Abstract
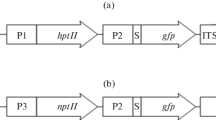
Genetic transformation of higher eukaryote mitochondria in vivo is an unresolved and important problem. For efficient expression of foreign genetic material in mitochondria, it is necessary to select regulatory elements that provide a high level of transcription and transcript stability. This work is aimed at studying the effectiveness of regulatory elements of mitochondrial genes flanking exogenous DNA using the phenomenon of natural competence of plant mitochondria. For this purpose, genetic constructs carrying the GFP gene under the control of the promoter regions of the RRN26 or COX1 genes and one of the two 3'-untranslated regions (3'-UTR) of mitochondrial genes were imported into isolated Arabidopsis mitochondria, followed by transcription in organello. It was shown that the level of GFP expression under the control of promoters of the RRN26 or COX1 genes in organello correlates with the level of transcription of these genes observed in vivo. At the same time, the presence of the tRNATrp sequence in the 3'-UTR leads to a higher level of the GFP transcript than the presence in this region of the 3'-UTR of the NAD4 gene containing the binding site of the MTSF1 protein. The results we obtained open prospects for creating a system for efficient transformation of the mitochondrial genome.




Similar content being viewed by others
REFERENCES
Larosa V., Remacle C. 2013. Transformation of the mitochondrial genome. Int. J. Dev. Biol. 57, 659–665. https://doi.org/10.1387/ijdb.130230cr
Remacle C., Larosa V., Salinas T., Hamel P., Subrahmanian N., Bonnefoy N., Kempken F. 2012. Transformation and nucleic acid delivery to mitochondria. In Genomics of Chloroplasts and Mitochondria. Advances in Photosynthesis and Respiration. 35. Bock R., Knoop V., Eds. Dordrecht: Springer. https://doi.org/10.1007/978-94-007-2920-9_19
Hammani K., Giegé P. 2014. RNA metabolism in plant mitochondria. Trends Plant Sci. 19, 380‒389. https://doi.org/10.1016/j.tplants.2013.12.008
Konstantinov Y.M., Dietrich A., Weber-Lotfi F., Ibrahim N., Klimenko E.S., Tarasenko V.I., Bolotova T.A., Koulintchenko M.V. 2016. DNA import into mitochondria. Biochemistry (Moscow). 81, 1044–1056. https://doi.org/10.1134/S0006297916100035
Koulintchenko M., Temperley R.J., Mason P.A., Dietrich A., Lightowlers R.N. 2006. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum. Mol. Genet. 15, 143–154. https://doi.org/10.1093/hmg/ddi435
Tarasenko T.A., Klimenko E.S., Tarasenko V.I., Koulintchenko M.V., Dietrich A., Weber-Lotfi F., Konstantinov Y.M. 2021. Plant mitochondria import DNA via alternative membrane complexes involving various VDAC isoforms. Mitochondrion. 60, 43‒58. https://doi.org/10.1016/j.mito.2021.07.006
Kühn K., Weihe A., Börner T. 2005. Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res. 33, 337–346. https://doi.org/10.1093/nar/gki179
Kühn K., Richter U., Meyer E., Delannoy E., de Longevialle A.F., O′Toole N., Börner T., Millar A., Small I., Whelan J. 2009. Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell. 21, 2762–2779. https://doi.org/10.1105/tpc.109.068536
Moller I.M., Rasmusson A.G., Van Aken O. 2021. Plant mitochondria—past, present and future. Plant J. 108, 912–959. https://doi.org/10.1111/tpj.15495
Holec S., Lange H., Kuhn K., Alioua M., Borner T., Gagliardi D. 2006. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 26, 2869–2876, https://doi.org/10.1128/MCB.26.7.2869-2876.2006
