-
PDF
- Split View
-
Views
-
Cite
Cite
Keke Luo, Yu Qin, Ting Ouyang, Xilan Wang, Aihua Zhang, Peng Luo, Xueli Pan, Let-7c-5p Regulates CyclinD1 in Fluoride-Mediated Osteoblast Proliferation and Activation, Toxicological Sciences, Volume 182, Issue 2, August 2021, Pages 275–287, https://doi.org/10.1093/toxsci/kfab054
Close - Share Icon Share
Abstract
Endemic fluorosis is caused by the intake of high environmental fluoride, which causes dental and skeletal fluorosis. Osteoblast proliferation and activation is closely related to skeletal fluorosis and is tightly regulated by the cell cycle. Several biological processes, including bone metabolism and osteoblast proliferation and activation, are regulated by a type of noncoding RNA called microRNAs (miRNAs). However, the understanding of miRNA functions in skeletal fluorosis is limited. Based on our previous miRNA sequencing results and bioinformatics analysis, we investigated the function of the miRNA let-7c-5p to regulate CyclinD1 in fluoride-induced osteoblast proliferation and activation. We designed population experiments as well as in vitro studies using 5-Ethynyl-2'-deoxyuridine (EdU), flow cytometry, immunofluorescence, dual-luciferase reporters, and chromatin immunoprecipitation. The population-based analysis showed a decrease in let-7c-5p expression as fluoride exposure increased. In addition, let-7c-5p levels were negatively correlated with CyclinD1 and Wnt9a (another let-7c-5p target). We verified in vitro that let-7c-5p participates in the fluoride-induced proliferation and activation of human osteoblasts by directly targeting CyclinD1. Furthermore, we demonstrated that let-7c-5p regulates CyclinD1 expression via the Wnt/β-catenin signaling pathway. This study demonstrated the participation of let-7c-5p in fluoride-induced proliferation and activation of human osteoblasts by regulation of CyclinD1 expression at the post-transcriptional and transcriptional levels.
Fluorine is an abundant element that exists as various compounds in nature and has a high reaction performance. Endemic fluorosis is a biogeochemical disease caused by high fluoride intake, and it is an important public health challenge worldwide. The threats of fluorosis have not been rooted out, and it is a serious health concern in 24 nations, including China (Srivastava and Flora, 2020). Based on the source of fluoride, endemic fluorosis is classified into 3 types: burning coal, drinking water, and drinking brick-tea (Wang and Huang, 1995). In China, the coal-burning endemic fluorosis was mostly reported in Guizhou, Sichuan, and Yunnan provinces (Hong et al., 2018). Excessive fluoride concentration in humans causes varying degrees of health concerns, mainly involving dental and skeletal fluorosis (Li et al., 2020a; Yuan et al., 2020). Skeletal fluorosis is the most harmful outcome, causing pain and reducing the patient’s quality of life. Therefore, it is necessary to take further measures for the prevention and control of skeletal fluorosis.
The cause of fluorosis is known, but its pathogenesis remains unclear. Previous studies have demonstrated that active osteogenesis and accelerated bone transformation are characteristic lesions of skeletal fluorosis (Jiang et al., 2018). Osteoblasts are the most crucial cells for bone formation, and the activation of osteoblast is closely related to osteogenesis (Hsiao et al., 2010; Zhang et al., 2007). Equally, the proliferation and activation of osteoblasts are tightly regulated by the cell cycle (Wu et al., 2019). The cell cycle is a highly precise, sequential, and complicated process whose stages determine the fate of the cell (Matson and Cook, 2017). The correct cooperation between cyclin, cyclin-dependent kinases (CDK), and CDK inhibitor (CKI) is very important to ensure the progression through cell cycle, and CyclinD1 is a key cell cycle regulatory factor that is mostly active during the G1 phase (Gao and Liu, 2019). In our previous studies on DNA methylation and histone acetylation of CyclinD1/CDK4/p16/p21 related to fluoride exposure, the expression of CyclinD1 was found to be positively correlated with fluoride exposure, even though there was no significant change in the CyclinD1 gene methylation and histone acetylation levels (Pan et al., 2020). However, how fluoride causes the upregulation of CyclinD1 is unknown.
Increasing evidence indicates that microRNA (miRNA) expression levels influence bone metabolism, including the proliferation and activation of osteoblasts (Heilmeier et al., 2016; Liao et al., 2019). miRNAs are 17–25 nucleotide long-noncoding RNA molecules that regulate biological processes, primarily by binding the 3′-untranslated regions (UTRs) of target mRNAs to alter gene expression (O’Hara et al., 2009). The deregulation of processes associated with miRNAs could induce pathophysiological conditions, including altered cell state regulation by targeting transcription factors (TCFs) and signaling molecules (Li et al., 2020b; Zhu et al., 2020). To date, the importance of miRNAs in fluorosis has been shown (Jiang et al., 2020; Raghunath et al., 2016). Thus, we hypothesized that the miRNAs are involved in the proliferation and activation of osteoblast in skeletal fluorosis via the regulation of CyclinD1.
