-
PDF
- Split View
-
Views
-
Cite
Cite
Yangbo Hu, Zakia Morichaud, Ayyappasamy Sudalaiyadum Perumal, Françoise Roquet-Baneres, Konstantin Brodolin, Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding, Nucleic Acids Research, Volume 42, Issue 16, 15 September 2014, Pages 10399–10408, https://doi.org/10.1093/nar/gku742
Close - Share Icon Share
Abstract
RbpA, a transcriptional activator that is essential for Mycobacterium tuberculosis replication and survival during antibiotic treatment, binds to RNA polymerase (RNAP) in the absence of promoter DNA. It has been hypothesized that RbpA stimulates housekeeping gene expression by promoting assembly of the σA subunit with core RNAP. Here, using a purified in vitro transcription system of M. tuberculosis, we show that RbpA functions in a promoter-dependent manner as a companion of RNAP essential for promoter DNA unwinding and formation of the catalytically active open promoter complex (RPo). Screening for RbpA activity using a full panel of the M. tuberculosis σ subunits demonstrated that RbpA targets σA and stress-response σB, but not the alternative σ subunits from the groups 3 and 4. In contrast to σA, the σB subunit activity displayed stringent dependency upon RbpA. These results suggest that RbpA-dependent control of RPo formation provides a mechanism for tuning gene expression during the switch between different physiological states, and in the stress response.
INTRODUCTION
Mycobacterium tuberculosis is one of the most successful human pathogens that can persist in human tissues for years in a dormant state which is not sensitive to a majority of antibiotics (1). Adaptation of the pathogen to the hostile environments that it meets inside host cells and its tolerance to drugs are regulated at the level of gene transcription (2,3). Transcription in bacteria is performed by the multisubunit DNA-dependent RNA polymerase (RNAP) holoenzyme composed of the catalytic core (E, subunits α2ββ’ω) and one of the σ subunits which are required for promoter-specific initiation of RNA synthesis (4,5). Recognition of the double-stranded DNA of the -10 and -35 consensus promoter elements by σ subunit domains 2 (σ2) and 4 (σ4) leads to the formation of the unstable ‘closed complex’ (RPc) between RNAP and promoter. RPc isomerizes into a transcriptionally competent ‘open complex’ (RPo) through several intermediate complexes (RPi) (6). During isomerization, a concerted action of the σ subunit and core RNAP triggers unwinding (melting) of ∼13 bp of the promoter DNA surrounding the transcription start site and makes the single-stranded DNA template available for initiation of RNA synthesis (7–10).
Each bacterial species has a characteristic library of σ subunits. The housekeeping (principal) σ subunit (σ70 in Escherichia coli or σA in M. tuberculosis) controls transcription of genes during exponential growth. Alternative σ subunits activate the transcription of specialized genes that are implicated in the stress response, virulence and the switch from exponential to stationary growth phase or to the persistent state (11,12). The M. tuberculosis genome encodes 12 alternative σ subunits, of which σB is a putative stationary phase subunit that is structurally similar to σA and orthologous to E. coli σS (13–15). Competition between σ subunits for binding to the core RNAP (σ-swapping) provides the basal regulatory mechanism for tuning bacterial gene expression in response to environmental signals (12). In addition, RNAP is regulated by a number of the non-DNA binding factors which interact with σ subunit and repress or activate transcription (reviewed by (16,17)). The molecular mechanisms that regulate activity of different σ subunits in E. coli are extensively studied, while the mechanisms employed by M. tuberculosis remain largely unknown.
M. tuberculosis RNA polymerase binding protein A (RbpA), which is present only in Actinomycetes species, has been assigned to the group of non-DNA binding factors (18,19). RbpA binds to the σ and β subunits of RNAP (20–23) and stimulates transcription dependent on the housekeeping σ subunit (19,23). RbpA is essential for M. tuberculosis growth and may play a critical role in control of pathogen physiological states because the rbpA gene was shown to be upregulated 8-fold during stationary phase, starvation, the stress response and rifampicin or vancomycin treatment (24,25). Yet, the role of RbpA in tolerance to rifampicin is not understood (22,23,26).
Three models describing the mechanism of action of RbpA are compatible with the available experimental data and include RbpA stimulating (1) holoenzyme assembly, (2) promoter complex formation or (3) promoter escape (23). To discriminate between these models, we developed a highly efficient in vitro transcription system for M. tuberculosis that allowed us to determine the step in transcription initiation regulated by the activator. Furthermore, we demonstrated that RbpA acts as promoter-specific and σ-selective activator controlling the activity of the σA and σB subunits of Mycobacterium.
