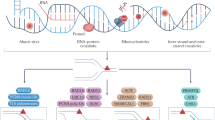
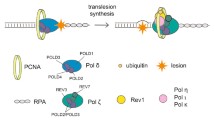
Abstract
Many chemotherapy drugs block tumor cell division by damaging DNA. DNA polymerases eta (Pol η), iota (Pol ι), kappa (Pol κ), REV1 of the Y-family and zeta (Pol ζ) of the B-family efficiently incorporate nucleotides opposite a number of DNA lesions during translesion DNA synthesis. Primase-polymerase PrimPol and the Pol α-primase complex reinitiate DNA synthesis downstream of the damaged sites using their DNA primase activity. These enzymes can decrease the efficacy of chemotherapy drugs, contribute to the survival of tumor cells and to the progression of malignant diseases. DNA polymerases are promising targets for increasing the effectiveness of chemotherapy, and mutations and polymorphisms in some DNA polymerases can serve as additional prognostic markers in a number of oncological disorders.


Similar content being viewed by others
Notes
* Xenograft – an experimental cancer model based on human tumor cells transplanted into animals (e.g., mice).
Abbreviations
- HR:
-
hazard ratio
- OR:
-
odds ratio
- Pol:
-
DNA-polymerase
- RIR:
-
REV1 interacting region
- UBM2:
-
ubiquitin-binding motif 2
- XP-V:
-
xeroderma pigmentosum variant
REFERENCES
Burgers, P. M. J., and Kunkel, T. A. (2017) Eukaryotic DNA replication fork, Annu. Rev. Biochem., 86, 417-438, doi: https://doi.org/10.1146/annurev-biochem-061516-044709.
Kunkel, T. A. (2009) Evolving views of DNA replication (in)fidelity, Cold Spring Harb. Symp. Quant. Biol., 74, 91-101, doi: https://doi.org/10.1101/sqb.2009.74.027.
Friedberg, E. C., Walker, G. C., Siede, W., Wood, R. D., Schultz, R. A., and Ellenberger, T. (2006) DNA Repair and Mutagenesis. Second Edition, ASM Press, Washington (DC).
Makarova, A. V., and Burgers, P. M. (2015) Eukaryotic DNA polymerase ζ, DNA Rep. (Amst), 29, 47-55, doi: https://doi.org/10.1016/j.dnarep.2015.02.012.
Rizzo, A. A., and Korzhnev, D. M. (2019) The Rev1-Pol ζ translesion synthesis mutasome: structure, interactions and inhibition, Enzymes, 45, 139-181, doi: https://doi.org/10.1016/bs.enz.2019.07.001.5.
Sale, J. E., Lehmann, A. R., and Woodgate, R. (2012) Y-family DNA polymerases and their role in tolerance of cellular DNA damage, Nat. Rev. Mol. Cell Biol., 13, 141-152, doi: https://doi.org/10.1038/nrm3289.
Yang, W., and Woodgate, R. (2007) What a difference a decade makes: insights into translesion DNA synthesis, Proc. Natl. Acad. Sci. USA, 104, 15591-15598, doi: https://doi.org/10.1073/pnas.0704219104.
Guilliam, T. A., Jozwiakowski, S. K., Ehlinger, A., Barnes, R. P., Rudd, S. G., Bailey, L. J., Skehel, M., Eckert, K. A., Chazin, W. J., and Doherty, A. J. (2015) Human PrimPol is a highly error-prone polymerase regulated by single-stranded DNA binding proteins, Nucleic Acids Res., 43, 1056-1068, doi: https://doi.org/10.1093/nar/gku1321.
Martínez-Jiménez, M. I., Lahera, A., and Blanco, L. (2017) Human PrimPol activity is enhanced by RPA, Sci. Rep., 7, 783, doi: https://doi.org/10.1038/s41598-017-00958-3.
Shilkin, E. S., Boldinova, E. O., Stolyarenko, A. D., Goncharova, R. I., Chuprov-Netochin, R. N., Khairullin, R. F., Smal, M. P., and Makarova, A. V. (2020) Translesion DNA synthesis and carcinogenesis, Biochemistry (Moscow), 85, 494-506, doi: https://doi.org/10.31857/S032097252004003X.
Siddik, Z. H. (2003) Cisplatin: mode of cytotoxic action and molecular basis of resistance, Oncogene, 22, 7265-7279, doi: https://doi.org/10.1038/sj.onc.1206933.
Gately, D., and Howell, S. (1993) Cellular accumulation of the anticancer agent cisplatin: a review, Br. J. Cancer, 67, 1171-1176, doi: https://doi.org/10.1038/bjc.1993.221.
Zhou, J., Kang, Y., Chen, L., Wang, H., Liu, J., Zeng, S., and Yu, L. (2020) The drug-resistance mechanisms of five platinum-based antitumor agents, Front. Pharmacol., 11, 1-17, doi: https://doi.org/10.3389/fphar.2020.00343.
Bancroft, D. P., Lepre, C. A., and Lippard, S. J. (1990) Platinum-195 NMR kinetic and mechanistic studies of cis- and trans-diamminedichloroplatinum(II) binding to DNA, J. Am. Chem. Soc., 112, 6860-6871, doi: https://doi.org/10.1021/ja00175a020.
Fichtinger-Schepman, A. M. J., Lohman, P. H. M., Berends, F., and van Oosterom, A. T. (1987) Cis-diamminedichloroplatinum(ii)-induced DNA adducts in peripheral leukocytes from seven cancer patients: quantitative immunochemical detection of the adduct induction and removal after a single dose of cis-diamminedichloroplatinum(ii), Cancer Res., 47, 3000-3004.