Our previous study employed miRNA-seq to measure miRNA levels in plasma samples from coal-burning endemic fluorosis patients. We identified 44 upregulated and 83 downregulated miRNAs (Wang et al., 2019). Here, we use bioinformatics to identify downregulated miRNAs that target CyclinD1, including miR-486-3p, miR-4755-5p, and let-7c-5p. These 3 miRNAs were all investigated and we only reported let-7c-5p research results in this study. Further bioinformatics analysis showed that let-7c-5p targets not only CyclinD1 but also Wnt9a. It is worth noting that some studies have shown that Wnt9a is also upstream of CyclinD1. Wnt9a is an important regulatory factor for organisms, which plays an important role in cell cycle, cell proliferation and differentiation (Bartoletti et al., 2020; Grainger et al., 2016; Richter et al., 2018). The classical Wnt/β-catenin signal pathway can promotes normal osteogenesis (Zhang et al., 2019), which is upregulated by fluoride exposure (Liu et al., 2012). However, in reviewing the literature, no data was found on the association between Wnt9a and CyclinD1 on fluorosis. Therefore, we speculated that let-7c-5p directly targets CyclinD1 or regulates its expression by affecting the Wnt/β-catenin pathway, thereby affecting osteoblast proliferation and activation.
To verify our hypothesis, in the current population analysis, we measured let-7c-5p and Wnt9a expression under fluoride exposure at different concentrations [CyclinD1 expression as well as the markers of osteoblast activation, alkaline phosphatase (ALP) activity and osteocalcin (BGP) content, had been detected in our previous study]. Further in vitro experiments were conducted to observe (1) let-7c-5p’s function(s) in osteoblast proliferation and activation, (2) let-7c-5p directly targeting CyclinD1 intervenes posttranscriptional regulation, (3) let-7c-5p targeting Wnt9a and then affecting Wnt9a/β-catenin pathway regulates CyclinD1 expression at the transcriptional level. The flowchart of experimental design is shown in Supplementary Figure 1.This study’s goal was to investigate the mechanism of CyclinD1 regulation by let-7c-5p and its role in fluoride-induced osteoblast proliferation and activation.
MATERIALS AND METHODS
Population study
A total 248 participants were enrolled into the present work, including 141 from Hehua Village, Zhijin County, Guizhou Province. This was a typical region subjected to coal-burning endemic fluorosis determined according to the endemic fluorosis area classification criteria (GB17018-2011, China), and all participants were underwent a physical examination by doctors from Guizhou Orthopedics Hospital. The 107 villagers with no fluorosis symptom came from Zhangguan Village, a nonendemic fluorosis region in Anshun City, Guizhou Province. Differences in lifestyle and economic level between villagers from Zhangguan Village and Hehua Village were not statistically significant.
This work had gained approval from the Ethical Committee of Guizhou Medical University. Each participant had provided the informed consent. Subjects who had histories of additional bone diseases, family cancer and recent infection were eliminated from this study. The well-designed questionnaires were adopted to record the demographic data and residence history from each participant. Morning urine samples were collected to test for fluoride exposure level. In addition, the fasting peripheral blood from each participant was also sampled and preserved within the EDTA tubes to isolate plasma and peripheral blood mononuclear cells (PBMCs). Whereas plasma was utilized to analyze miRNA level, and PBMCs were adopted to analyze the Wnt9a level.
Urine fluoride measurements
An electrode selective for fluoride ions (PXJ-1B ion meter) was used to examine urinary fluoride (UF) levels, which is the standard approach in the China health industry (WS/T 89–2015, China). UF was normalized against urinary creatinine (Cr), which was measured using alkaline picrate (Nanjing Jiancheng Biotech Company, Nanjing, China). In China, the mean UF content is 1.6 mg/l among the general population (WS/T 256–2005, China). After adjusting for mean urinary Cr, the fluoride exposure reference value was calculated at 1.96 mg/g Cr. As a result, subjects were sorted into 3 groups according to UF values: <1.96, 1.96–3.92, and ≥3.92 mg/g Cr (The n for each group was 117, 65, and 66, respectively).
Cell culture and NaF treatments
Human bone fragments were collected from 3 healthy donors who underwent fracture surgery. Two were males aged 2 and 26, and the third was a female aged 23 years old. The methods for human osteoblast isolation, culture, purification, and identification were shown in our previous study (Ming et al., 2019). Based on our former results that 72 h of 1200 μmol/l NaF exposure showed greater cell viability of human osteoblasts (Gao et al., 2020), we treated osteoblasts similarly with either 0, 600, or 1200 µmol/l of NaF. The Cell Bank (Institute of Biological Sciences, Shanghai, China) provided the 293T cell line, which was cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS, HyClone, Utah) and used for the dual-luciferase reporter assays as tool cell.
Prediction of miRNAs targeting CyclinD1
miRNAs that targeted CyclinD1 were predicted using 4 different algorithms (miRDB, miRWalk, Targetscan, miRTarBase) in miRWalk 3.0 database (http://mirwalk.umm.uni-heidelberg.de). Criteria below were adopted to select miRNAs in later study: differentially down-regulated miRNAs (|log2FC |≥1 and p ≤ .05) upon our miRNA sequencing results (Wang et al., 2019); miRNAs selected based on 2 or more of the above 4 algorithms, and the binding site was 3′-UTR. The miRNA let-7c-5p was selected based on predicted results, and the target gene Wnt9a was predicted by the Targetscan algorithm.
Osteoblast transfection
Human osteoblasts were transfected with 50 nmol/l of a let-7c-5p mimic or mimic negative control (mimic NC) for 24 h using the riboFECT CP Reagent (RiboBio, Guang zhou, China). Transfections were conducted per manufacturer’s protocols prior to exposure to either 0 or 1200 μmol/l NaF.
Tests of cell viability and proliferation
Cell viability was measured using the Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan). Briefly, osteoblasts were plated at 6000 cells/well into 96-well plates using the hemocytometer. Cells were incubated in CCK-8 solution that had been diluted with FBS-free DMEM for another 2 h at 37°C with 5% CO2 conditions. The relative absorbance (OD) was measured at 450 nm with a Max200 microplate reader (Bio-Tek, Winooski, Vermont).