MATERIALS AND METHODS
Proteins and DNA templates
Recombinant M. tuberculosis RNAP core enzyme containing 6×His-tag at the C-terminus of the β′ subunit was expressed in E. coli BL21(DE3) cells from the pMR4 plasmid and purified as described in the supplementary file (Supplementary Figure S1). The plasmids used for expression of σ subunits are listed in Supplementary Table S1. The pSR01 plasmid coding for σA and pSR5 plasmid coding for σF were a generous gift from Dr Sébastien Rodrique (27). The σ subunits were expressed in E. coli BL21(ED3) and purified from soluble fraction (σA, σC and σH) or from insoluble fraction by Ni2+-agarose affinity chromatography. M. tuberculosis RbpA protein was purified as described before (23). The C-terminal truncation of RbpA was a generous gift from Dr H. O'Hare and Dr A. Bortoluzzi. The DNA fragments bearing the M. tuberculosis promoters were amplified from genomic DNA (23) using corresponding primers (Supplementary Table S2). The forward primers were labeled with fluorescein at the 5′-end. The H37Rv genomic DNA was obtained from BEI Resources. The H37Ra genomic DNA was purchased from ATCC. lacUV5 and sinP3 promoter DNA fragments were prepared as described (10,23). Synthetic galP1AA promoter with the substitutions A-16T-17T-18G-19 to T-16G-17C-18T-19 inactivating the galP2 promoter (28) and substitutions G-8G-9 to A-8A-9 was prepared by annealing two oligonucleotides (Supplementary Table S2). sinP3 promoter with the substitution C-13 → T and sigAP promoter with the substitution T-13 → C were prepared by annealing of two oligonucleotides (Supplementary Table S2) followed by PCR amplification with the primers used for amplification of the corresponding wild type promoter DNA fragments (ref. (23) and Supplementary Table S2).
In vitro transcription and electrophoretic mobility shift assays
Transcription was performed in 5 μl of transcription buffer (TB, 20 mM Tris–HCl pH 7.9, 50 mM NaCl, 5 mM MgSO4, 1 mM dithiothreitol (DTT), 0.1 mM ethylenediaminetetraacetic acid (EDTA) and 5% glycerol). The RNAP holoenzyme was assembled by mixing 600 nM σ subunit with 200 nM core RNAP and incubation for 5 min at 37°C. RbpA at 600 nM or at the concentrations indicated on the figures was added to the mixtures and incubated for 5 min. Promoter DNA fragment (15 nM) was added and incubated at 37°C for 10 min. Transcription was initiated by the addition of 50 μM adenosine triphosphate (ATP), guanosine triphosphate (GTP), cytidine triphosphate (CTP) and 3 μCi [α-32P] uridine triphosphate (UTP) and carried out for 5 min at 37 °C. The reactions were stopped by adding 8 M urea and the synthesized RNA products were analyzed on denaturing (7 M urea) 18% polyacrylamide gelelectrophoresis (PAGE). For the electrophoretic mobility shift assays (EMSA) experiments, the fluorescein-labeled promoter fragments were mixed with RNAP or RNAP-RbpA complex, prepared as described above, in 10 μl TB and incubated for 10 min at 37°C. Then, 20 μg/ml of poly (dA-dT) was added and incubated for 5 min at 37°C. Afterwards, samples were resolved on 5% native 0.5×TBE-PAGE. Gels were scanned by Typhoon 9200 Imager (GE Healthcare) and quantified using ImageQuant software (Molecular Dynamics).
DNase I footprinting and KMnO4 probing
For DNase I footprinting experiments, fluorescein-labeled promoter DNA fragment (40 nM) was mixed with 400 nM RNAP in 50 μl TB. RbpA was added before promoter complex formation as described for in vitro transcription. The samples were treated with 2 U/ml DNAse I (Promega) for 1 min at 37°C. The reactions were stopped by addition of 10 mM EDTA (pH 8.0) and 400 ng poly(dA-dT). For KMnO4 probing, RNAP-promoter complexes were formed as described for DNase I footprinting. The samples were treated with 5 mM KMnO4 for 30 s at 37°C. Reactions were quenched by addition of 1/2 volume of 1 M 2-mercaptoethanol, 1.5 M Na(CH3COO) pH 7.0. Samples were treated with 0.5 M piperidine and DNA fragments were analyzed on 8% sequencing gel.
RbpA labeling and native gel analysis
Purified RbpA protein was conjugated with the sulfhydryl-reactive dye, DyLight633 maleimide (Thermo Scientific), at the single cysteine site (C56) according to the manufacturer protocol. Briefly, 1 mg of RbpA protein was incubated with the 20 μl maleimide-activated dye in the conjugation buffer (0.1 M phosphate, 0.15 M NaCl, 2 mM EDTA, pH 7.4) at 24°C for 2 h. The excess dye reagent was then removed from the sample by dialysis. Protein was concentrated to 4 mg/ml by Ultracel-10 membrane filter unit (Millipore) and stored at −20°C in 50% glycerol. For the native gel analysis, the labeled RbpA (1.6 μM) was incubated with 2.4 μM of indicated σ subunit in 10 μl TB at 37°C for 15 min. The complexes were analyzed on 5–10% native PAGE in Tris–Glycine buffer. Gels were scanned by Typhoon 9200 Imager (GE Healthcare) and quantified using ImageQuant software (Molecular Dynamics).