Perrin R., Meyer E.H., Zaepfel M., Kim Y.J., Mache R., Grienenberger J.M., Gualberto J.M., Gagliardi D. 2004. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 279, 25440–25446. https://doi.org/10.1074/jbc.M401182200
Haïli N., Arnal N., Quadrado M., Amiar S., Tcherkez G., Dahan J., Briozzo P., Colas des Francs-Small C., Vrielynck N., Mireau H. 2013. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 41, 6650–6663. https://doi.org/10.1093/nar/gkt337
Ruwe H., Wang G., Gusewski S., Schmitz-Linneweber C. 2016. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 44, 7406–7417. https://doi.org/10.1093/nar/gkw466
Forner J., Weber B., Thuss S., Wildum S., Binder S. 2007. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: T-elements contribute to 5′ and 3′‑end formation. Nucleic Acids Res. 35, 3676–3692. https://doi.org/10.1093/nar/gkm270
MacIntosh G.C., Castandet B. 2020. Organellar and secretory ribonucleases: major players in plant RNA homeostasis. Plant Physiol. 183, 1438–1452. https://doi.org/10.1104/pp.20.00076
Dombrowski S., Brennicke A., Binder S. 1997. 3′-Inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators. EMBO J. 16, 5069–5076. https://doi.org/10.1093/emboj/16.16.5069
Kuhn J., Tengler U., Binder S. 2001. Transcript lifetime is balanced between stabilizing stem-loop structures and degradation promoting polyadenylation in plant mitochondria. Mol. Cell. Biol. 21, 731–742. https://doi.org/10.1128/MCB.21.3.731-742.2001
Wang C., Aubé F., Planchard N., Quadrado M., Dargel-Graffin C., Nogué F., Mireau H. 2017. The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3′ end of its 5′-half intron. Nucleic Acids Res. 45, 6119‒6134 https://doi.org/10.1093/nar/gkx162
Wang C., Blondel L., Quadrado M., Dargel-Graffin C., Mireau H. 2022. Pentatricopeptide repeat protein MIT-CHONDRIAL STABILITY FACTOR 3 ensures mitochondrial RNA stability and embryogenesis. Plant Physiol. 190, 669‒681. https://doi.org/10.1093/plphys/kiac309
Koulintchenko M., Konstantinov Y., Dietrich A. 2003. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 22, 1245–1254. https://doi.org/10.1093/emboj/cdg128
Sweetlove L.J., Taylor N.L., Leaver C.J. 2007. Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol. Biol. 372, 125–136. https://doi.org/10.1007/978-1-59745-365-3_9
Bradford M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Douce R., Neuburger M. 1989. The uniqueness of plant mitochondria. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40, 371–414. https://doi.org/10.1146/annurev.pp.40.060189.002103
Tarasenko T.A., Subota I.Yu., Tarasenko V. I., Konstantinov Y.M., Koulintchenko M.V. 2020. Plant mitochondrial subfractions have different ability to import DNA. Theor. Exp. Plant Physiol. 32, 5–18. https://doi.org/10.1007/s40626-020-00167-w
Tarasenko T.A., Tarasenko V.I., Koulintchenko M.V., Klimenko E.S., Konstantinov Y.M. 2019. DNA import into plant mitochondria: complex approach for in organello and in vivo studies. Biochemistry (Moscow). 84, 817–828. https://doi.org/10.1134/S0006297919070113
Farré J.C., Araya A. 2001. Gene expression in isolated plant mitochondria: High fidelity of transcription, splicing and editing of a transgene product in electroporated organelles. Nucleic Acids Res. 29, 2484–2491. https://doi.org/10.1093/nar/29.12.2484
Tarasenko V.I., Katyshev A.I., Yakovleva T.V., Garnik E.Y., Chernikova V.V., Konstantinov Y.M., Koulintchenko M.V. 2016. RPOTmp, an Arabidopsis RNA polymerase with dual targeting, plays an important role in mitochondria, but not in chloroplasts. J. Exp. Bot. 67, 5657–5669. https://doi.org/10.1093/jxb/erw327