Osteoblast proliferation was measured via 5-Ethynyl-2'-deoxyuridine (EdU) incorporation with the Cell-Light EdU Apollo-567 In Vitro Imaging Kit (RiboBio, China). Briefly, 50 mmol/l of EdU was added to cells prior to a 4 h incubation at 37°C. The cells were next fixed in 4% paraformaldehyde then permeabilized with 0.5% Triton X-100 for 15 and 10 min, respectively. The treated cells were then incubated for 30 min with the Apollo reaction cocktail followed by a 30 min incubation with Hoechst 33342 for nuclear staining. The cells were visualized and imaged under a fluorescent microscope (Olympus, Tokyo, Japan).
Cell cycle determination
The cell cycle distribution was determined by the cell cycle and apoptosis kit (Beyotime, Shanghai, China). Cell proportion at each cell cycle stage was determined by the ACEA NovoCyte flow cytometer (Agilent Biosciences, California) and analyzed by Novoexpress.
Reverse transcription and quantitative real-time PCR
Total RNA was extracted from either human plasma or osteoblasts to measure miRNA levels using the miRNeasy serum/plasma kit (QIAGEN, Valencia, California) or TRIzol reagent (ThermoFisher Scientific, Waltham, Massachusetts), respectively. Total RNA (1 μg) was reverse transcribed to cDNA with the miDETECTA Track miRNA quantitative real-time PCR (qRT-PCR) Kit (RiboBio, Guangzhou, China) and analyzed in a CFX96 RT-PCR machine (Bio-Rad, Hercules, California). Each miRNA level was normalized to cel-miR-39 (QIAGEN) expression. To calculate relative miRNA expression levels, the 2−ΔΔCT method was applied.
To determine the mRNA levels of CyclinD1 of osteoblast and Wnt9a in PBMCs and osteoblast, TRIzol (ThermoFisher Scientific) was used to extract total RNA from human PBMCs or osteoblasts. Reverse transcription was conducted with the PrimeScript RT reagent kit (TaKaRa, Dalian, China). CyclinD1 and Wnt9a mRNA levels were measured in a CFX96 RT-PCR machine (Bio-Rad) with SYBR Green (TaKaRa, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control and the relative mRNA expression levels were calculated with the method. Supplementary Table 1 lists the primer sequences of the evaluated genes.
Enzyme-linked immunosorbent assay
Total PBMC protein was extracted using lysis buffer (Beyotime, China). The Wnt9a protein levels was determined with human Wnt9a enzyme-linked immunosorbent assay (ELISA) kits (Langton Biotechnology Co., Ltd., China) using a Max200 microplate reader (Bio-Tek, California). The activity of ALP and BGP content in cell culture supernatants were measured with standard micronutrient enzyme method and ELISA Kit (Shanghai Yanhui Biotechnology Co., Ltd., China).
Western blotting
Osteoblast total protein was extracted with RIPA lysis buffer (Beyotime, Shanghai, China) with phenylmethylsulphonyl fluoride. Besides, the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China) were used to isolate nuclear and cytoplasmic proteins per manufacturer protocols. Lysate proteins were measured with a BCA kit (Beyotime, China). Proteins were separated with sodium dodecylsulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Millipore, California). A 5% nonfat milk TBS-Tween 20 solution was used to block the membranes for 2 h followed by primary antibody incubation. The membranes were last incubated with an HRP-conjugated goat antirabbit secondary antibody (1:5000) for 2 h. Enhanced chemiluminescence assay kits (ECL, Millipore) were used to detect bands, and the Image Lab software (Bio-Rad) was adopted to measure the band OD values. There were 6 primary antibodies: anti-CyclinD1 (1:2000), anti-Wnt9a (1:1000), anti-GAPDH (1:2000), anti-β-actin (1:2000; Beyotime, China), anti-β-catenin (1:2000), and anti-Histone H1 (1:1000; CST, Massachusetts).
Dual-luciferase reporter assay
Bioinformatics analysis predicted that the 3′UTR-2214-2220 (CyclinD1 1) and the 3′UTR-2873-2879 (CyclinD1 2) base positions of CyclinD1 and that the Wnt9a 3′UTR-2746-2753 base position all have let-7c-5p complementary pairing sites. Plasmids (pmirGLO-CyclinD1 1/pmirGLO-mutant (mut) -CyclinD1 1, pmir-CyclinD1 2/pmirGLO-mut-CyclinD1 2, and pmirGLO-Wnt9a/pmirGLO-mut-Wnt9a) containing the 3′-UTR promoters of the CyclinD1 1, CyclinD1 2 and Wnt9a genes, respectively, were provided by Yile Biotech (Shanghai, China). The plasmids were transfected into 293T cells along with either the let-7c-5p mimic or mimic NC, with the riboFECT CP Transfection Kit (RiboBio, China). In line with Promega instructions, cells were collected at 24 h later using Dual-luciferase Reporter Assay system (Promega, Madison, Wisconsin) to determine luciferase activities, and GloMax 20/20 luminometer (Promega) was used to record results.
Immunofluorescence
Human osteoblasts fixed with 4% paraformaldehyde were incubated in anti-β-catenin (1:100; CST) antibodies overnight at 4°C followed by the Cy3-conjugated goat-anti-rabbit secondary antibody (1:50; Beyotime, China) at 37°C in the dark for 1 h. Cell nuclei were counterstained with 4,6-diamino-2-phenyl indole (Solarbio, Beijing, China). A confocal microscope (Carl Zeiss, Oberkochen, Germany) was used to capture images.