RESULTS
RbpA stimulates transcription by RNAP containing either the σA or σB subunit
To explore the role of RbpA in regulation of M. tuberculosis transcription, we designed a plasmid co-overexpressing the M. tuberculosis RNAP core subunits α, β, β′ and ω (Supplementary Figure S1). The plasmid allowed us to achieve high expression levels of core RNAP in E. coli cells, and in vivo-assembled enzyme was more stable and displayed higher specific activity than RNAP that was assembled in vitro from individually expressed subunits (23,27). To define the full range of σ subunits that are regulated by RbpA, all 13 σ subunits from M. tuberculosis were expressed, purified and assembled with the core RNAP (Supplementary Figure S1). The ability of the σ subunits to drive promoter-specific transcription initiation was tested in multiple-round transcription assays using DNA fragments (≈100 bp in length) bearing M. tuberculosis promoters that were reported to be recognized by their corresponding σ subunits (Figure 1A, Supplementary Figure S2, Supplementary Table S3). Transcription was performed either in the presence or in the absence of RbpA, and core RNAP alone was used to control the specificity of the initiation reaction. All σ subunits except the σG and σK, supported transcription initiation by the RNAP holoenzyme, while no or little RNA synthesis was detected when using core RNAP alone. The lack of detectable activity for σG and σK may be caused by misfolding of the recombinant proteins, or the inability of these σ subunits to recognize the tested promoters without auxiliary transcription activators. The transcription patterns varied significantly between σs and displayed a large amount of RNA products that were shorter than the expected 40–50 nucleotides (nt) run-off products that could form due to transcription pausing or arrest. Noticeably, the σC- and σH-containing RNAPs displayed defects in promoter escape and produced mainly short RNA products. The σB subunit and, to a lesser extent, σA were only weakly active in initiation of RNA synthesis from the sigAP and rrnAP3 promoters without RbpA (Figure 1A, Supplementary Figure S2A). The addition of RbpA strongly stimulated transcription initiated by the σB-containing RNAP (≥10-fold), while the activity of none of the other alternative σ subunits was increased in the presence of the activator. The activation level for σA-RNAP was weaker (∼3-fold) and corresponded to the one previously reported for the in vitro assembled holoenzyme (23).
Screening for RbpA-targeted σ subunits. (A) The [32P]-RNA products synthesized in the multiple-round transcription assay from the indicated promoters. Transcription was carried by RNAP holoenzymes containing the indicated σ subunit in the presence or absence of RbpA. For each σ panel, the RNA products that are marked by an asterisk were quantified and normalized to the signal obtained in the presence of RbpA. Results of the quantification are presented as bar graphs and are shown below each panel. (B) EMSA of the promoter complexes formed in the presence or absence of RbpA under the same conditions as in panel A. (C) Scheme showing the organization of the M. tuberculosis σ subunits, with numbers representing the evolutionarily conserved regions, and arrows indicating the interactions with the promoter elements. (D) Screening for the interactions between RbpA and free σ subunits using native gel electrophoresis. Free RbpA labeled with the DyLight633 dye (RbpA-DL) migrated at the bottom of the gel. All σ subunits were added at 2.4 μM, and RbpA was added at 1.6 μM.
RbpA stimulates stable promoter complex formation by the σB-RNAP
Previously, we showed that RbpA stimulates the formation of stable promoter complexes by the σA-RNAP (23). To explore if RbpA can affect promoter binding of RNAPs containing alternative σ subunits, we performed EMSA using the above-mentioned promoter DNA fragments that were end-labeled with fluorescein (Figure 1B, Supplementary Figure S2 showing complete images of the gels). To suppress non-specific DNA binding, the promoter complexes were challenged by adding poly(dA–dT) as a competitor. No formation of competitor-resistant complexes between the σB-RNAP and sigAP promoter was detected without RbpA, which was in agreement with the results of the transcription assay. Meanwhile, all RNAPs containing alternative σs, that were active in transcription, formed competitor-resistant complexes at their corresponding promoters (Figure 1B). The addition of RbpA induced the formation of competitor-resistant promoter complexes of σB-RNAP but did not affect complexes formed by RNAPs bearing other alternative σ subunits. The competitor-resistant complex of σA-RNAP at the rrnAP3 promoter was formed even without RbpA, but its formation was stimulated if RbpA was present in the reaction (23). Surprisingly, we observed no stable complex formation between σA-RNAP and the sigAP promoter fragment, even in the presence of RbpA (Supplementary Figure S2B), which was in striking contrast to the stimulation effect that was observed using the transcription assay (Supplementary Figure S2A). Therefore, σA-RNAP and σB-RNAP recognized the same promoter sequence but formed structurally different open complexes. Together, the results of the transcription and EMSA assays suggest that RbpA only stimulates the activity of the structurally similar σ70-like group 1 and group 2 σ subunits (Figure 1C) and has a strong bias for the stress-response σB subunit.
