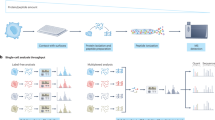
Abstract
Recent technical advances in genomic technology have led to the explosive growth of transcriptome-wide studies at the level of single cells. The review describes the first steps of the single cell proteomics that has originated soon after development of transcriptomics methods. The first studies on the shotgun proteomics of single cells that used liquid chromatography/mass spectrometry have been already published. In these works, the cells were separated by the methods used in transcriptomics studies (e.g., cell sorting) and analyzed by modified mass spectrometry with tandem mass tags. The new proteogenomics approach involving integration of single cell transcriptomics and proteomics data will provide better understanding of the mechanisms of cell interactions in normal development and disease.
Similar content being viewed by others
Abbreviations
- ADAR:
-
adenosine deaminase
- RNA:
-
dependent
- FACS:
-
fluorescence-activated cell sorting
- FISSEQ:
-
in situ fluorescence RNA sequencing
- NGS:
-
next generation sequencing
- SCoPE-MS:
-
Single Cell ProtEomics by Mass Spectrometry
- TMT:
-
tandem mass tag
- UMI:
-
unique molecular identifier
References
Horgan, R. P., and Kenny, L. C. (2011) “Omic” technologies: genomics, transcriptomics, proteomics and metabolomics, Obstet. Gynaecol., 13, 189–195, doi: 10.1576/toag.13.3.189.27672.
Geyer, P. E., Voytik, E., Treit, P. V., Doll, S., Kleinhempel A., Niu, L., Muller, J. B., Buchholtz, M., Bader, J. M. Teupser, D., Holdt, L. M., and Mann, M. (2019) Plasma proteome profiling to detect and avoid sample-related biases in biomarker studies, EMBO Mol. Med, doi: 10.15252/emmm.201910427.
Banfalvi, G. (2011) Overview of cell synchronization Methods Mol. Biol., 761, 1–23, doi: 10.1007/978-1-61779-182-6-1.
Emmert-Buck, M. R., Bonner, R. F., Smith, P. D. Chuaqui, R. F., Zhuang, Z., Goldstein, S. R., Weiss, R. A. and Liotta, L. A. (1996) Laser capture microdissection Science, 274, 998–1001, doi: 10.1126/science.274.5289.998.
Ziegenhain, C., Vieth, B., Parekh, S., Reinius, B. Guillaumet-Adkins, A., Smets, M., Leonhardt, H., Heyn H., Hellmann, I., and Enard, W. (2017) Comparative analysis of single-cell RNA sequencing methods, Mol. Cell65, 631–643, doi: 10.1016/j.molcel.2017.01.023.
Lee, J. H., Daugharthy, E. R., Scheiman, J., Kalhor, R. Yang, J. L., Ferrante, T. C., Terry, R., Jeanty, S. S. F., Li C., Amamoto, R., Peters, D. T., Turczyk, B. M. Marblestone, A. H., Inverso, S. A., Bernard, A., Mali, P. Rios, X., Aach, J., and Church, G. M. (2014) Highly multiplexed subcellular RNA sequencing in situ, Science, 343 1360–1363, doi: 10.1126/science.1250212.
Lee, J. H., Daugharthy, E. R., Scheiman, J., Kalhor, R. Ferrante, T. C., Terry, R., Turczyk, B. M., Yang, J. L., Lee H. S., Aach, J., Zhang, K., and Church, G. M. (2015) Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues, Nat. Protoc.10, 442–458, doi: 10.1038/nprot.2014.191.
Picelli, S., Faridani, O. R., Bjorklund, A. K., Winberg, G. Sagasser, S., and Sandberg, R. (2014) Full-length RNA-seq from single cells using Smart-seq2, Nat. Protoc., 9, 171–181, doi: 10.1038/nprot.2014.006.
Valihrach, L., Androvic, P., and Kubista, M. (2018) Platforms for single-cell collection and analysis, Int. J. Mol. Sci., 19, E807, doi: 10.3390/ijms19030807.
Zheng, G. X. Y., Terry, J. M., Belgrader, P., Ryvkin, P. Bent, Z. W., et al. (2017) Massively parallel digital transcriptional profiling of single cells, Nat. Commun., 8, 14049 doi: 10.1038/ncomms14049.
Islam, S., Zeisel, A., Joost, S., La Manno, G., Zajac, P. Kasper, M., Lonnerberg, P., and Linnarsson, S. (2014) Quantitative single-cell RNA-seq with unique molecular identifiers, Nat. Methods, 11, 163–166, doi: 10.1038/nmeth.2772.
