ya (Upper Miocene Flora of the Southern Primorye), Leningrad: Nauka, 1974.
Korman, D.B., Osnovy protivoopukholevoi khimioterapii (Fundamentals of Antitumor Chemotherapy), Moscow: Prakticheskaya Meditsina, 2006.
Wani, M.C., Taylor, H.L., Wall, M.E., et al., Plant antitumor agents: 4. The isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia, J. Am. Chem. Soc., 1971, vol. 93, no. 9, pp. 2325–2327. doi 10.1021/ja00738a045
Kolesnikov, B.P., About a shrub form of Japanese yew (Taxus cuspidata Sieb. et Zucc. ex Endl.), Vestn. Dal’nevost. Fil. Sib. Otd. Akad. Nauk SSSR, 1935, pp. 31–45.
Vasil’ev, N.G., Ivliev, L.A., and Khavkina, N.V., Japanese yew (Taxus cuspidata Sieb. et Zucc. ex Endl.) and its regeneration on Petrov Island (Primorskii krai), in Lesovedenie v Primorskom krae (Forest Science in Primorskii krai), Vladivostok: Biologo-Pochvennyi Institut Dal’nevostochnogo Oltdeleniya Akademii Nauk SSSR, 1969, pp. 37–50.
Voroshilova, G.I., Morphological and anatomical structure of leaf and timber of yew—Taxus cuspidata Siebold et Zucc., in Redkie i ischezayushchie drevesnye rasteniya yuga Dal’nego Vostoka (Rare and Endangered Arboreal Plants of the South of the Far East), Vladivostok: Dal’nevostochnyi Nauchnyi Tsentr Akademii Nauk SSSR, 1978, pp. 129–132.
Prisyazhnyuk, N.P., Habitat and status of Taxus cuspidata Sieb. et Zucc. ex End1. populations in the Lazovsky Reserve of Primorskii krai, Rast. Resur., 1986, vol. 22, no. 4, pp. 487–492.
Belikovich, A.V. and Galanin, A.V., Japanese yew, in Rastitel’nyi mir Sikhote-Alinskogo biosfernogo zapovednika: raznoobrazie, dinamika, monitoring (Plant World of the Sikhote-Alin Biosphere Reserve: Diversity, Dynamics, Monitoring), Vladivostok: Biologo-Pochvennyi Institut Dal’nevostochnogo Oltdeleniya Akademii Nauk SSSR, 2000, pp. 103–107.
Chubar’, E.A., Taxus cuspidata (Taxaceae) on the islands of the Far Eastern State Marine Reserve (Peter the Great Bay, the Sea of Japan), Bot. Zh., 1999, vol. 84, no. 6, pp. 82–94.
Chubar’, E.A., Biological features and structure of populations of Taxus cuspidata (Taxaceae) on the islands of Peter the Great Bay of the Sea of Japan, Bot. Zh., 2016, vol. 101, no. 1, pp. 13–39.
Potenko, V.V., Inheritance of allozymes and genetic variation in natural population of Japanese yew in Petrov Island, Russia, For. Genet., 2001, vol. 8, no. 4, pp. 307–313.
Chung, M.G., Oh, G.S., and Chung, J.M., Allozyme variation in Korean populations of Taxus cuspidata (Taxaceae), Scand. J. For. Res., 1999, vol. 14, pp. 103–110. doi 10.1080/02827589950152827
Lee, S.W., Choi, W.Y., Kim, W.W., and Kim, Z.S., Genetic variation of Taxus cuspidata Sieb. et Zucc. in Korea, Silv. Genet., 2000, vol. 49, no. 3, pp. 124–130.