RbpA binds σB but none of the other alternative σ subunits
RbpA has been shown to bind to free σA and σB subunits (20,21). To test if the ability of RbpA to activate transcription correlates with its ability to bind σ, we tested the interactions between RbpA and free σ subunits using native gel electrophoresis (Figure 1D). To monitor RbpA-σ complex formation RbpA was labeled with the DyLight633 dye at the single Cys56 residue whose modification does not influence RbpA activity (23,26). Each of the 12 M. tuberculosis σs was mixed with RbpA in the presence of BSA to reduce non-specific binding, and the resulting complexes were resolved on a native gel. In agreement with the results of the transcription assay, we observed that RbpA formed stable complexes with σA and σB, but not with any of the other alternative σ subunits. Because the concentrations of σs were equal (2.4 μM) in all samples but only 20% of RbpA (at 1.6 μM) was bound to σA compared with 100% that was bound to σB, we concluded that RbpA has higher affinity for the latter subunit. Therefore, the ability of RbpA to activate transcription correlated with its ability to bind free σ subunits and indicated that the interaction with σ may be a part of the activation mechanism. In support of this conclusion, RbpA lacking its C-terminal domain, which is required to interact with σ subunits (20), was unable to stimulate promoter complex formation (Supplementary Figure S2N).
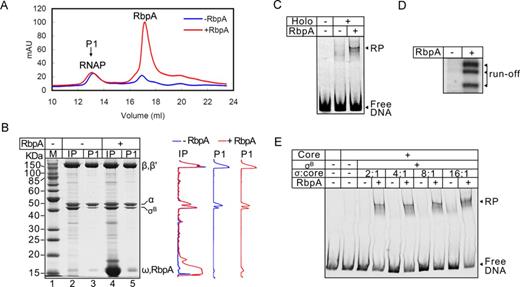
Stabilization of the σB-RNAP holoenzyme by RbpA is not the basis of transcription activation
Previously, we proposed that the basis of RbpA-mediated transcription activation was an increased affinity of σ for the core, which leads to stabilization of the RNAP holoenzyme (23). Because RbpA binds σ and the core, it may affect the stability of the holoenzyme by bridging the partners. Therefore, the lack of a stable promoter complex without RbpA can be explained by low stability of the σB-RNAP holoenzyme. To explore this idea, we compared the stability of a σB-RNAP holoenzyme that was assembled with or without RbpA using chromatography with a Superose-6 gel-filtration column (Figure 2A). The fractions containing RNAP holoenzyme (Figure 2A, peak ‘P1’) were pooled and analyzed using SDS-PAGE (Figure 2B). Quantification of the gel showed that RbpA increased the retention of σB in the holoenzyme (Figure 2B, gel profiles on the right), which was in agreement with the previous result for the σA subunit (23). Additionally, RbpA co-eluted with RNAP suggesting that it was stably bound to the holoenzyme. However, even without RbpA, the RNAP holoenzyme exhibited 60% binding to σB compared with the one assembled in the presence of RbpA suggesting that RbpA is not obligatory for holoenzyme assembly. To test if the assembled σB-RNAP holoenzyme that was collected using gel-filtration was responsive to RbpA, EMSA assay and a run-off transcription assay using the sigAP promoter were performed (Figure 2C and D). The experiment showed that purified σB-RNAP holoenzyme could not form a stable promoter complex and initiate transcription using the sigAP promoter, while addition of RbpA stimulated both events. Because RbpA activates transcription more than it stimulates formation of the holoenzyme (>10-fold versus 1.7-fold correspondingly) we proposed that stabilization of the holoenzyme is not a major cause of transcription activation. If the function of RbpA is to compensate for the low affinity of σ for the core, then it could be bypassed by increasing the concentration of σ. To test this assumption, we performed EMSA using the sigAP promoter in the presence of different amounts of σB (Figure 2E). No stimulation of promoter complex formation was observed in the absence of RbpA, even when 16-fold excess of σB (3.2 μM) over the RNAP core (200 nM) was used. Thus, we concluded that activation of σB-dependent transcription is not caused by increased affinity of σB for the core RNAP, but is due to a RbpA-mediated conformational change in the holoenzyme, σ subunit, or both that stimulates promoter complex formation.
RbpA stabilizes the σB-RNAP holoenzyme. (A) Superose-6 elution profile of the σB-RNAP holoenzyme assembled in the presence or absence of RbpA. P1 indicates the peak corresponding to the RNAP core and holoenzyme. (B) SDS-PAGE of the RNAP holoenzyme that was assembled in the presence (lanes 4 and 5) or absence of RbpA (lanes 2 and 3) and fractionated on a Superose-6 column as shown in panel A. Samples of the input (IP) before fractionation and of the pooled peak fractions (P1) are shown. Profiles on the right show the scan of lanes 2 and 3 (black) and 4 and 5 (gray). (C) Effects of RbpA on promoter complex formation by the pre-assembled σB-RNAP (from the panel B, lane 3). (D) Run-off RNA products that were synthesized by the pre-assembled σB-RNAP in a single round of transcription from the sigAP promoter DNA. (E) EMSA of the σB-RNAP complexes with the sigAP promoter formed in the presence of increasing concentrations of σB (0.4, 0.8, 1.6, 3.2 μM). RbpA (800 nM) was added where indicated.