Plooy, A. C. M., Fichtinger-Schepman, A. M. J., Schutte, H. H., Van Dijk, M., and Lohman, P. H. M. (1985) The quantitative detection of various Pt-DNA-adducts in chinese hamster ovary cells treated with cisplatin: application of immunochemical techniques, Carcinogenesis, 6, 561-566, doi: https://doi.org/10.1093/carcin/6.4.561.
Cummings, J., Spanswick, V. J., and Smyth, J. F. (1995) Re-evaluation of the molecular pharmacology of mitomycin C, Eur. J. Cancer, 31, 1928-1933, doi: https://doi.org/10.1016/0959-8049(95)00364-9.
Tomasz, M., (1995) Mitomycin C: small, fast and deadly (but very selective), Chem. Biol., 2, 575-579, doi: https://doi.org/10.1016/1074-5521(95)90120-5.
Umar, G. S., Lipman, R., Cummings, J., and Tomasz, M. (1997) Mitomycin cDNA adducts generated by DT-diaphorase. revised mechanism of the enzymatic reductive activation of mitomycin C, Biochemistry, 36, 14128-14136, doi: https://doi.org/10.1021/bi971394i.
Warren, A. J., Maccubbin, A. E., and Hamilton, J. W. (1998) Detection of mitomycin C DNA adducts in vivo by 32P-postlabeling: time course for formation and removal of adducts and biochemical modulation, Cancer Res., 58, 453-461.
Tomasz, M., Lipman, R., Chowdary, D., Pawlak, J., Verdine, G., and Nakanishi, K. (1987) Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA, Science, 235, 1204-1208, doi: https://doi.org/10.1126/science.3103215.
Dong, Q., Barsky, D., Colvin, M. E., Melius, C. F., Ludeman, S. M., Moravek, J. F., Colvin, O. M., Bigner, D. D., Modrich, P., and Friedman, H. S. (1995) A structural basis for a phosphoramide mustard-induced DNA interstrand cross-link at 5′-d(GAC), Proc. Natl. Acad. Sci. USA, 92, 12170-12174, doi: https://doi.org/10.1073/pnas.92.26.12170.
Rink, S. M., Solomon, M. S., Taylor, M. J., Hopkins, P. B., Rajur, S. B., and McLaughlin, L. W. (1993) Covalent structure of a nitrogen mustard-induced DNA interstrand cross-link: an N7-to-N7 Linkage of deoxyguanosine residues at the duplex sequence 5′-d(GNC), J. Am. Chem. Soc., 115, 2551-2557, doi: https://doi.org/10.1021/ja00060a001.
Rink, S. M., and Hopkins, P. B. (1995) A mechlorethamine-induced DNA interstrand cross-link Bends duplex DNA, Biochemistry, 34, 1439-1445, doi: https://doi.org/10.1021/bi00004a039.
Brandes, A. A., Bartolotti, M., Tosoni, A., and Franceschi, E. (2016) Nitrosoureas in the management of malignant gliomas, Curr. Neurol. Neurosci. Rep., 16, 13, doi: https://doi.org/10.1007/s11910-015-0611-8.
Nikolova, T., Roos, W. P., Krämer, O. H., Strik, H. M., and Kaina, B. (2017) Chloroethylating nitrosoureas in cancer therapy: DNA damage, repair and cell death signaling, Biochim. Biophys. Acta Rev. Cancer, 1868, 29-39, doi: https://doi.org/10.1016/j.bbcan.2017.01.004.
Chen, F. X., Bodell, W. J., Liang, G., and Gold, B. (1996) Reaction of N-(2-chloroethyl)-N-nitrosoureas with DNA: effect of buffers on DNA adduction, cross-linking, and cytotoxicity, Chem. Res. Toxicol., 9, 208-214, doi: https://doi.org/10.1021/tx950097g.
Denny, B. J., Tsang, L. L. H., Slack, J. A., Wheelhouse, R. T., and Stevens, M. F. G. (1994) NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA, Biochemistry, 33, 9045-9051, doi: https://doi.org/10.1021/bi00197a003.
Friedman, H. S., Kerby, T., and Calvert, H. (2000) Temozolomide and treatment of malignant glioma, Clin. Cancer Res., 6, 2585-2597.
Tentori, L., and Graziani, G. (2002) Pharmacological strategies to increase the antitumor activity of methylating agents, Curr. Med. Chem., 9, 1285-1301, doi: https://doi.org/10.2174/0929867023369916.
Tsesmetzis, N., Paulin, C. B. J., Rudd, S. G., and Herold, N. (2018) Nucleobase and nucleoside analogues: resistance and re-sensitisation at the level of pharmacokinetics, pharmacodynamics and metabolism, Cancers (Basel), 10, 240, doi: https://doi.org/10.3390/cancers10070240.
Berdis, A. J. (2017) Inhibiting DNA polymerases as a therapeutic intervention against cancer, Front. Mol. Biosci., 4, 1-12, doi: https://doi.org/10.3389/fmolb.2017.00078.
Mini, E., Nobili, S., Caciagli, B., Landini, I., and Mazzei, T. (2006) Cellular pharmacology of gemcitabine, Ann. Oncol., 17, 7-12, doi: https://doi.org/10.1093/annonc/mdj941.