Cheng, B.-B., Zheng, Y.-Q., and Sun, Q.-W., Genetic diversity and population structure of Taxus cuspidata in the Changbai Mountains assessed by chloroplast DNA sequences and microsatellite markers, Biochem. Syst. Ecol., 2015, vol. 63, pp. 157–164. doi 10.1016/j.bse.2015.10.009
Wheeler, N.C., Jech, K.S., Masters, S.A., et al., Genetic variation and parameter estimates in Taxus brevifolia (Pacific yew), Can. J. For. Res., 1995, vol. 25, pp. 1913–1927. doi 10.1139/x95-207
Lewandowski, A., Burczyk, J., and Mejnartowicz, L., Genetic structure of English yew (Taxus baccata L.) in the Wierzchlas Reserve: implications for genetic conservation, For. Ecol. Manage., 1995, vol. 73, pp. 221–227. doi 10.1016/0378-1127(94)03477-E
González-Martínez, S.C., Dubreuil, M., Riba, M., et al., Spatial genetic structure of Taxus baccata L. in the western Mediterranean Basin: past and present limits to gene movement over a broad geographic scale, Mol. Phylogenet. Evol., 2010, vol. 55, pp. 805–815. doi 10.1016/j.ympev.2010.03.001
Dubreuil, M., Riba, M., Santiago, C., et al., Genetic effects of chronic habitat fragmentation revisited: strong genetic structure in a temperate tree, Taxus baccata (Taxaceae), with great dispersal capability, Am. J. Bot., 2010, vol. 97, no. 2, pp. 303–310. doi 10.3732/ajb.0900148
Senneville, S., Beaulieu, J., Daoust, G., et al., Evidence for low genetic diversity and metapopulation structure in Canada yew (Taxus canadensis): considerations for conservation, Can. J. For. Res., 2001, vol. 31, pp. 110–116. doi 10.1139/cjfr-31-1-110
Poudel, R.C., Möller, M., Li, D.-Z., et al., Genetic diversity, demographical history and conservation aspects of the endangered yew tree Taxus contorta (syn. Taxus fuana) in Pakistan, Tree Genet. Genomes, 2014, vol. 10, no. 3, pp. 653–665. doi 10.1007/s11295-014-0711-7
Gao, L.M., Möller, M., Zhang, X.-M., et al., High variation and strong phylogeographic pattern among cpDNA haplotypes in Taxus wallichiana (Taxaceae) in China and North Vietnam, Mol. Ecol., 2007, vol. 16, pp. 4684–4698. doi 10.1111/j.1365-294X.2007.03537.x
Liu, J., Möller, M., Provan, J., et al., Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot, New Phytol., 2013, vol. 199, no. 3, pp. 1093–1108. doi 10.1111/nph.12336
Zhang, D.-Q. and Zhou, N., Genetic diversity and population structure of the endangered conifer Taxus wallichiana var. mairei (Taxaceae) revealed by Simple Sequence Repeat (SSR) markers, Biochem. Syst. Ecol., 2013, vol. 49, pp. 107–114. doi 10.1016/j.bse.2013.03.030
Miao, Y.C., Lang, X.D., Zhang, Z.Z., and Su, J.R., Phylogeography and genetic effects of habitat fragmentation on endangered Taxus yunnanensis in southwest China as revealed by microsatellite data, Plant Biol., 2014, vol. 16, pp. 365–374. doi 10.1111/plb.12059
Collins, D., Mill, R.R., and Möller, M., Species separation of Taxus baccata, T. canadensis, and T. cuspidata (Taxaceae) and origins of their reputed hybrids from RAPD and cpDNA data, Am. J. Bot., 2003, vol. 90, no. 2, pp. 175–182. doi 10.3732/ajb.90.2.175
So, S., Hwang, Y., Lee, C., et al., Taxonomic position of Taxus cuspidata var. latifolia endemic to Ulleung Island, Korean J. Pl. Taxon., 2013, vol. 43, no. 1, pp. 46–55. doi 10.11110/kjpt.2013.43.1.46
Hao, D.C., Xiao, P.G., Huang, B., et al., Interspecific relationships and origins of Taxaceae and Cephalotaxaceae revealed by partitioned Bayesian analyses of chloroplast and nuclear DNA sequences, Plant Syst. Evol., 2008, vol. 276, pp. 89–104. doi 10.1007/s00606-008-0069-0
Hao, D.C., Huang, B., and Yang, L., Phylogenetic relationships of the genus Taxus inferred from chloroplast intergenic spacer and nuclear coding DNA, Biol. Pharm. Bull., 2008, vol. 31, no. 2, pp. 260–265.