RbpA is required for promoter binding and RPo formation
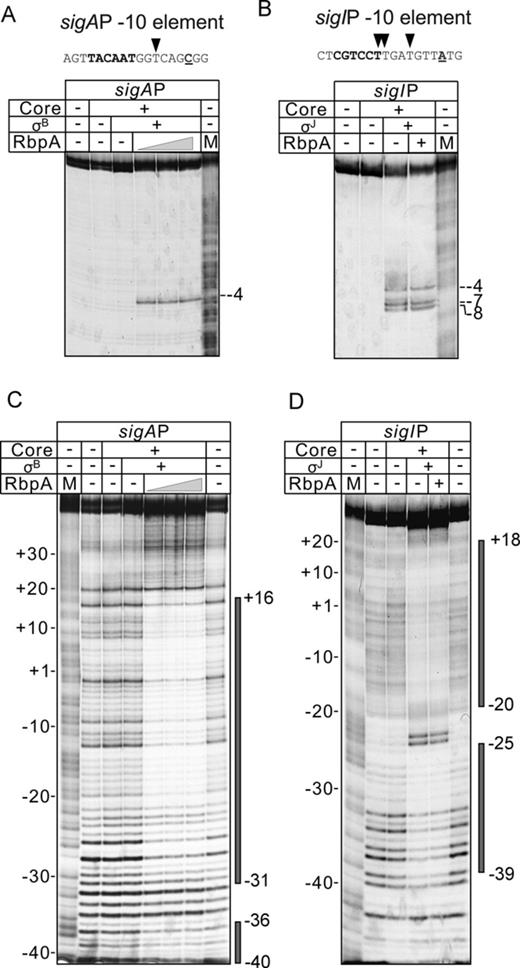
To determine which of the steps on the pathway to the open promoter complex (RPo) formation is targeted by RbpA, DNA–protein interactions in the σB-RNAP promoter complexes were probed with KMnO4 and DNaseI at equilibrium conditions in the absence of competitor. As a control we followed binding of σJ-RNAP to the sigI promoter, which is non-responsive to RbpA. First, we tested whether σB-RNAP could form RPo at the -10/-35 consensus sigAP promoter. Promoter DNA melting was monitored by probing the accessibility of the thymine at position -4 (T-4) of the non-template DNA strand to KMnO4 (Figure 3A). The experiment showed that T-4 was accessible to KMnO4 only when RbpA was present in the reaction. Therefore, even at equilibrium conditions (without competitor) at 37°C, the σB-RNAP was unable to melt promoter DNA without RbpA. RNAP assembled with the σJ subunit formed RPo at the sigIP promoter in the absence of RbpA. Furthermore, the sigIP promoter melting, marked by the presence of unpaired thymines at positions -4, -7 and -8 of the non-template DNA strand, was not affected by addition of the activator, which was in agreement with the results of the transcription and EMSA assays (Figure 3B).
RbpA is required for the sigAP promoter binding and melting by σB-RNAP. (A) KMnO4 probing of the sigAP promoter complex formed by σB-RNAP and (B) the sigIP promoter complex formed by σJ-RNAP. The sequences of the melted promoter regions of the sigAP and sigIP promoters are shown at the top of the panels. The -10 promoter element is indicated in bold, and the transcription start site base is underlined. The thymines of the non-template DNA strand that were accessible to KMnO4 are indicated by triangles. RbpA was added at 400, 800 and 1600 nM (panel A) or at 1600 nM (panel B). M: A+G sequencing marker. The positions of the thymines that were reactive to KMnO4 in the open promoter complex are indicated. (C) DNase I footprinting of the sigAP promoter complex with σB-RNAP and (D) the sigIP promoter complex with σJ-RNAP. Promoter DNA was labeled on the non-template strand. RbpA was added at the concentrations listed in panels A and B. The promoter regions that were protected from DNase I by RNAP are indicated by gray boxes on the right side of the panels.
The lack of detectable promoter melting does not exclude that RNAP can bind to a promoter and form RPc or RPi complexes. Thus, the closed promoter complexes, which do not have a melted DNA region, were detected on several E. coli promoters using DNase I footprinting (29–32). To test if M. tuberculosis σB-RNAP could also bind to the sigAP promoter in the absence of RbpA, we performed DNase I footprinting (Figure 3C) under the same conditions as those used for KMnO4 probing. No protection of the promoter DNA from DNase I was detected without RbpA, while addition of the activator resulted in protection of positions -40 to +16. The RNAP holoenzyme containing the σJ subunit protected positions -39 to +18 of the promoter DNA equally well with or without RbpA (Figure 3D). The lack of a detectable footprint at the sigAP promoter without RbpA indicates that closed promoter complex is highly unstable and cannot be detected by DNase I due to its short life time. Thus, we concluded that σB-RNAP is ineffective in promoter binding and isomerization to RPo at the sigAP promoter and requires RbpA to accomplish this process.