Liang, X., Wu, Q., Luan, S., Yin, Z., He, C., Yin, L., Zou, Y., Yuan, Z., Li, L., Song, X., He, M., Lv, C., and Zhang, W. (2019) A comprehensive review of topoisomerase inhibitors as anticancer agents in the past decade, Eur. J. Med. Chem., 171, 129-168, doi: https://doi.org/10.1016/j.ejmech.2019.03.034.
Minotti, G., Menna, P., Salvatorelli, E., Cairo, G., and Gianni, L. (2004) Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity, Pharmacol. Rev., 56, 185-229, doi: https://doi.org/10.1124/pr.56.2.6.
Masutani, C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K., and Hanaoka, F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta, Nature, 399, 700-704, doi: https://doi.org/10.1038/21447.
Alt, A., Lammens, K., Chiocchini, C., Lammens, A., Pieck, J. C., Kuch, D., Hopfner, K. P., and Carell, T. (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta, Science, 318, 967-970, doi: https://doi.org/10.1126/science.1148242.
Ouzon-Shubeita, H., Baker, M., Koag, M.-C., and Lee, S. (2019) Structural basis for the bypass of the major oxaliplatin-DNA adducts by human DNA polymerase η, Biochem. J., 476, 747-758, doi: https://doi.org/10.1042/BCJ20180848.
Ummat, A., Rechkoblit, O., Jain, R., Roy Choudhury, J., Johnson, R. E., Silverstein, T. D., Buku, A., Lone, S., Prakash, L., Prakash, S., and Aggarwal, A. K. (2012) Structural basis for cisplatin DNA damage tolerance by human polymerase η during cancer chemotherapy, Nat. Struct. Mol. Biol., 19, 628-632, doi: https://doi.org/10.1038/nsmb.2295.
Vaisman, A., Masutani, C., Hanaoka, F., and Chaney, S. G. (2000) Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta, Biochemistry, 39, 4575-4580, doi: https://doi.org/10.1021/bi000130k.
Zhao, Y., Biertümpfel, C., Gregory, M. T., Hua, Y. J., Hanaoka, F., and Yang, W. (2012) Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin, Proc. Natl. Acad. Sci. USA, 109, 7269-7274, doi: https://doi.org/10.1073/pnas.1202681109.
Shachar, S., Ziv, O., Avkin, S., Adar, S., Wittschieben, J., Reissner, T., Chaney, S., Friedberg, E. C., Wang, Z., Carell, T., Geacintov, N., and Livneh, Z. (2009) Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals, EMBO J., 28, 383-393, doi: https://doi.org/10.1038/emboj.2008.281.
Srivastava, A. K., Han, C., Zhao, R., Cui, T., Dai, Y., Mao, C., Zhao, W., Zhang, X., Yu, J., and Wang, Q. E. (2015) Enhanced expression of DNA polymerase eta contributes to cisplatin resistance of ovarian cancer stem cells, Proc. Natl. Acad. Sci. USA, 112, 4411-4416.
Li, X.-Q., Ren, J., Chen, P., Chen, Y.-J., Wu, M., Wu, Y., Chen, K., and Li, J. (2018) Co-inhibition of Pol η and ATR sensitizes cisplatin-resistant non-small cell lung cancer cells to cisplatin by impeding DNA damage repair, Acta Pharmacol. Sin., 39, 1359-1372, doi: https://doi.org/10.1038/aps.2017.187.
Zhang, J., Sun, W., Ren, C., Kong, X., Yan, W., and Chen, X. (2019) A PolH transcript with a short 3′UTR enhances PolH expression and mediates cisplatin resistance, Cancer Res., 79, 3714-3724, doi: https://doi.org/10.1158/0008-5472.CAN-18-3928.
Ceppi, P., Novello, S., Cambieri, A., Longo, M., Monica, V., Iacono, M. L., Giaj-Levra, M., Saviozzi, S., Volante, M., and Papotti, M. (2009) Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy, Clin. Cancer Res., 15, 1039-1045, doi: https://doi.org/10.1158/1078-0432.CCR-08-1227.
Teng, K., Qiu, M., Li, Z., Luo, H., Zeng, Z., Luo, R., Zhang, H., Wang, Z., Li, Y., and Xu, R. (2010) DNA polymerase η protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy, J. Transl. Med., 8, 126, doi: https://doi.org/10.1186/1479-5876-8-126.
Albertella, M. R., Green, C. M., Lehmann, A. R., and O’Connor, M. J. (2005) A role for polymerase η in the cellular tolerance to cisplatin-induced damage, Cancer Res., 65, 9799-806, doi: https://doi.org/10.1158/0008-5472.CAN-05-1095.
Sumiyoshi, M., Soda, H., Sadanaga, N., Taniguchi, H., Ikeda, T., Maruta, H., Dotsu, Y., Ogawara, D., Fukuda, Y., and Mukae, H. (2017) Alert regarding cisplatin-induced severe adverse events in cancer patients with xeroderma pigmentosum, Intern. Med., 56, 979-982, doi: https://doi.org/10.2169/internalmedicine.56.7866.
Chen, Y.-W., Cleaver, J. E., Hanaoka, F., Chang, C.-F., and Chou, K.-M. (2006) A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents, Mol. Cancer Res., 4, 257-265, doi: https://doi.org/10.1158/1541-7786.MCR-05-0118.
Rechkoblit, O., Choudhury, J. R., Buku, A., Prakash, L., Prakash, S., and Aggarwal, A. K. (2018) Structural basis for polymerase η-promoted resistance to the anticancer nucleoside analog cytarabine, Sci. Rep., 8, 12702, doi: https://doi.org/10.1038/s41598-018-30796-w.