Zhang, X., Li, T., Liu, F., Chen, Y., Yao, J., Li, Z. Huang, Y., and Wang, J. (2019) Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems, Mol. Cell, 73, 130–142, doi: 10.1016/j.molcel.2018.10.020.
Soldatov, R., Kaucka, M., Kastriti, M. E., Petersen, J. Chontorotzea, T., et al. (2019) Spatiotemporal structure of cell fate decisions in murine neural crest, Science, 364 9536, doi: 10.1126/science.aas9536.
La Manno, G., Soldatov, R., Zeisel, A., Braun, E. Hochgerner, H., Petukhov, V., Lidschreiber, K., Kastriti M. E., Lonnerberg, P., Furlan, A., Fan, J., Borm, L. E. Liu, Z., van Bruggen, D., Guo, J., He, X., Barker, R. Sundstrom, E., Castelo-Branco, G., Cramer, P. Adameyko, I., Linnarsson, S., and Kharchenko, P. V. (2018) RNA velocity of single cells, Nature, 560, 494–498 doi: 10.1038/s41586-018-0414-6.
Burgess, D. J. (2018) Full speed ahead for single-cell analysis, Nat. Rev. Genet., 19, 668–669, doi: 10.1038/s41576018-0049-3.
Hodge, R. D., Bakken, T. E., Miller, J. A., Smith, K. A. Barkan, E. R., et al. (2019) Conserved cell types with divergent features in human versus mouse cortex, Nature, 573 61–68, doi: 10.1038/s41586-019-1506-7.
Khrameeva, E., Kurochkin, I., Han, D., Guijarro, P. Kanton, S., Santel, M., Qian, Z., Rong, S., Mazin, P. Bulat, M., Efimova, O., Tkachev, A., Guo, S., Sherwood C. C., Camp, J. G., Paabo, S., Treutlein, B., and Khaitovich, P. (2019) Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains bioRxiv, doi: 10.1101/764936.
Shekhar, K., and Menon, V. (2019) Identification of cell types from single-cell transcriptomic data, Methods Mol. Biol., 1935, 45–77, doi: 10.1007/978-1-4939-9057-3-4.
Archakov, A., Ivanov, Y., Lisitsa, A., and Zgoda, V. (2009) Biospecific irreversible fishing coupled with atomic force microscopy for detection of extremely low-abundant proteins Proteomics, 9, 1326–1343, doi: 10.1002/pmic.200800598.
Aymoz, D., Wosika, V., Durandau, E., and Pelet, S. (2016) Real-time quantification of protein expression at the singlecell level via dynamic protein synthesis translocation reporters Nat. Commun., 7, 11304, doi: 10.1038/ncomms11304.
Fulwyler, M. J. (1965) Electronic separation of biological cells by volume, Science, 150, 910–911, doi: 10.1126/science.150.3698.910.
Picot, J., Guerin, C. L., Le Van Kim, C., and Boulanger, C. M. (2012) Flow cytometry: retrospective, fundamentals and recent instrumentation, Cytotechnology, 64, 109–130 doi: 10.1007/s10616-011-9415-0.
Hughes, A. J., Spelke, D. P., Xu, Z., Kang, C.-C., Schaffer D. V., and Herr, A. E. (2014) Single-cell western blotting Nat. Methods, 11, 749–755, doi: 10.1038/nmeth.2992.
Bendall, S. C., Simonds, E. F., Qiu, P., Amir, el-A. D. Krutzik, P. O., Finck, R., Bruggner, R. V., Melamed, R. Trejo, A., Ornatsky, O. I., Balderas, R. S., Plevritis, S. K. Sachs, K., Pe’er, D., Tanner, S. D., and Nolan, G. P. (2011) Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum Science, 332, 687–696, doi: 10.1126/science.1198704.
Palii, C. G., Cheng, Q., Gillespie, M. A., Shannon, P. Mazurczyk, M., Napolitani, G., Price, N. D., Ranish, J. A., Morrissey, E., Higgs, D. R., and Brand, M. (2019) Single-cell proteomics reveal that quantitative changes in co-expressed lineage-specific transcription factors determine cell fate, Cell Stem Cell, 24, 812–820, doi: 10.1016/j.stem.2019.02.006.