The requirement of RbpA for RPo formation is dependent on the promoter sequence
The inability of σB-RNAP to form RPo without RbpA may arise from the low affinity of σB to the -10 and -35 elements of promoter. Indeed, the sequences of the -10 element of the sigAP (5′-TACAAT-3′) and the rrnAP3 (5′-TAGACT-3′) promoters differ from the -10 consensus sequence 5′-TATAAT-3′. Furthermore, the -35 element of the sigAP promoter (5′-TGTACT-3′) displays only weak similarity to the -35 consensus 5′-TTGACA-3′. To examine whether the variations in promoter sequence can modulate the efficiency of RPo formation, we performed EMSA using ‘strong’ E. coli promoters: lacUV5 and the ‘extended -10’ galP1AA (derivative of galP1) containing the perfect -10 consensus sequence. We reasoned that using of the ‘extended -10’ promoter, which does not contain the -35 element, allows to neglect the impact of upstream interaction between the σ domain 4 and the -35 element on RPo formation. However, stable complex formation with both promoters showed RbpA-dependence (Supplementary Figure S2O). Also, no melting of the galP1AA promoter was detected by the KMnO4 probing performed in the absence of RbpA (Supplementary Figure S2P). These results suggest that neither a perfect match to the -10 consensus nor the presence of the extended -10 motif ‘TG’ can stimulate RPo formation or suppress the requirement in RbpA. Interestingly, the hybrid enzyme comprising the core RNAP of E. coli and σB formed stable promoter complex with the lacUV5 promoter in the absence of RbpA, suggesting that requirement in RbpA for RPo formation depends on the interplay between σB and core RNAP (Supplementary Figure S3A).
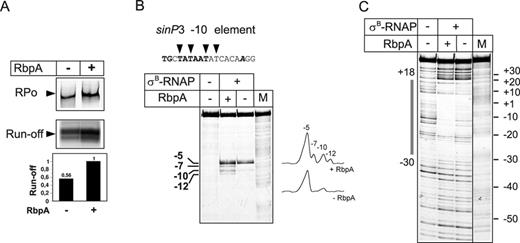
We showed previously that σA-RNAP, without RbpA, was unable to form stable promoter complexes with the ‘extended -10’ consensus sinP3 promoter from B. subtilis (23). Strikingly, in contrast to σA-RNAP, the σB-RNAP formed a competitor-resistant complex with the sinP3 promoter and successfully initiated transcription without RbpA, although RbpA stimulated both reactions approximately 2-fold (Figure 4A). The KMnO4 probing of the sinP3 promoter complex, formed by σB-RNAP without RbpA, showed that thymine at position -5 of the non-template strand was accessible to KMnO4 and a weak reactivity was observed at the position -10 (Figure 4B). In addition, a clear protection of the sinP3 promoter from DNaseI was observed (Figure 4C) suggesting that RbpA is not essential for RPo formation at the sinP3 promoter. Noticeably, addition of RbpA stimulated unwinding of the upstream part of transcription bubble (thymines at positions -7, -10, -12) and enhanced DNaseI protection, indicating that activator still contributes to the promoter melting and stabilization of the complex. These data suggest that σB-RNAP activity is restricted to a limited set of promoters while RbpA broadens the range of promoters recognized by RNAP.
RbpA is dispensable for RPo formation at the sinP3 promoter. (A) Formation of the complexes between σB-RNAP and the sinP3 promoter in the presence or absence of RbpA was tested by EMSA (‘RPo’ panel) and a multiple-round transcription assay (‘run-off’ panel). Quantification of the run-off RNA products is presented as a bar graph below the panel. Values were normalized to the signal obtained in the presence of RbpA. (B) KMnO4 probing and (C) DNase I footprinting of the sinP3 promoter complexes formed by σB-RNAP. Promoter DNA was labeled on the non-template strand. DNA sequence of the open region of the sinP3 promoter is shown at the top of the panel B. The -10 promoter element is indicated in bold, and the transcription start site base is in bold italic. The thymines of the non-template DNA strand that were accessible to KMnO4 are indicated by triangles. M: A+G sequencing marker. The promoter region that was protected from DNase I by RNAP is indicated by a gray box on the left side of the panel C.
Cytosine at position -13 (C-13) was shown to be required for efficient recognition of promoters by the E. coli stress-response σS(33) which is orthologous to σB. SinP3 promoter carries C at position -13 while T is found at the corresponding position of the sigAP promoter. To explore if C-13 is responsible for weak dependence of the sinP3 promoter on RbpA, we constructed two mutant templates: sinP3 with the substitution C-13 → T (sinP3mut) and sigAP with the substitution A-13 → to C (sigAPmut) (Supplementary Figure S4A). The activity of the mutant promoters was tested in EMSA and transcription assays with the σB-RNAP (Supplementary Figure S4B, C and D). The assays showed that the mutant sinP3 promoter was still active in transcription in the absence of RbpA, yet an overall activity was reduced to 70% of that observed with the wild type template. The mutant sigAP promoter was unable to support transcription in the absence of RbpA. No effect of the substitutions on the promoter activity was observed in the presence of RbpA. These results suggest that C-13, while contributes to recognition of sinP3 promoter by σB-RNAP, does not obviate the requirement for RbpA in transcription initiation.