Rechkoblit, O., Johnson, R. E., Buku, A., Prakash, L., Prakash, S., and Aggarwal, A. K. (2019) Structural insights into mutagenicity of anticancer nucleoside analog cytarabine during replication by DNA polymerase η, Sci. Rep., 9, 16400, doi: https://doi.org/10.1038/s41598-019-52703-7.
Yoon, J.-H., Choudhury, R. J., Prakash, L., and Prakash, S. (2019) Translesion synthesis DNA polymerases η, ι, and υ promote mutagenic replication through the anticancer nucleoside cytarabine, J. Biol. Chem., 294, 19048-19054, doi: https://doi.org/10.1074/jbc.RA119.011381.
Yasui, M., Suzuki, N., Laxmi, Y. S., and Shibutani, S. (2006) Translesion synthesis past tamoxifen-derived DNA adducts by human DNA polymerases eta and kappa, Biochemistry, 45, 12167-12174, doi: https://doi.org/10.1021/bi0608461.
McDonald, J. P., Rapić-Otrin, V., Epstein, J. A., Broughton, B. C., Wang, X., Lehmann, A. R., Wolgemuth, D. J., and Woodgate, R. (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase eta, Genomics, 60, 20-30, doi: https://doi.org/10.1006/geno.1999.5906.
Smith, L. A., Makarova, A. V., Samson, L., Thiesen, K. E., Dhar, A., and Bessho, T. (2012) Bypass of a psoralen DNA interstrand cross-link by DNA polymerases β, ι, and κ in vitro, Biochemistry, 51, 8931-8938, doi: https://doi.org/10.1021/bi3008565.
Minko, I. G., Harbut, M. B., Kozekov, I. D., Kozekova, A., Jakobs, P. M., Olson, S. B., Moses, R. E., Harris, T. M., Rizzo, C. J., and Lloyd, R. S. (2008) Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links, J. Biol. Chem., 283, 17075-17082, doi: https://doi.org/10.1074/jbc.M801238200.
Ho, V., Guainazzi, A., Derkunt, S. B., Enoiu, M., and Schärer, O. D. (2011) Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases, Nucleic Acids Res., 39, 7455-7464, doi: https://doi.org/10.1093/nar/gkr448.
Jha, V., and Ling, H. (2018) Structural basis for human DNA polymerase kappa to bypass cisplatin intrastrand cross-link (Pt-GG) lesion as an efficient and accurate extender, J. Mol. Biol., 430, 1577-1589, doi: https://doi.org/10.1016/j.jmb.2018.04.023.
Kanemaru, Y., Suzuki, T., Sassa, A., Matsumoto, K., Adachi, N., Honma, M., Numazawa, S., and Nohmi, T. (2017) DNA polymerase kappa protects human cells against MMC-induced genotoxicity through error-free translesion DNA synthesis, Genes Environ., 39, 6, doi: https://doi.org/10.1186/s41021-016-0067-3.
Takeiri, A., Wada, N. A., Motoyama, S., Matsuzaki, K., Tateishi, H., Matsumoto, K., Niimi, N., Sassa, A., Grúz, P., Masumura, K., Yamada, M., Mishima, M., Jishage, K. I., and Nohmi, T. (2014) In vivo evidence that DNA polymerase kappa is responsible for error-free bypass across DNA cross-links induced by mitomycin C, DNA Rep., 24, 113-121, doi: https://doi.org/10.1016/j.dnarep.2014.09.002.
Williams, H. L., Gottesman, M. E., and Gautier, J. (2012) Replication-independent repair of DNA interstrand crosslinks, Mol. Cell, 47, 140-147, doi: https://doi.org/10.1016/j.molcel.2012.05.001.
Zhuo, M., Gorgun, M. F., and Englander, E. W. (2018) Translesion synthesis DNA polymerase kappa is indispensable for DNA repair synthesis in cisplatin exposed dorsal root ganglion neurons, Mol. Neurobiol., 55, 2506-2515, doi: https://doi.org/10.1007/s12035-017-0507-5.
Shao, M., Jin, B., Niu, Y., Ye, J., Lu, D., and Han, B. (2014) Association of POLK polymorphisms with platinum-based chemotherapy response and severe toxicity in non-small cell lung cancer patients, Cell Biochem. Biophys., 70, 1227-1237, doi: https://doi.org/10.1007/s12013-014-0046-x.
Goričar, K., Kovač, V., and Dolžan, V. (2014) Polymorphisms in translesion polymerase genes influence treatment outcome in malignant mesothelioma, Pharmacogenomics, 15, 941-950, doi: https://doi.org/10.2217/pgs.14.14.
Ye, J., Chu, T., Li, R., Niu, Y., Jin, B., Xia, J., Shao, M., and Han, B. (2015) Pol ζ polymorphisms are associated with platinum based chemotherapy response and side effects among non-small cell lung cancer patients, Neoplasma, 62, 833-839, doi: https://doi.org/10.4149/neo_2015_101.
Goričar, K., Kovač, V., Jazbec, J., Zakotnik, B., Lamovec, J., and Dolžan, V. (2015) Translesion polymerase genes polymorphisms and haplotypes influence survival of osteosarcoma patients, OMICS, 19, 180-185, doi: https://doi.org/10.1089/omi.2014.0159.