Marcon, E., Jain, H., Bhattacharya, A., Guo, H., Phanse S. et al. (2015) Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation, Nat. Methods, 12, 725–731, doi: 10.1038/nmeth.3472.
Coscia, F., Watters, K. M., Curtis, M., Eckert, M. A. Chiang, C. Y., Tyanova, S., Montag, A., Lastra, R. R. Lengyel, E., and Mann, M. (2016) Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status, Nat. Commun., 7 12645, doi: 10.1038/ncomms12645.
Kaur, P., and O’Connor, P. B. (2007) Quantitative determination of isotope ratios from experimental isotopic distributions, Anal. Chem., 79, 1198–1204, doi: 10.1021/ac061535z.
Ho, B., Baryshnikova, A., and Brown, G. W. (2018) Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome, Cell Syst., 6 192–205, doi: 10.1016/j.cels.2017.12.004.
Siwiak, M., and Zielenkiewicz, P. (2013) Transimulation–protein biosynthesis web service, PLoS One, 8, e73943, doi: 10.1371/journal.pone.0073943.
Virant-Klun, I., Leicht, S., Hughes, C., and Krijgsveld, J. (2016) Identification of maturation-specific proteins by single-cell proteomics of human oocytes, Mol. Cell. Proteomics15, 2616–2627, doi: 10.1074/mcp.M115.056887.
Sun, L., Dubiak, K. M., Peuchen, E. H., Zhang, Z., Zhu G., Huber, P. W., and Dovichi, N. J. (2016) Single cell proteomics using frog (Xenopus laevis) blastomeres isolated from early stage embryos, which form a geometric progression in protein content, Anal. Chem., 88, 6653–6657, doi: 10.1021/acs.analchem.6b01921.
Moroz, L. L. (2018) Neurosystematics and periodic system of neurons: model vs reference species at single-cell resolution, ACS Chem. Neurosci., 9, 1884–1903, doi: 10.1021/acschemneuro.8b00100.
Chesnokova, E., Zuzina, A., Bal, N., Vinarskaya, A. Roshchin, M., Artyuhov, A., Dashinimaev, E., Aseyev, N. Balaban, P., and Kolosov, P. (2019) Experiments with snails add to our knowledge about the role of aPKC subfamily kinases in learning, Int. J. Mol. Sci., 20, 2117, doi: 10.3390/ijms20092117.
Thompson, A., Schafer, J., Kuhn, K., Kienle, S., Schwarz J., Schmidt, G., Neumann, T., Johnstone, R. Mohammed, A. K. A., and Hamon, C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS, Anal. Chem., 75, 1895–1904, doi: 10.1021/ac0262560.
Budnik, B., Levy, E., Harmange, G., and Slavov, N. (2018) SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation Genome Biol., 19, 161, doi: 10.1186/s13059-018-1547-5.
Huffman, R. G., Chen, A., Specht, H., and Slavov, N. (2019) DO-MS: data-driven optimization of mass spectrometry methods, J. Proteome Res., 18, 2493–2500, doi: 10.1021/acs.jproteome.9b00039.
Chen, A. T., Franks, A., and Slavov, N. (2019) DART-ID increases single-cell proteome coverage, PLOS Comput. Biol., 15, e1007082, doi: 10.1371/journal.pcbi.1007082.
Specht, H., Emmott, E., Perlman, D. H., Koller, A., and Slavov, N. (2019) High-throughput single-cell proteomics quantifies the emergence of macrophage heterogeneity bioRxiv, doi: 10.1101/665307.
Dou, M., Clair, G., Tsai, C.-F., Xu, K., Chrisler, W. B. Sontag, R. L., Zhao, R., Moore, R. J., Liu, T., Pasa-Tolic L., Smith, R. D., Shi, T., Adkins, J. N., Qian, W.-J., Kelly R. T., Ansong, C., and Zhu, Y. (2019) High-throughput single cell proteomics enabled by multiplex isobaric labeling in a nanodroplet sample preparation platform, Anal. Chem., 91, 13119–13127, doi: 10.1021/acs.analchem.9b03349.
Zhu, Y., Piehowski, P. D., Zhao, R., Chen, J., Shen, Y. Moore, R. J., Shukla, A. K., Petyuk, V. A., CampbellThompson, M., Mathews, C. E., Smith, R. D., Qian, W.J., and Kelly, R. T. (2018) Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells, Nat. Commun., 9, 882, doi: 10.1038/s41467-018-03367-w.
