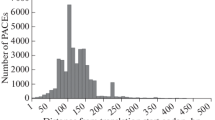
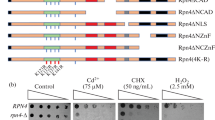
Abstract
In eukaryotes, the ubiquitin-proteasome proteolytic system is involved in metabolizing most of cell proteins and in major regulation pathways. Although its structure and functioning are fairly well studied, little is known about the regulation of gene expression in this system. The regulation of proteasome gene expression has been described only for Saccharomyces cerevisiae and includes the proteasome-associated transcription factor Rpn4p and its binding site, the proteasome-associated control element (PACE). There are two questions concerning the role of Rpn4p as a transcription factor: whether Rpn4p regulates the PACE-containing genes of the protein ubiquitination system and what contribution Rpn4p makes to the stress-induced changes in proteasome mRNA levels. Semiquantitative RT-PCR showed that deletion of S. cerevisiae RPN4 decreased the RAD6, RAD23, and CDC48 mRNA levels, while the UBI4 mRNA level increased. Stress factors, such as heat shock or the alkylating agent methyl methanesulfonate, induced Rpn4p-dependent transcriptional upregulation of RPT4 and RPN5. At the same time, methyl methanesulfonate downregulated the CDC48 expression in the wild-type strain. Apparently, Rpn4p acts both as an activator and a repressor of transcription of the ubiquitinproteasome genes under normal and stress conditions.
Similar content being viewed by others
References
Voges D., Zwickl P., Baumeister W. 1999. The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068.
Coux O., Tanaka K., Goldberg A.L. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847.
Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479.
Varshavsky A. 1997. The ubiquitin system. Trends Biochem. Sci. 22, 383–387.
Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 404, 770–774.
Ferrel K., Wilkinson C.R., Dubiel W., Gordon C. 2000. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Cell Biol. 25, 83–88.
Baboshina O.V., Haas A.L. 1996. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2-EPF and RAD6 are recognized by the 26S proteasome subunit 5. J. Biol. Chem. 271, 2823–2831.
Thrower J.S., Hoffman L., Rechsteiner M., Pickart C. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102.
Glickmann M., Rubin D.M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumaister W., Fried V.A., Finley D. 1998. A subcomplex of the proteasome regulator particle required for ubiquitin-conjugate degradation and related to the COP9-Signalosome and eIF3. Cell. 94, 615–623.
Lam Y., Lowson T.G., Velayutham M., Zweier J.L., Pickart C.M. 2002. A protaasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 18, 763–767.
Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., Koonin E.V., Deshaies R.J. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 298, 611–615.
Groll M., Ditzel L., Lowe J., Stock D., Bochtler M., Bartunik H.D., Huber R. 1997. Structure of 20S proteasome from yeast at a 2.4 Å resolution. Nature. 386, 463–471.
Lowe J., Stock D., Jap B., Zwickl P., Baumeister W., Huber R. 1995. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 268, 533–539.
Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. 1999. Rpn4 acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450, 27–34.
Kapranov A.B., Kuryatova M.V., Preobrazhenskaya O.V., Tutyaeva V.V., Stucka R., Feldmann H., Karpov V.L. 2001. Isolation and identification of PACE-binding protein Rpn4, a new transcriptional activator regulating 26S-proteasomal and other genes. Mol. Biol. 35, 420–431.
Fujimuro M., Tanaka K., Yokosawa H., Toh-e A. 1998. Son1p is a component of the 26S proteasome of the yeast Saccharomyces cerevisiae. FEBS Lett. 423, 149–154.
Glickman M.H., Ciechanover A. 2002. Ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82, 373–428.
Prinz S., Avila-Campillo I., Aldridge C., Srinivasan A., Dimitrov K., Siegel A.F., Galitski T. 2004. Control of yeast filamentous-form growth by modules in an integrated molecular network. Genome Res. 14, 380–390.
Xie Y., Varshavsky A. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: A negative feedback circuit. Proc. Natl. Acad. Sci. USA. 98, 3056–3061.
Nelson M.K., Kurihara T., Silver P.A. 1993. Extragenic suppressors of mutations in the cytoplasmic C terminus of Sec63 define five genes in Saccharomyces cerevisae. Genetics. 134, 159–173 nerevisae. Genetics J. 134, 159–173.
Johnston E. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456.
Ng D.T., Spear E.D., Walter P. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150, 77–88.
Ju D., Wang L., Mao X., Xie Y. 2004. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem. Biophys. Res. Commun. 321, 51–57.
London M.K., Keck B.I., Ramos P.C., Dohmen R.J. 2004. Regulatory mechanisms controlling biogenesis of ubiquitin and the proteasome. FEBS Lett. 567, 259–264.
Owsianik G., Balzil L., Ghislain M. 2002. Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol. Microbiol. 43, 1295–1308.
Hahn J.S., Neef D.W., Thiele D.J. 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 60, 240–251.
Schmitt M.E., Brown T.A., Trumpower B.L. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091–3092.
Jelinsky S.A., Estep P., Church G.M., Samson L.D. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell. Biol. 20, 8157–8167.
Sung P., Berleth E., Pickart C., Prakash S., Prakash L. 1991. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 10, 2187–2193.
Prakash S., Sung P., Prakash L. 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27, 33–70.
Jelinsky S.A., Leona D.S. 1999. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA. 96, 1486–1491.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © D.S. Karpov, S.A. Osipov, O.V. Preobrazhenskaya, V.L. Karpov, 2008, published in Molekulyarnaya Biologiya, 2008, Vol. 42, No. 3, pp. 518–525.
Rights and permissions
About this article
Cite this article
Karpov, D.S., Osipov, S.A., Preobrazhenskaya, O.V. et al. Rpn4p is a positive and negative transcriptional regulator of the ubiquitin-proteasome system. Mol Biol 42, 456–462 (2008). https://doi.org/10.1134/S0026893308030151
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893308030151