Zheng, Y., Deng, Z., Yin, J., Wang, S., Lu, D., Wen, X., Li, X., Xiao, D., Hu, C., Chen, X., Zhang, W., Zhou, H., and Liu, Z. (2017) The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy, Int. J. Cancer, 141, 2336-2347, doi: https://doi.org/10.1002/ijc.30921.
Wang, J., Wang, X., Zhao, M., Choo, S. P., Ong, S. J., Ong, S. Y., Chong, S. S., Teo, Y. Y., and Lee, C. G. (2014) Potentially functional SNPs (pfSNPs) as novel genomic predictors of 5-FU response in metastatic colorectal cancer patients, PLoS One, 9, e111694, doi: https://doi.org/10.1371/journal.pone.0111694.
Huang, K. K., Jang, K. W., Kim, S., Kim, H. S., Kim, S.-M., Kwon, H. J., Kim, H. R., Yun, H. J., Ahn, M. J., and Park, K. U. (2016) Exome sequencing reveals recurrent REV3L mutations in cisplatin-resistant squamous cell carcinoma of head and neck, Sci. Rep., 6, 19552, doi: https://doi.org/10.1038/srep19552.
Xu, H. L., Gao, X. R., Zhang, W., Cheng, J. R., Tan, Y. T., Zheng, W., Shu, X. O., and Xiang, Y. B. (2013) Effects of polymorphisms in translesion DNA synthesis genes on lung cancer risk and prognosis in Chinese men, Cancer Epidemiol., 37, 917-922, doi: https://doi.org/10.1016/j.canep.2013.08.003.
Baranovskiy, A. G., Lada, A. G., Siebler, H. M., Zhang, Y., Pavlov, Y. I., and Tahirov, T. H. (2012) DNA polymerase δ and ζ switch by sharing accessory subunits of DNA polymerase δ, J. Biol. Chem., 287, 17281-17287, doi: https://doi.org/10.1074/jbc.M112.351122.
Lee, Y. S., Gregory, M. T., and Yang, W. (2014) Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass, Proc. Natl. Acad. Sci. USA, 111, 2954-2959, doi: https://doi.org/10.1073/pnas.1324001111.
Makarova, A. V., Stodola, J. L., and Burgers, P. M. (2012) A four-subunit DNA polymerase ζ complex containing Pol δ accessory subunits is essential for PCNA-mediated mutagenesis, Nucleic Acids Res., 40, 11618-11626, doi: https://doi.org/10.1093/nar/gks948.
Rizzo, A. A., Vassel, F.-M., Chatterjee, N., D’Souza, S., Li, Y., Hao, B., Hemann, M. T., Walker, G. C., and Korzhnev, D. M. (2018) Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex, Proc. Natl. Acad. Sci. USA, 115, 8191-8200, doi: https://doi.org/10.1073/pnas.1801149115.
Adachi, M., Ijichi, K., Hasegawa, Y., Ogawa, T., Nakamura, H., Yasui, Y., Fukushima, M., and Ishizaki, K. (2008) Hypersensitivity to cisplatin after hRev3 mRNA knockdown in head and neck squamous cell carcinoma cells, Mol. Med. Rep., 1, 695-698, doi: https://doi.org/10.3892/mmr_00000015.
Chen, X., Zhu, H., Ye, W., Cui, Y., and Chen, M. (2019) MicroRNA-29a enhances cisplatin sensitivity in non-small cell lung cancer through the regulation of REV3L, Mol. Med. Rep., 19, 831-840, doi: https://doi.org/10.3892/mmr.2018.9723.
Lin, X., Trang, J., Okuda, T., Stephen, B., and Howell, S. B. (2006) DNA polymerase zeta accounts for the reduced cytotoxicity and enhanced mutagenicity of cisplatin in human colon carcinoma cells that have lost DNA mismatch repair, Clin. Cancer Res., 12, 563-568, doi: https://doi.org/10.1158/1078-0432.CCR-05-1380.
Song, L., McNeil, E. M., Ritchie, A.-M., Astell, K. R., Gourley, C., and Melton, D. W. (2017) Melanoma cells replicate through chemotherapy by reducing levels of key homologous recombination protein RAD51 and increasing expression of translesion synthesis DNA polymerase ζ, BMC Cancer, 17, 864, doi: https://doi.org/10.1186/s12885-017-3864-6.
Wang, H., Zhang, S.-Y., Wang, S, Lu, J., Wu, W., Weng, L., Chen, D., Zhang, Y., Lu, Z., Yang, J., Chen, Y., Zhang, X., Chen, X., Xi, C., Lu, D., and Zhao, S. (2009) REV3L confers chemoresistance to cisplatin in human gliomas: the potential of its RNAi for synergistic therapy, Neuro Oncol., 11, 790-802, doi: https://doi.org/10.1215/15228517-2009-015.
Wang, W., Sheng, W., Yu, C., Cao, J., Zhou, J., Wu, J., Zhang, H., and Zhang, S. (2015) REV3L modulates cisplatin sensitivity of non-small cell lung cancer H1299 cells, Oncol. Rep., 34, 1460-1468, doi: https://doi.org/10.3892/or.2015.4121.
Wu, F., Lin, X., Okuda, T., and Howell, S. B. (2004) DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance, Cancer Res., 64, 8029-8035, doi: https://doi.org/10.1158/0008-5472.CAN-03-3942.
Zhu, X., Zou, S., Zhou, J., Zhu, H., Zhang, S., Shang, Z., Ding, W.-Q., Wu, J., and Chen, Y. (2016) REV3L, the catalytic subunit of DNA polymerase ζ, is involved in the progression and chemoresistance of esophageal squamous cell carcinoma, Oncol. Rep., 35, 1664-1670, doi: https://doi.org/10.3892/or.2016.4549.