Chromatin immunoprecipitation
The Simple chromatin immunoprecipitation (ChIP) Plus Enzymatic Chromatin IP Kit (CST) was used to perform the ChIP assays per manufacturer protocols. Briefly, cells were incubated for 20 min in 1% formaldehyde to cross-link proteins and DNA. After enzyme digestion and sonication, with 2% of the supernatant as input, genomic DNA-protein complexes with either anti-β-catenin antibody (20 μl) or 1 of 2 control antibodies, anti-Histone H3 antibody (10 μl) or rabbit IgG (2 μl, all from CST). The cross-links were then reversed and the samples were treated with proteinase K for 2 h at 65°C. DNA was cleaned on spin columns prior to PCR. The CFX96 RT-PCR machine (Bio-Rad) was employed to amplify immunoprecipitated and input DNA content using the ChIP1 and ChIP2 primers. ChIP1 was located in the transcriptional regulatory region of CyclinD1 while ChIP2 was located in the coding region of CyclinD1. All results were standardized to the input DNA. See Supplementary Table 2 for the primer sequences of the evaluated genes.
Statistical analysis
SPSS 22.0 was used for all statistical analyses. The 1-way ANOVA and Chi-square test were used to analyze the demographic data. Medians (interquartile range) were calculated from population studies. The Kruskal-Wallis and Mann-Whitney U test were used for the statistical analysis and comparison between 2 groups, respectively. A Spearman correlation analysis was used to analyze correlations. Data associated with in vitro experiments are presented as the mean ± SD. Comparisons among groups were analyzed by ANOVA, and intergroup relationships were analyzed with the LSD test. A p-value < .05 (2-tailed) indicated statistical significance.
RESULTS
Subject Demographic Data
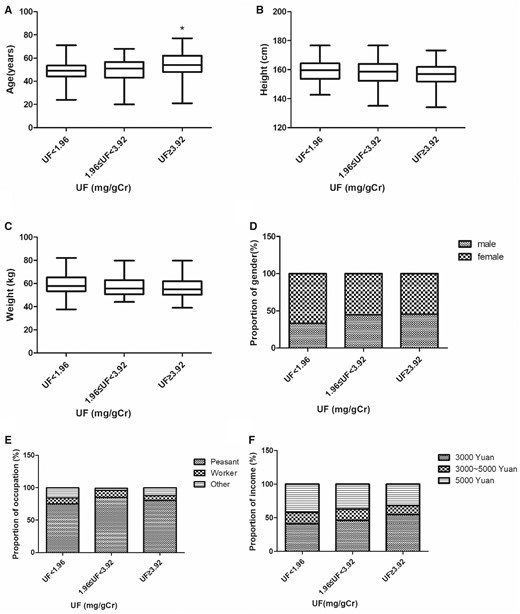
The study enrolled 248 subjects; 141 were from a fluorosis endemic area and 107 from nonfluorosis endemic areas, with 98 males and 150 females. All subjects were placed into 1 of 3 groups according to their UF levels. There were no significant difference in gender, height, weight, occupation, and income among 3 groups (Figure 1). It was confirmed in our previous study that the markers of osteogenic activation ALP and BGP expression increased along with UF values in fluoride-exposed population, indicating that osteoblasts activated by fluoride exposure were present in high UF participants.
Baseline characteristics of the study population with different fluoride exposures. Differences in age (A), height (B), weight (C), gender (D), occupation (E), and income (F) were analyzed among 3 groups. *p < .05 versus urinary fluoride < 1.96 (mg/g Cr) group.
Low Let-7c-5p Levels and High CyclinD1 and Wnt9a Levels Were Detected in the Fluoride Exposure Groups
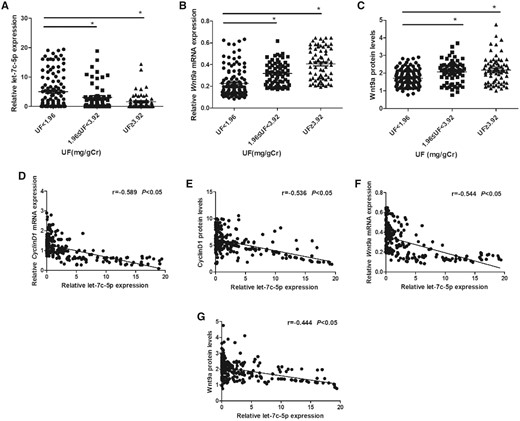
Based on the prior miRNA-seq data and the above-mentioned criteria, let-7c-5p expression in plasma was measured using qRT-PCR and its association with fluoride exposure determined. At high UF concentrations, let-7c-5p was downregulated (Figure 2A), matching our prior miRNA-seq data. Our previous study showed that CyclinD1 mRNA and protein levels increased as the concentration of UF increased (Gao et al., 2020). In this study, qRT-PCR and ELISA assays were used to measure Wnt9a expression in PBMCs obtained from the fluoride exposure group. As UF content increased, Wnt9a expression increased (Figs. 2B and 2C). Additionally, spearman correlation was used to investigate the association of let-7c-5p with CyclinD1 or Wnt9a expression level. There was a negative correlation between let-7c-5p and both CyclinD1 mRNA (r = −0.589, Figure 2D) and protein (r = −0.536, Figure 2E) levels. Similar results were observed for Wnt9a mRNA (r = −0.544, Figure 2F) and protein (r = −0.444, Figure 2G) levels. These findings suggest that let-7c-5p may participate in the increase in CyclinD1 and Wnt9a levels.