Kühn K., Bohne A.V., Liere K., Weihe A., Börner T. 2007. Arabidopsis phage-type RNA polymerases: accurate in vitro transcription of organellar genes. Plant Cell. 19, 959–971. https://doi.org/10.1105/tpc.106.046839
Binder S., Hatzack F., Brennicke A. 1995. A novel pea mitochondrial in vitro transcription system recognizes homologous and heterologous mRNA and tRNA promoters. J. Biol. Chem. 270, 22182‒2218. https://doi.org/10.1074/jbc.270.38.22182
Rovira A.G., Smith A.G. 2019. PPR proteins—orchestrators of organelle RNA metabolism. Physiol. Plant. 166, 451‒459. https://doi.org/10.1111/ppl.12950
Hanic-Joyce P.J., Gray M.W. 1991. Accurate transcription of a plant mitochondrial gene in vitro. Mol. Cell Biol. 11, 2035‒2039. https://doi.org/10.1128/mcb.11.4.2035-2039.1991
Attardi G., Chomyn A., King M.P., Kruse B., Polosa P.L., Murdter N.N. 1990. Regulation of mitochondrial gene expression in mammalian cells. Biochem. Soc. Trans. 18, 509‒513. https://doi.org/10.1042/bst0180509
Micol V., Fernández-Silva P., Attardi G. 1997. Functional analysis of in vivo and in organello footprinting of HeLa cell mitochondrial DNA in relationship to ATP and ethidium bromide effects on transcription. J. Biol. Chem. 272, 18896‒18904. https://doi.org/10.1074/jbc.272.30.18896
Kotrys A.V., Szczesny R.J. 2019. Mitochondrial gene expression and beyond-novel aspects of cellular physiology. Cells. 9, 17. https://doi.org/10.3390/cells9010017
Newton K.J., Winberg B., Yamato K., Lupold S., Stern D.B. 1995. Evidence for a novel mitochondrial promoter preceding the cox2 gene of perennial teosintes. EMBO J. 14, 585‒593. https://doi.org/10.1002/j.1460-2075.1995.tb07034.x
Xiao S., Zang J., Pei Y., Liu J., Liu J., Song W., Shi Z., Su A., Zhao J., Chen H. 2020. Activation of mitochondrial orf355 gene expression by a nuclear-encoded DREB transcription factor causes cytoplasmic male sterility in maize. Mol. Plant. 13, 1270‒1283. https://doi.org/10.1016/j.molp.2020.07.002
Tarasenko V.I., Subota I.Yu., Kobzev V.F., Konstantinov Yu.M. 2005. Isolation of mitochondrial DNA-binding proteins specific to the maize cox1 promoter. Mol. Biol. (Moscow). 39 (3), 350‒356. )https://doi.org/10.1007/s11008-005-0049-1
Rapp W.D., Stern D.B. 1992. A conserved 11 nucleotide sequence contains an essential promoter element of the maize mitochondrial atp1 gene. EMBO J. 11, 1065‒1073. https://doi.org/10.1002/j.1460-2075.1992.tb05145.x
ACKNOWLEDGMENTS
The equipment of the Bioanalytika Center for Collective Use, Siberian Institute of Plant Physiology and Biochemistry of the Siberian Branch of the Russian Academy of Sciences (Irkutsk) was used in the work.
Funding
The study was supported by the Russian Science Foundation grant No. 22-74-00114, https://rscf.ru/project/22-74-00114/.
Author information
Authors and Affiliations
Contributions
V.I. Tarasenko and T.A. Tarasenko contributed equally to this study.
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest. This article does not contain a description of studies performed by the authors involving humans or using animals as objects.
Rights and permissions
About this article
Cite this article
Tarasenko, V.I., Tarasenko, T.A., Gorbenko, I.V. et al. Differential Expression of a Foreign Gene in Arabidopsis Mitochondria In Organello. Mol Biol 57, 447–456 (2023). https://doi.org/10.1134/S0026893323030123
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893323030123