Roos, W. P., Tsaalbi-Shtylik, A., Tsaryk, R., Güvercin, F., de Wind, N., and Kaina, B. (2009) The translesion polymerase Rev3L in the tolerance of alkylating anticancer drugs, Mol. Pharmacol., 76, 927-934, doi: https://doi.org/10.1124/mol.109.058131.
Doles, J., Oliver, T. G., Cameron, E. R., Hsu, G., Jacks, T., Walker, G. C., and Hemann, M. T. (2010) Suppression of Rev3, the catalytic subunit of pol zeta, sensitizes drug-resistant lung tumors to chemotherapy, Proc. Natl. Acad. Sci. USA, 107, 20786-20791, doi: https://doi.org/10.1073/pnas.1011409107.
Yang, L., Shi, T., Liu, F., Ren, C., Wang, Z., Li, Y., Tu, X., Yang, G., and Cheng, X. (2015) REV3L, a promising target in regulating the chemosensitivity of cervical cancer cells, PLoS One, 10, e0120334, doi: https://doi.org/10.1371/journal.pone.0120334.
Xie, K., Doles, J., Hemann, M. T., and Walker, G. C. (2010) Error-prone translesion synthesis mediates acquired chemoresistance, Proc. Natl. Acad. Sci. USA, 107, 20792-20797, doi: https://doi.org/10.1073/pnas.1011412107.
Gu, C., Luo, J., Lu, X., Tang, Y., Ma, Y., Yun, Y., Cao, J., Cao, J., Huang, Z., and Zhou, X. (2019) REV7 confers radioresistance of esophagus squamous cell carcinoma by recruiting PRDX2, Cancer Sci., 110, 962, doi: https://doi.org/10.1111/cas.13946.
Okina, S., Yanagisawa, N., Yokoyama, M., Sakurai, Y., Numata, Y., Umezawa, A., Higashihara, M., and Murakumo, Y. (2015) High expression of REV7 is an independent prognostic indicator in patients with diffuse large B-cell lymphoma treated with rituximab, Int. J. Hematology, 102, 662-669, doi: https://doi.org/10.1007/s12185-015-1880-3.
Masuda, Y., and Kamiya, K. (2002) Biochemical properties of the human REV1 protein, FEBS Lett., 520, 88-92, doi: https://doi.org/10.1016/s0014-5793(02)02773-4.
Choi, J. Y., Lim, S., Kim, E. J., Jo, A., and Guengerich, F. P. (2010) Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1, J. Mol. Biol., 404, 34-44, doi: https://doi.org/10.1016/j.jmb.2010.09.015.
Choi, J. Y., and Guengerich, F. P. (2008) Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1, J. Biol. Chem., 283, 23645-23655, doi: https://doi.org/10.1074/jbc.M801686200.
Lin, X., Okuda, T., Trang, J., and Howell, S. B. (2006) Human REV1 modulates the cytotoxicity and mutagenicity of cisplatin in human ovarian carcinoma cells, Mol. Pharmacol., 69, 1748-1754, doi: https://doi.org/10.1124/mol.105.020446.
Okuda, T., Lin, X., Trang, J., and Howell, S. B. (2005) Suppression of hREV1 expression reduces the rate at which human ovarian carcinoma cells acquire resistance to cisplatin, Mol. Pharmacol., 67, 1852-1860, doi: https://doi.org/10.1124/mol.104.010579.
Rusz, O., Pál, M., Szilágyi, É., Rovó, L., Varga, Z., Tomisa, B., Fábián, G., Kovács, L., Nagy, O., Mózes, P., Reisz, Z., Tiszlavicz, L., Deák, P., and Kahán, Z. (2017) The expression of checkpoint and DNA repair genes in head and neck cancer as possible predictive factors, Pathol. Oncol. Res., 23, 253-264, doi: https://doi.org/10.1007/s12253-016-0088-z.
Actis, M. L., Ambaye, N. D., Evison, B. J., Shao, Y., Vanarotti, M., Inoue, A., McDonald, E. T., Kikuchi, S., Heath, R., Hara, K., Hashimoto, H., and Fujii, N. (2016) Identification of the first small-molecule inhibitor of the REV7 DNA repair protein interaction, Bioorg. Med. Chem., 24, 4339-4346, doi: https://doi.org/10.1016/j.bmc.2016.07.026.
Coggins, G. E., Maddukuri, L., Penthala, N. R., Hartman, J. H., Eddy, S., Ketkar, A., Crooks, P. A., and Eoff, R. L. (2013) N-aroyl indole thiobarbituric acids as inhibitors of DNA repair and replication stress response polymerases, ACS Chem. Biol., 8, 1722-1729, doi: https://doi.org/10.1021/cb400305r.
Dash, R. C., Ozen, Z., McCarthy, K. R., Chatterjee, N., Harris, C. A., Rizzo, A. A., Walker, G. C., Korzhnev, D. M., and Hadden, M. K. (2019) Virtual pharmacophore screening identifies small-molecule inhibitors of the Rev1-CT/RIR protein–protein interaction, Chem. Med. Chem., 14, 1610-1617, doi: https://doi.org/10.1002/cmdc.201900307.
Dash, R. C., Ozen, Z., Rizzo, A. A., Lim, S., Korzhnev, D. M., and Hadden, M. K. (2018) Structural approach to identify a lead scaffold that targets the translesion synthesis polymerase Rev1, J. Chem. Inf. Model., 58, 2266-2277, doi: https://doi.org/10.1021/acs.jcim.8b00535.