Low let-7c-5p levels and high CyclinD1 and Wnt9a levels were detected in the fluoride exposure groups. A, The let-7c-5p plasma contents of subjects with diverse urinary fluoride (UF) levels. The PBMC mRNA and protein levels for Wnt9a (B, C), isolated from subjects with diverse UF levels (n = 248), *p < .05 versus UF < 1.96 (mg/g Cr) group. The negative correlations between let-7c-5p and either CyclinD1 (D, E) or Wnt9a (F, G) expression in the participants.
NaF Treatment Decreased Let-7c-5p Level and Overexpression of Let-7c-5p Reduces Fluoride-Induced Osteoblast Proliferation and Activation
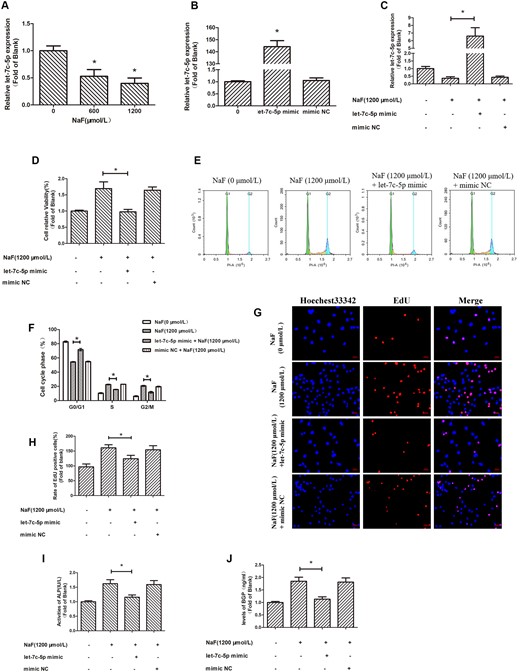
To verify the relationship of fluoride exposure with let-7c-5p level, we treated osteoblasts for 72 h with NaF at 0, 600, or 1200 µmol/l and the expression of let-7c-5p in human osteoblasts was measured. The result showed that let-7c-5p expression was decreased along with NaF exposure (Figure 3A). From our previous studies, NaF-induced osteoblast proliferation and activation in vitro has been confirmed in human osteoblasts (Gao et al., 2020), but the role of let-7c-5p in this is unknown. First, the let-7c-5p levels in osteoblasts were measured 24 h post-transfection. The expression of let-7c-5p in the mimic group was higher than that in the mimic NC group, which indicated an effective transfection (Figure 3B). Further, to determined how let-7c-5p expression and osteoblast proliferation and activation by NaF are related, osteoblasts were treated for 72 h with 0 or 1200 μmol/l of NaF post-transfection with the let-7c-5p mimic. Osteoblasts transfected with the let-7c-5p mimic upregulated let-7c-5p expression compared with those only treated with NaF (Figure 3C). The viability of osteoblasts transfected with let-7c-5p mimic was inhibited compared with only 1200 μmol/l NaF treatment group (Figure 3D). Meanwhile, overexpressing let-7c-5p increased the proportion of G0/G1 phase osteoblasts but decreased those in the S and G2/M phases (Figs. 3E and 3F). Osteoblast proliferation rates were also reduced (Figs. 3G and 3H). In addition, ALP and BGP values in the let-7c-5p mimic-transfected osteoblasts were decreased (Figs. 3I and 3J). As demonstrated by these findings, let-7c-5p overexpression reduces osteoblast proliferation and activation induced by NaF.
NaF treatment decreased let-7c-5p level and overexpression of let-7c-5p reduces fluoride-induced osteoblast proliferation and activation. Osteoblasts treated for 72 h with NaF at 0, 600, or 1200 μmol/l. A, The let-7c-5p expression was detected with qRT-PCR, with cel-miR-39 as the reference (*p < .05 versus 0 μmol/l NaF). B, Osteoblasts were transfected with either the let-7c-5p mimic or mimic NC for 24 h. After transfection, qRT-PCR with cel-miR-39 as the reference was used to measure let-7c-5p expression (*p < .05 versus mimic NC group). C, Osteoblasts were transfected with the let-7c-5p mimic prior to 1200 μmol/l NaF treatment. qRT-PCR was used to measure let-7c-5p expression. D, A plot of cell viability measured with a CCK-8 assay. E, The cell cycle stages determined by flow cytometry. F, The percentage in G0/G1, S, or G2/M phase of osteoblasts. G, Osteoblast proliferation was detected with the EdU incorporation assay and the nuclei stain Hoechst 33342 (blue). Bar = 50 µm. H, The proportion of EdU-positive cells was determined with ImageJ software. Osteoblast (I) ALP activity and (G) BGP content from various groups. Values are the mean ± SD. *p < .05 versus 1200 μmol/l NaF.