Schoof, E. M., Rapin, N., Savickas, S., Gentil, C. Lechman, E., Haile, J. S., auf dem Keller, U., Dick, J. E. and Porse, B. T. (2019) A quantitative single-cell proteomics approach to characterize an acute myeloid leukemia hierarchy, bioRxiv, doi: 10.1101/745679.
Johansson, H. J., Socciarelli, F., Vacanti, N. M., Haugen M. H., Zhu, Y., Siavelis, I., Fernandez-Woodbridge, A. Aure, M. R., Sennblad, B., Vesterlund, M., Branca, R. M. Orre, L. M., Huss, M., Fredlund, E., Beraki, E., Garred O., Boekel, J., Sauer, T., Zhao, W., Nord, S., Hoglander, E. K., Jans, D. C., Brismar, H., Haukaas, T. H., Bathen, T. F. Schlichting, E., Naume, B., Consortia Oslo Breast Cancer Research Consortium (OSBREAC), Luders, T., Borgen E., Kristensen, V. N., Russnes, H. G., Lingjærde, O. C. Mills, G. B., Sahlberg, K. K., Borresen-Dale, A.-L., and Lehtio, J. (2019) Breast cancer quantitative proteome and proteogenomic landscape, Nat. Commun., 10, 1600, doi: 10.1038/s41467-019-09018-y.
Dimitrakopoulos, L., Prassas, I., Diamandis, E. P. Nesvizhskii, A., Kislinger, T., Jaffe, J., and Drabovich, A. (2016) Proteogenomics: opportunities and caveats, Clin. Chem., 62, 551–557, doi: 10.1373/clinchem.2015.247858.
Smith, L. M., Kelleher, N. L., and Consortium for Top Down Proteomics (2013) Proteoform: a single term describing protein complexity, Nat. Methods, 10, 186–187 doi: 10.1038/nmeth.2369.
Simoes, A. E., Pereira, D. M., Amaral, J. D., Nunes, A. F. Gomes, S. E., Rodrigues, P. M., Lo, A. C., D’Hooge, R. Steer, C. J., Thibodeau, S. N., Borralho, P. M., and Rodrigues, C. M. (2013) Efficient recovery of proteins from multiple source samples after Trizol® or Trizol®LS RNA extraction and long-term storage, BMC Genomics, 14, 181 doi: 10.1186/1471-2164-14-181.
Mun, D. G., Bhin, J., Kim, S., Kim, H., Jung, J. H., et al. (2019) Proteogenomic characterization of human earlyonset gastric cancer, Cancer Cell, 35, 111–124, doi: 10.1016/j.ccell.2018.12.003.
Poirion, O., Zhu, X., Ching, T., and Garmire, L. X. (2018) Using single nucleotide variations in single-cell RNA-seq to identify subpopulations and genotype-phenotype linkage, Nat. Commun., 9, 4892, doi: 10.1038/s41467-01807170-5.
Levitsky, L. I., Kliuchnikova, A. A., Kuznetsova, K. G. Karpov, D. S., Ivanov, M. V., Pyatnitskiy, M. A., Kalinina O. V., Gorshkov, M. V., and Moshkovskii, S. A. (2019) Adenosine-to-inosine RNA editing in mouse and human brain proteomes, Proteomics, 19, e1900195, doi: 10.1002/pmic.201900195.
Ximerakis, M., Lipnick, S. L., Innes, B. T., Simmons, S. K., Adiconis, X., Dionne, D., Mayweather, B. A., Nguyen L., Niziolek, Z., Ozek, C., Butty, V. L., Isserlin, R. Buchanan, S. M., Levine, S. S., Regev, A., Bader, G. D. Levin, J. Z., and Rubin, L. L. (2019) Single-cell transcriptomic profiling of the aging mouse brain, Nat. Neurosci.22, 1696–1708, doi: 10.1038/s41593-019-0491-3.
Funding
The work was supported by the Russian Science Foundation (project 17–15–01229).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval. This paper contains no description of studies using humans or animals performed by any of the authors.
Additional information
Conflict of interest. The authors declare no conflict of interest in financial of any other sphere.
Published in Russian in Biokhimiya, 2020, Vol. 85, No. 2, pp. 165-173.
Rights and permissions
About this article
Cite this article
Moshkovskii, S.A., Lobas, A.A. & Gorshkov, M.V. Single Cell Proteogenomics — Immediate Prospects. Biochemistry Moscow 85, 140–146 (2020). https://doi.org/10.1134/S0006297920020029
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297920020029