Ozen, Z., Dash, R. C., McCarthy, K. R., Chow, S. A., Rizzo, A. A., Korzhnev, D. M., and Hadden, M. K. (2018) Small molecule scaffolds that disrupt the Rev1-CT/RIR protein–protein interaction, Bioorg. Med. Chem., 26, 4301-4309, doi: https://doi.org/10.1016/j.bmc.2018.07.029.
Sail, V., Rizzo, A. A., Chatterjee, N., Dash, R. C., Ozen, Z., Walker, G. C., Korzhnev, D. M., and Hadden, M. K. (2017) Identification of small molecule translesion synthesis inhibitors that target the Rev1-CT/RIR protein−protein interaction, ACS Chem. Biol., 12, 1903-1912, doi: https://doi.org/10.1021/acschembio.6b01144.
Vanarotti, M., Evison, B. J., Actis, M. L., Inoue, A., McDonald, E. T., Shao, Y., Heath, R. J., and Fujiia, N. (2018) Small-molecules that bind to the ubiquitin-binding motif of REV1 inhibit REV1 interaction with K164-monoubiquitinated PCNA and suppress DNA damage tolerance, Bioorg. Med. Chem., 26, 2345-2353, doi: https://doi.org/10.1016/j.bmc.2018.03.028.
Wojtaszek, J. L., Chatterjee, N., Najeeb, J., Ramos, A., Lee, M., Bian, K., Xue, J. Y., Fenton, B. A., Park, H., Li, D., Hemann, M. T., Hong, J., Walker, G. C., and Zhou, P. (2019) A small molecule targeting mutagenic translesion synthesis improves chemotherapy, Cell, 178, 152-159, doi: https://doi.org/10.1016/j.cell.2019.05.028.
Yamanaka, K., Dorjsuren, D., Eoff, R. L., Egli, M., Maloney, D. J., Jadhav, A., Simeonov, A., Stephen, R., and Lloyd, R. S. (2012) A comprehensive strategy to discover inhibitors of the translesion synthesis DNA polymerase κ, PLoS One, 7, e45032, doi: https://doi.org/10.1371/journal.pone.0045032.
Zafar, M. K., Maddukuri, L., Ketkar, A., Penthala, N. R., Reed, M. R., Eddy, S., Crooks, P. A., and Eoff, R. L. (2018) A small-molecule inhibitor of human DNA polymerase η potentiates the effects of cisplatin in tumor cells, Biochemistry, 57, 1262-1273, doi: https://doi.org/10.1021/acs.biochem.7b01176.
Dumstorf, C. A., Mukhopadhyay, S., Krishnan, E., Haribabu, B., and McGregor, W. G. (2009) REV1 is implicated in the development of carcinogen-induced lung cancer, Mol. Cancer Res., 7, 247-254, doi: https://doi.org/10.1158/1541-7786.MCR-08-0399.
Lakhin, A. V., Kazakov, A. A., Makarova, A. V., Pavlov, Y. I., Efremova, A. S., Shram, S. I., Tarantul, V. Z., and Gening, L. V. (2012) Isolation and characterization of high affinity aptamers against DNA polymerase iota, Nucleic Acids Ther., 22, 49-57, doi: https://doi.org/10.1089/nat.2011.0324.
Zafar, M. K., and Eoff, R. L. (2017) Translesion DNA synthesis in cancer: molecular mechanisms and therapeutic opportunities, Chem. Res. Toxicol., 30, 1942-1955, doi: https://doi.org/10.1021/acs.chemrestox.7b00157.
Choi, J.-S., Kim, S., Motea, E., and Berdis, A. (2017) Inhibiting translesion DNA synthesis as an approach to combat drug resistance to DNA damaging agents, Oncotarget, 20, 40804-40816, doi: https://doi.org/10.18632/oncotarget.17254.
García-Gómez, S., Reyes, A., Martínez-Jiménez, M. I., Chocrón, E. S., Mourón, S., Terrados, G., Powell, C., Salido, E., Méndez, J., Holt, I. J., and Blanco, L. (2013) PrimPol, an archaic primase/polymerase operating in human cells, Mol. Cell, 52, 541-553, doi: https://doi.org/10.1016/j.molcel.2013.09.025.
Bianchi, J., Rudd, S. G., Jozwiakowski, S. K., Bailey, L. J., Soura, V., Taylor, E., Stevanovic, I., Green, A. J., Stracker, T. H., Lindsay, H. D., and Doherty, A. J. (2013) PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication, Mol. Cell, 52, 566-573, doi: https://doi.org/10.1016/j.molcel.2013.10.035.
Kobayashi, K., Guilliam, T. A., Tsuda, M., Yamamoto, J., Bailey, L. J., Iwai, S., Takeda, S., Doherty, A. J., and Hirota, K. (2016) Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides, Cell Cycle, 15, 1997-2008, doi: https://doi.org/10.1080/15384101.2016.1191711.
Quinet, A., Tirman, S., Jackson, J., Šviković, S., Lemaçon, D., Carvajal-Maldonado, D., González-Acosta, D., Vessoni, A. T., Cybulla, E., Wood, M., Tavis, S., Batista, L. F. Z., Méndez, J., Sale, J. E., and Vindigni, A. (2020) PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells, Mol. Cell, 77, 461-474, doi: https://doi.org/10.1016/j.molcel.2019.10.008.