The fact that σB-RNAP recognized the same promoters as σA-RNAP and σ70-RNAP suggests that these σs have similar promoter consensus sequence specificity. In support to this conclusion, alignment of the σ subunits regions 2, 3 and 4 revealed that residues important for recognition of the DNA bases of the -10 and -35 elements are identical between σA, σB and σ70 (Supplementary Figure S3B).
RbpA remains bound to RNAP in the promoter complex
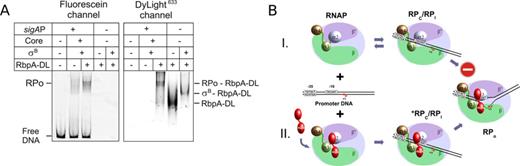
Two scenarios could describe the fate of RbpA during initiation: (1) RbpA binds to RNAP transiently and dissociates as soon as the active promoter complex is assembled (chaperone-like function), or (2) RbpA remains bound to RNAP in the promoter complex as a true transcription initiation factor. To determine which model is correct, we performed EMSA using a fluorescein-labeled sigAP promoter and DyLight633-labeled RbpA (RbpADL) (Figure 5A). This approach allowed for simultaneous detection of the binding of promoter DNA (fluorescein channel) and RbpA (DyLight633 channel) to RNAP. RbpADL induced open complex formation by the σB-RNAP with the same efficiency as the unlabeled RbpA (Figure 5A, fluorescein channel). Scanning of the gel using the DyLight633 channel revealed that nearly all RbpA was shifted and co-localized with the RNAP-sigAP promoter complex. This finding supported the idea that RbpA is an integral component of the initiation complex and does not dissociate after open complex formation. In support of this view, ChIP analysis performed on Streptomyces showed that RbpA co-localized with RNAP at promoter regions in vivo (21).
RbpA remains bound to RNAP in the promoter complex. (A) EMSA of the complexes between σB-RNAP and sigAP promoter DNA that were performed either in the absence or presence of 800 nM of DyLight633-labeled RbpA (RbpA-DL). The gel was scanned either at an emission wavelength of 526 nm to detect the DNA fragment (fluorescein channel) or at 670 nm to detect RbpA (DyLight633 channel). (B) A model of the RbpA action. RNAP is presented schematically as a light blue ellipse with the β subunit in green, the β′ subunit in violet. The σ subunit domains 4 (brown), 3 (green) and 2 (blue) are represented as ellipsoids. RbpA is shown by two red ellipsoids representing two structural domains of the protein (20). The sigAP promoter DNA is represented by black lines with the sequences of the -10 and -35 elements in boxes. Transcription start site (+1) is indicated by arrow.
DISCUSSION
In the previous work we proposed that RbpA helps the σA subunit to compete with the alternative σs for binding to core RNAP by stimulating assembly of the σA-RNAP holoenzyme (23). Our present study demonstrated that RbpA induces formation of the catalytically competent RPo complex by the RNAP containing the stress-response σB subunit. In addition, the assembly of the σB with core RNAP was also stimulated by RbpA but did not contribute significantly to the activation of transcription. Based on these results we suggest that control of the σ factors activity by RbpA occurs through the stimulation of RPo formation but not through the stimulation of the assembly of a particular σ with core RNAP.
RbpA comprises two structural domains connected by a flexible linker (20,21). The structured core domain interacts with the β subunit (22,23) while the C-terminal domain binds to the σ subunit domain 2 (20,21). We propose a model where RbpA interaction with σ and β subunits remodels RNAP holoenzyme structure, induces an optimal fit between the σ subunit and promoter consensus elements and thus facilitates the isomerization from the RPc to RPo (Figure 5B). In support to this model, the requirement in RbpA for RPo formation can be bypassed by changing promoter DNA sequence or by assembly of σB with heterologous core RNAP from E. coli. Considering that the efficiency of the RPc to RPo conversion is modulated by a promoter DNA sequences, promoter spacing and numerous transcriptional activators (6,34), we expect that a requirement in RbpA for initiation may be limited to a subset of Mycobacterium promoters. Our finding that RbpA is dispensable for the RPo formation at the sinP3 promoter supports this hypothesis. An intriguing question for the future studies will be to define promoter DNA motifs modulating RbpA activity and to explore its interplay with other transcription factors.
RbpA increases promoter melting potential of the group 1 and group 2 σ subunits of M. tuberculosis
An unexpected finding of our study is that M. tuberculosis RNAP containing the principal-like σB subunit is deficient in promoter melting and stable RPo formation at promoters bearing a perfect or nearly perfect -10 consensus element or the ‘extended -10’ element (sigAP, lacUV5, galP1cons). This finding contrasts with the ability of E. coli RNAP containing the orthologous σS to efficiently unwind promoters with perfect consensus elements (35,36). The molecular basis of this deficiency is likely in the inability of the σB-RNAP to undergo conformational changes required to form RPo. The activity of the σA-RNAP was also stimulated by RbpA, but it did not display a strong requirement for the activator as was observed for σB-RNAP. Based on the above observations, we propose that σB-RNAP is under ‘stringent’ control by RbpA, at least at a set of housekeeping promoters, while σA-RNAP is under ‘relaxed’ control. In contrast to the principal group 1 and group 2 σs, the group 3 and group 4 alternative σs of M. tuberculosis are ‘melting proficient’ and promote stable promoter complex formation in the absence of RbpA. Therefore, RNAPs containing alternative σs are ready to be engaged at promoters and initiate transcription as soon as the holoenzyme is assembled.