Hao, D.C., Huang, B.L., Chen, S.L., and Mu, J., Evolution of the chloroplast trnL–trnF region in the gymnosperm lineages Taxaceae and Cephalotaxaceae, Biochem. Genet., 2009, vol. 47, pp. 351–369. doi 10.1007/s10528-009-9233-7
Liu, J., Provan, J., Gao, L.-M., and Li, D.-Z., Sampling strategy and potential utility of indels for DNA barcoding of closely related plant species: a case study in Taxus, Int. J. Mol. Sci., 2012, vol. 13, no. 7, pp. 8740–8751. doi 10.3390/ijms13078740
Zhang, Y., Ma, J., Yang, B., et al., The complete chloroplast genome sequence of T. chinensis var. mairei (Taxaceae): loss of an inverted repeat region and comparative analysis with related species, Gene, 2014, vol. 540, pp. 201–209. doi 10.1016/j.gene.2014.02.037
Anderson, E.D. and Owens, J.N., Megagametophyte development, fertilization, and cytoplasmic inheritance in Taxus brevifolia, Int. J. Pl. Sci., 1999, vol. 160, pp. 459–469.
Artyukova, E.V., Kholina, A.B., Kozyrenko, M.M., and Zhuravlev, Yu.N., Analysis of genetic variation in rare endemic species Oxytropis chankaensis Jurtz. (Fabaceae) using RAPD markers, Russ. J. Genet., 2004, vol. 40, no. 7, pp. 710–716.
Taberlet, P., Gielly, L., Pautou, G., and Bouvet, J., Universal primers for amplification of three non-coding regions of chloroplast DNA, Plant Mol. Biol., 1991, vol. 17, no. 5, pp. 1105–1109.
Shaw, J., Lickey, E.B., Beck, J.T., et al., The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis, Am. J. Bot., 2005, vol. 92, pp. 142–166. doi 10.3732/ajb.92.1.142
Bonfeld, J.K., Smith, K.F., and Staden, R., A new DNA sequence assembly program, Nucleic Acids Res., 1995, vol. 23, pp. 4992–4999. doi 10.1093/nar/23.24.4992
Gouy, M., Guindon, S., and Gascuel, O., SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building, Mol. Biol. Evol., 2010, vol. 27, pp. 221–224. doi 10.1093/molbev/msp259
Excoffier, L. and Lischer, H.E.L., Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows, Mol. Ecol. Resour., 2010, vol. 10, pp. 564–567. doi 10.1111/j.1755-0998.2010.02847.x
Librado, P. and Rozas, J., DnaSP v5: a software for comprehensive analysis of DNA polymorphism data, Bioinformatics, 2009, vol. 25, no. 11, pp. 1451–1452. doi 10.1093/bioinformatics/btp187
Bandelt, H.-J., Forster, P., and Röhl, A., Median-joining networks for inferring intraspecific phylogenies, Mol. Biol. Evol., 1999, vol. 16, no. 1, pp. 37–48.
Gaudeul, M., Taberlet, P., and Till-Bottraud, I., Genetic diversity in an endangered alpine plant,Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers, Mol. Ecol., 2000, vol. 9, pp. 1625–1637. doi 10.1046/j.1365-294x.2000.01063.x
Nybom, H., Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants, Mol. Ecol., 2004, vol. 13, pp. 1143–1155. doi 10.1111/j.1365-294X.2004.02141.x
Golubeva, L.V. and Karaulova, L.P., Rastitel’nost’ i klimatostratigrafiya pleistotsena i golotsena Dal’nego Vostoka (Vegetation and Climatic Stratigraphy of the Pleistocene and Holocene of the Far East), Moscow: Nauka, 1983.
Hamrick, J.L. and Godt, M.J.W., Effects of life history traits on genetic diversity in plant species, Philos. Trans. R. Soc., B, 1996, vol. 351, pp. 1291–1298. doi 10.1098/rstb.1996.0112
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © M.M. Kozyrenko, E.V. Artyukova, E.A. Chubar, 2017, published in Genetika, 2017, Vol. 53, No. 8, pp. 911–921.
Rights and permissions
About this article
Cite this article
Kozyrenko, M.M., Artyukova, E.V. & Chubar, E.A. Genetic diversity and population structure of Taxus cuspidata Sieb. et Zucc. ex Endl. (Taxaceae) in Russia according to data of the nucleotide polymorphism of intergenic spacers of the chloroplast genome. Russ J Genet 53, 865–874 (2017). https://doi.org/10.1134/S1022795417070079
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795417070079