Hatano, Y., Tamada, M., Matsuo, M., and Hara, A. (2020) Molecular trajectory of BRCA1 and BRCA2 mutations, Front. Oncol., 10, 361, doi: https://doi.org/10.3389/fonc.2020.00361.
Tokarsky, E. J., Wallenmeyer, P. C., Phi, K. K., and Suo, Z. (2017) Significant impact of divalent metal ions on the fidelity, sugar selectivity, and drug incorporation efficiency of human primPol, DNA Rep., 49, 51-59, doi: https://doi.org/10.1016/j.dnarep.2016.11.003.
Liu, J., Lee, W., Jiang, Z., Chen, Z., Jhunjhunwala, S., Haverty, P. M., Gnad, F., Guan, Y., Gilbert, H. N., Stinson, J., Klijn, C., Guillory, J., Bhatt, D., Vartanian, S., Walter, K., Chan, J., Holcomb, T., Dijkgraaf, P., Johnson, S., Koeman, J., Minna, J. D., Gazdar, A. F., Stern, H. M., Hoeflich, K. P., Wu, T. D., Settleman, J., de Sauvage, F. J., Gentleman, R. C., Neve, R. M., Stokoe, D., Modrusan, Z., Seshagiri, S., Shames, D. S., and Zhang, Z. (2012) Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events, Genome Res., 22, 2315-2327, doi: https://doi.org/10.1101/gr.140988.112.
Díaz-Talavera, A., Calvo, P. A., González-Acosta, D., Díaz, M., Sastre-Moreno, G., Blanco-Franco, L., Guerra, S., Martínez-Jiménez, M. I., Méndez, J., and Blanco, L. (2019) A cancer-associated point mutation disables the steric gate of human PrimPol, Sci. Rep., 9, 1121, doi: https://doi.org/10.1038/s41598-018-37439-0.
Taylor, M. R. G., and Yeeles, J. T. P. (2018) The initial response of a eukaryotic replisome to DNA damage, Mol. Cell, 70, 1067-1080, doi: https://doi.org/10.1016/j.molcel.2018.04.022.
Lopes, M., Foiani, M., and Sogo, J. M. (2006) Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions, Mol. Cell, 21, 15-27, doi: https://doi.org/10.1016/j.molcel.2005.11.015.
Svoboda, D. L., and Vos, J. M. (1995) Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation, Proc. Natl. Acad. Sci. USA, 92, 11975-11979, doi: https://doi.org/10.1073/pnas.92.26.11975.
Veaute, X., Mari-Giglia, G., Lawrence, C. W., and Sarasin, A. (2000) UV lesions located on the leading strand inhibit DNA replication but do not inhibit SV40 T-antigen helicase activity, Mutat. Res., 459, 19-28, doi: https://doi.org/10.1016/s0921-8777(99)00052-x.
Elvers, I., Johansson, F., Groth, P., Erixon, K., and Helleday, T. (2011) UV stalled replication forks restart by re-priming in human fibroblasts, Nucleic Acids Res., 39, 7049-7057, doi: https://doi.org/10.1093/nar/gkr420.
Hedglin, M., and Benkovic, S. J. (2017) Eukaryotic translesion DNA synthesis on the leading and lagging strands: unique detours around the same obstacle, Chem. Rev., 117, 7857-7877, doi: https://doi.org/10.1021/acs.chemrev.7b00046.
Schiavone, D., Jozwiakowski, S. K., Romanello, M., Guilbaud, G., Guilliam, T. A., Bailey, L. J., Sale, J. E., and Doherty, A. J. (2016) PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells, Mol. Cell, 61, 161-169, doi: https://doi.org/10.1016/j.molcel.2015.10.038.
Pilzecker, B., Buoninfante, O. A., Pritchard, C., Blomberg, O. S., Huijbers, I. J., van den Berk, P. C. M., and Jacobs, H. (2016) PrimPol prevents APOBEC/AID family mediated DNA mutagenesis, Nucleic Acids Res., 44, 4734-4744, doi: https://doi.org/10.1093/nar/gkw123.
Han, T., Goralski, M., Capota, E., Padrick, S. B., Kim, J., Xie, Y., and Nijhawan, D. (2016) The anti-tumor toxin CD437 is a direct inhibitor of DNA polymerase α, Nat. Chem. Biol., 12, 511-515, doi: https://doi.org/10.1038/nchembio.2082.
Abdel-Samad, R., Aouad, P., Gali-Muhtasib, H., Sweidan, Z., Hmadi, R., Kadara, H., D’Andrea, E. L., Fucci, A., Pisano, C., and Darwiche, N. (2018) Mechanism of action of the atypical retinoid ST1926 in colorectal cancer: DNA damage and DNA polymerase α, Am. J. Cancer Res., 8, 39-55.
Funding
This research was financially supported by the Russian Foundation for Basic Research (projects nos. komfi 17-00-00264, AVM; Bel-a 18-54-00024, AVM), by the Belarusian Republican Foundation for Basic Research (project no. B18R-094, MPS) [chapters on DNA polymerases of the Y-family]; and by the Russian Science Foundation (project no. 18-14-00354, AVM) [chapter on reinitiation of DNA synthesis and PrimPol].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Shilkin, E.S., Boldinova, E.O., Stolyarenko, A.D. et al. Translesion DNA Synthesis and Reinitiation of DNA Synthesis in Chemotherapy Resistance. Biochemistry Moscow 85, 869–882 (2020). https://doi.org/10.1134/S0006297920080039
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920080039