Regulation of σB activity by RbpA and the stress response
The σB subunit is responsible for the expression of genes during stationary phase, starvation and the stress response (14,24–25,37–39). The expression of the rbpA gene is also upregulated at these physiological states and induced by antibiotics (19,25). The strong dependence of σB-RNAP activity on RbpA leads to a proposal that RbpA is a principal regulator of the σB-dependent stress response in Mycobacterium. Also, this finding supports the view that role of RbpA in tolerance to rifampicin is indirect (23) and may be linked to the stress response leading to inactivation or elimination of the drug (2). Because, according to our results, σB displayed the same promoter sequence specificity as σA, we predict that the genes that are controlled by σA could be efficiently transcribed by σB-RNAP in the presence of RbpA. The σB–RbpA pair might serve as an alternative system to support the expression of housekeeping genes during conditions when σA gene expression is repressed or σA is sequestered. Noticeably, none of the DNA templates that were used in our work and recognized by σB-RNAP displayed any similarity to the sequence, 5′-NNGNNG-3′, which was suggested as a -10 consensus for σB-dependent promoters (14). That discrepancy may reflect a high ‘flexibility’ in the usage of the -10 consensus by σB or its erroneous attribution.
RbpA differs from other global regulators of transcription
Previously we proposed that RbpA functions similar to the Crl protein (23), which was found in γ-Proteobacteria and belongs to a small group of transcription factors that do not bind DNA (16). Indeed, similar to RbpA, Crl stimulates the activity of the E. coli stationary phase σS by binding the region 2 and increasing the affinity of σS for core RNAP (40,41). However, our current study revealed that RbpA acts though a different mechanism because stabilization of the holoenzyme by RbpA is not a basis for the RbpA-driven stimulation of transcription. The mode of RbpA action also differs from the recently described transcription factor CarD, which was suggested to be a global regulator of transcription in M. tuberculosis (42). In contrast to RbpA, CarD was shown to bind to double-stranded DNA and proposed to act through β1-lobe interactions by stimulating RPo formation (43).
Lastly, our results delineate RbpA as an essential co-factor of M. tuberculosis RNAP and a global regulator of the expression of housekeeping genes in Mycobacteria and likely in other Actinomycetes. A challenge for future studies will be to define the full set of genomic targets of RbpA and to explore its role in pathogenesis, tolerance to antibiotics and bacterial fitness.
We thank Dr M. Buck for critical reading of the manuscript and Dr H. O'Hare and Dr A. Bortoluzzi for providing a sample of the RbpA mutant.
FUNDING
CNRS and INSERM; Fondation pour la Recherche Médicale (FRM) [to Y.H.]; ERASMUS MUNDUS Svaagata fellowship [to A.S.P.].
Conflict of interest statement. None declared.
Present address: Yangbo Hu, Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan 430071, China.



![Screening for RbpA-targeted σ subunits. (A) The [32P]-RNA products synthesized in the multiple-round transcription assay from the indicated promoters. Transcription was carried by RNAP holoenzymes containing the indicated σ subunit in the presence or absence of RbpA. For each σ panel, the RNA products that are marked by an asterisk were quantified and normalized to the signal obtained in the presence of RbpA. Results of the quantification are presented as bar graphs and are shown below each panel. (B) EMSA of the promoter complexes formed in the presence or absence of RbpA under the same conditions as in panel A. (C) Scheme showing the organization of the M. tuberculosis σ subunits, with numbers representing the evolutionarily conserved regions, and arrows indicating the interactions with the promoter elements. (D) Screening for the interactions between RbpA and free σ subunits using native gel electrophoresis. Free RbpA labeled with the DyLight633 dye (RbpA-DL) migrated at the bottom of the gel. All σ subunits were added at 2.4 μM, and RbpA was added at 1.6 μM.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/nar/42/16/10.1093_nar_gku742/1/m_gku742fig1.jpeg?Expires=1716442294&Signature=Y6-GDWZSf10sjbxm8lYq-QGr5rqIsKeCuYnECtEkE2CmJD0wWUKf1uAiYfcdzUXjdlhPFLGJH-c4xwrHTPUJ8WOrHiOHsO1zwHRjHoRW~5zlfKDZPdDPx8SY0Yj3v6f9QW1L79IbLyxjPmWqHZWTqUcrlaQxsKNh7zvho9FxM8PCAZDiluyglnuKPMwr9DTVlb6UQE797EWrqq1b3Z4tsY7xsw9ojX87G~fCiffbPr2~PYElYi5MfsL4CQxpyzZBvzzGSNWJeJkwh3Trp3-uFbxVenqEOM-vVBaN-usP4aM2tV-oGx2OxNW2tF9RMuZqxv-6te1TZKfoU13XaZpM3Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

Comments