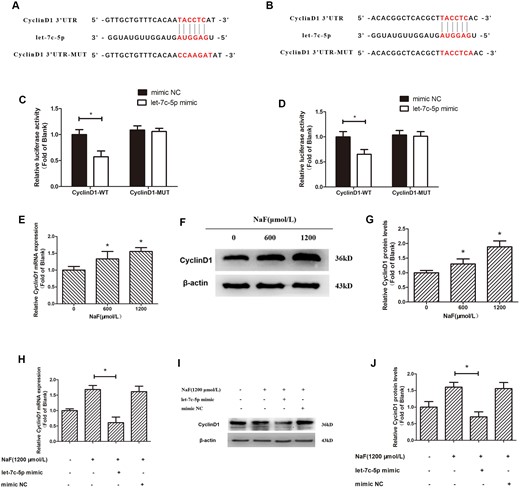
Downregulation of Let-7c-5p Regulates NaF-Induced Upregulation of CyclinD1 by Directly Targeting Its 3’UTR
TargetScan was used to show that the possible target sites of let-7c-5p in the 3’UTR of CyclinD1 (Figs. 4A and 4B). CyclinD1 1- or CyclinD1 2–3′UTRs containing a let-7c-5p binding site was cloned into the downstream in luciferase open reading frame. The resulting plasmids, and either the let-7c-5p mimic or mimic NC, were transfected into 293T cells. The resulting wild-type constructs markedly reduced luciferase activities compared with the mutated constructs (Figs. 4C and 4D).
Downregulation of let-7c-5p regulates NaF-induced upregulation of CyclinD1 by directly targeting its 3′UTR. A and B, The dual-luciferase vectors were wild-type and mutant versions of the CyclinD1 3′UTR. The candidate binding site for let-7c-5p was detected in the wild-type (WT)-UTR construct, but not in MUT-UTR. Wild-type or mutant constructs were co-transfected with either the mimic NC or let-7c-5p mimic into 293 T cells. C and D, Luciferase activity measurements. *p < .05 versus CyclinD1-3′UTR-WT + mimic NC. E, Cells treated for 72 h with NaF at 0, 600, or 1200 μmol/l. The CyclinD1 mRNA level was detected by qRT-PCR; GAPDH was the reference. F and G, CyclinD1 protein expression was detected with a Western blotting and β-actin was the reference. Values are the mean ± SD. *p < .05 versus 0 μmol/l NaF. H, Osteoblasts were transfected with the let-7c-5p mimic prior to 1200 μmol/l NaF treatment. CyclinD1 mRNA expression was measured by qRT-PCR; GAPDH was reference. I and J, Western blotting detecting CyclinD1 protein expression, with β-actin as the reference. Values are the mean ± SD. *p < .05 versus 1200 μmol/l NaF.
In our previous studies (Gao et al., 2020; Li et al., 2020d), we have found that NaF elevated CyclinD1 levels in osteoblasts, which is verified in this study (Figs. 4E–G). To determine how let-7c-5p expression and CyclinD1 upregulation by NaF are related, osteoblasts were treated for 72 h with 0 or 1200 μmol/l of NaF post-transfection with the let-7c-5p mimic. The NaF-induced increases in CyclinD1 mRNA (Figure 4H) and protein (Figs. 4I and 4J) levels were suppressed in cells transfected with let-7c-5p mimic. These data demonstrate the involvement of let-7c-5p in the increase of CyclinD1 expression induced by NaF in osteoblasts and indicate the posttranscriptional regulation of CyclinD1 by let-7c-5p in osteoblasts.
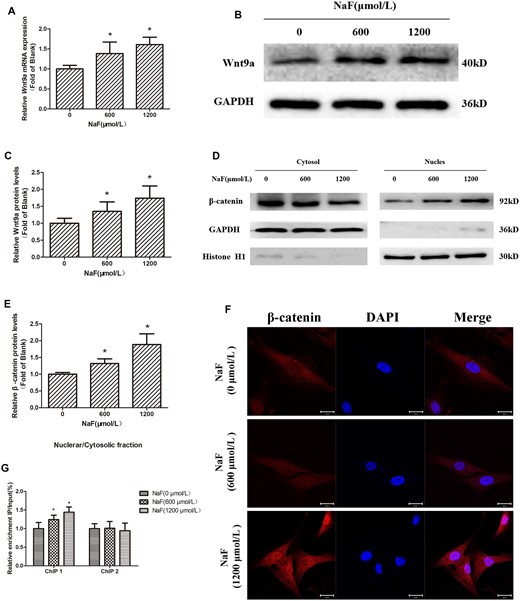
NaF Activates Wnt9a/β-Catenin/CyclinD1 Axis
The above-mentioned population study showed that an increase in Wnt9a level was accompanied by an increase in fluoride exposure. To further explore how fluoride exposure and Wnt9a/β-catenin pathway activation are related, qRT-PCR and Western blotting were used to measure Wnt9a expression after treated with NaF at 0, 600, and 1200 μmol/l for 72 h. As the NaF content increased, Wnt9a expression (Figs. 5A–C) also increased. Subsequently, β-catenin nuclear translocation was detected using Western blotting and immunofluorescence. The amount of β-catenin present in the nuclear increased, while its levels decreased in the cytoplasm (Figs. 5D and 5E). Immunofluorescence showed that as the concentration of NaF increased, the amount of β-catenin entering the nucleus also increased (Figure 5F). β-catenin is a multifunctional protein that translocates to the nucleus where it associates with TCF-4 to transcriptionally activate target genes. To determine the role of β-catenin binding within the CyclinD1 transcriptional regulatory region in NaF-induced CyclinD1 upregulation, we conducted ChIP assays to measure β-catenin binding levels. The ratios of precipitated DNA to input DNA were used to calculate relative precipitated fold enrichment. It was found that as the NaF content increased, β-catenin was enriched in the CyclinD1 transcriptional regulatory region; however, there was no significant difference in β-catenin binding to the coding region of CyclinD1 (Figure 5G). These results showed that NaF-mediated upregulation of CyclinD1 by activating the Wnt9a/β-catenin pathway.
NaF activates the Wnt9a/β-catenin/CyclinD1 axis. Cells were treated for 72 h with 0, 600, or 1200 μmol/l NaF. A, Wnt9a mRNA expression was measured with qRT-PCR; GAPDH was reference. B and C, Western blotting detecting Wnt9a protein expression, with GAPDH as the reference. D and E, Western blotting of β-catenin expressed in the cytosol and nucleus; GAPDH and Histone H1 were references. F, Cellular β-catenin expression and localization as measured by confocal laser scanning microscopy using an anti-β-catenin antibody. Bars = 20 μm. G, The enrichment levels of β-catenin in the transcriptional regulatory region and coding region of CyclinD1 were measured by chromatin immunoprecipitation assays, and the results were normalized to the input DNA. Values are the mean ± SD.*p < .05 versus 0 μmol/l NaF.
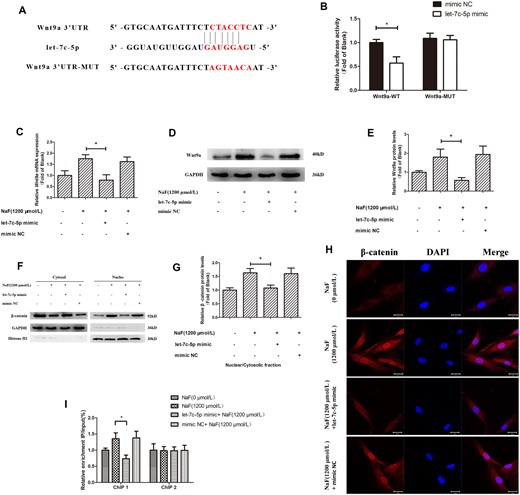
Downregulation of Let-7c-5p Regulated the NaF-Induced Upregulation of CyclinD1 by Wnt9a/β-Catenin Pathway
Further bioinformatics analyses suggested that let-7c-5p also targets Wnt9a, which has a complimentary binding site for let-7c-5p in the 3′-UTR (Figure 6A). The dual-luciferase reporter showed that wild-type promoter constructs markedly reduced luciferase activity, which was not observed in the mutated constructs (Figure 6B). According to these findings, the Wnt9a gene is a let-7c-5p target.
Downregulation of let-7c-5p regulates NaF-induced upregulation of CyclinD1 by Wnt9a/β-catenin pathway. Osteoblasts were transfected with the mimic NC or let-7c-5p mimic. A, The dual-luciferase vectors of wild-type and mutant Wnt9a 3′UTR. The candidate binding site sequence for let-7c-5p was detected in the WT-UTR construct, but not in MUT-UTR. Mutant or wild-type constructs were cotransfected with either the mimic NC or let-7c-5p mimic into 293T cells. B, Luciferase activity measurements. *p < .05 versus Wnt9a-3′UTR-WT + mimic NC group. C, After transfection and 1200 μmol/l NaF treatment, Wnt9a mRNA level in osteoblasts was measured with qRT-PCR; GAPDH was reference. D and E, Western blotting detecting Wnt9a protein expression, with GAPDH as the reference. F and G, β-catenin protein expression in the nucleus and cytoplasm was determined by Western blotting; GAPDH and Histone H1 were references. H, The expression and localization of β-catenin were measured by confocal laser scanning microscopy using an anti-β-catenin antibody. Bars = 20 μm. I, The enrichment levels of β-catenin in the transcriptional regulatory region and coding region of CyclinD1 were measured by chromatin immunoprecipitation assays; the results were normalized to the input DNA. Values are the mean ± SD. *p < .05 versus 1200 μmol/l NaF.
To further evaluate the requirement of let-7c-5p downregulation for NaF-induced Wnt9a/β-catenin signaling, we also observed how let-7c-5p overexpression effected NaF-induced Wnt9a expression in osteoblasts. The results showed that NaF-induced increases in osteoblast Wnt9a mRNA (Figure 6C) and protein (Figs. 6D and 6E) levels were instead suppressed following transfection with the let-7c-5p mimic, compared with 1200 μmol/l NaF alone. In addition, Western blotting and immunofluorescence showed that let-7c-5p overexpression inhibits β-catenin entry to the nucleus (Figs. 6F–H). ChIP assays showed a decrease in the β-catenin enrichment in the transcriptional regulatory region of CyclinD1 with let-7c-5p overexpression; there was no significant difference in the β-catenin binding to the CyclinD1 coding region (Figure 6I). As indicated by these findings, the overexpression of let-7c-5p can inhibit Wnt9a levels, β-catenin nuclear translocation, and reduce the enrichment of β-catenin in the transcriptional regulatory region of CyclinD1. These data confirmed that downregulation of let-7c-5p is involved in the NaF-induced upregulation of CyclinD1 via the Wnt9a/β-catenin pathway at the transcriptional level.
DISCUSSION
Fluoride is a persistent environmental contaminant released from several environmental and industrial activities that negatively impacts human health by causing fluorosis (Yadav et al., 2019). Many countries across the world have populations at a serious risk of fluorosis, and endemic fluorosis is a serious public health threat to China (Zhou et al., 2012). The widely accepted mechanism of fluorosis pathogenesis includes oxidative stress (Li et al., 2020c), endoplasmic reticulum stress (Li et al., 2019), and epigenetics (Pramanik and Saha, 2017). However, the epigenetic mechanism needs further exploration. Identifying the epigenetic mechanism of fluorosis would open new avenues for effective fluorosis prophylactics and therapies. Fluoride can induce skeletal fluorosis, which is accompanied by abnormal osteoblast proliferation and activation. Our previous studies have shown that fluoride exposure was positively correlated with CyclinD1 expression, in which miRNA exerts an influence (Gao et al., 2020; Pan et al., 2020).
Posttranscriptional genetic regulation can be mediated by miRNAs, which are vital to biological activities including cell cycle and cell growth (Cai et al., 2019; Martos et al., 2015; Shin and Chu, 2014). At the same time, the possible role of miRNAs was evaluated during diagnosis, treatment, and prognosis (Puik et al., 2017). We used previous miRNA sequences data and bioinformatics analyses to identify the miRNA let-7c-5p, which targets CyclinD1, for further study. The role of let-7c-5p in cataract (Cao et al., 2021), lung cancer (Petrek et al., 2021), and neuroendocrine tumors (Bösch et al., 2019) have been reported. However, let-7c-5p has not been studied in fluorosis. Interestingly, further bioinformatics analysis showed that let-7c-5p also targets Wnt9a in the Wnt/β-catenin pathway. However, the role of Wnt9a has not been studied in fluorosis. Several studies reported that miRNAs can regulate cell states by targeting TCFs and signaling molecules in signal pathways (Suzuki, 2018; Zhang et al., 2019). Therefore, we speculate that let-7c-5p participates in fluoride-induced proliferation and activation of human osteoblasts by regulating the expression of CyclinD1 at posttranscriptional and transcriptional levels.
We first verified the expression of let-7c-5p in the fluoride-exposure population with high ALP and BGP levels, and found that let-7c-5p expression decreased with the increase of fluoride exposure. We also found that let-7c-5p and CyclinD1 are negatively correlated, which suggested that let-7c-5p downregulation may be related to CyclinD1 upregulation. Besides, increased expression of Wnt9a, another let-7c-5p target gene, was also observed in the fluoride-exposed population and inversely correlated with let-7c-5p.
To define how let-7c-5p, CyclinD1 and Wnt9a interact, we performed in vitro experiments with human primary osteoblasts. Based on our own research that fluoride induces osteoblast proliferation and activation, we further demonstrated overexpressing let-7c-5p inhibited the osteoblast proliferation and activation induced by fluoride. Moreover, we confirmed a direct interaction between the let-7c-5p and CyclinD1 3′UTR, as well as that overexpressing let-7c-5p downregulates CyclinD1 transcription and translation level. These findings verify the participation of let-7c-5p in the fluoride induction of osteoblast proliferation and activation by post-transcriptional regulation of CyclinD1 expression.
Activating the Wnt/β-catenin signaling transduction pathway accelerates osteogenesis and bone transformation (Liu et al., 2019; Wang et al., 2017), and evidence suggests that fluoride ingestion regulates Wnt/β-catenin signaling in mouse odontoblast differentiation (Kao et al., 2020). However, literature reviews did not find data describing the association between Wnt9a/β-catenin pathway and fluorosis. Therefore, the expression of Wnt9a in the fluoride-exposure population and NaF-treated osteoblasts were first detected. Meanwhile, we confirmed Wnt9a was activated during fluoride exposure. Some research has demonstrated that Wnt pathway activation impedes degradation of the core factor β-catenin, and Wnt9a expression promotes β-catenin entry to the nucleus from the cytoplasm (Ling et al., 2017; Zhou et al., 2009). Accordingly, Western blotting and immunostaining detected the migration of β-catenin into the nucleus, which increased with NaF content. It is known that when β-catenin accumulates to meet a certain threshold in the cytoplasm, it migrates into the nucleus and binds to TCFs to initiate gene transcription (Xia et al., 2019). Herein, a ChIP assay demonstrated that β-catenin could bind to the transcriptional regulatory region of CyclinD1 in osteoblasts and that NaF promoted this interaction. These results verified the function of the Wnt9a/β-catenin/CyclinD1 axis in the occurrence and development of fluorosis.
Further, to elucidate let-7c-5p regulation of Wnt9a/β-catenin pathway, we demonstrated a direct interaction between the let-7c-5p and Wnt9a 3′UTR using a luciferase assay. Subsequently overexpression of let-7c-5p in osteoblasts decreased Wnt9a expression, β-catenin accumulation in the nucleus, as well as β-catenin enrichment in the transcriptional regulatory region of CyclinD1. We draw the conclusion that let-7c-5p can target Wnt9a to transcriptionally regulate CyclinD1 through the Wnt9a/β-catenin pathway.
Collectively, this work supports the hypothesis that let-7c-5p can regulate the expression of CyclinD1 at post-transcriptional and transcriptional levels, thereby inducing proliferation and activation of osteoblasts. This study also shows, for the first time, that the Wnt9a/β-catenin/CyclinD1 axis is important to fluorosis development. let-7 was found as the first human miRNA, so an in-depth understanding of the function of let-7 family will help to develop new treatments or prevent many disease (Fu et al., 2017; Wang et al., 2018, 2020). Therefore, understanding the function of let-7c-5p in skeletal fluorosis may help manage endemic fluorosis as a public health threat. Nonetheless, skeletal fluorosis development is mediated by multiple miRNAs that constitute a complex regulatory network. Further studies are needed to identify the miRNA that plays a predominant role and understand how miRNAs interact to cause skeletal fluorosis.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
National Natural Science Foundation of China (No. 81660524) and the First-class Discipline Construction Project in Guizhou Province-Public Health and Preventive Medicine (No. 2017[85]).



Comments