-
PDF
- Split View
-
Views
-
Cite
Cite
Yang Wang, Xue-Jia Hu, Xu-Dong Zou, Xian-Hui Wu, Zhi-Qiang Ye, Yun-Dong Wu, WDSPdb: a database for WD40-repeat proteins, Nucleic Acids Research, Volume 43, Issue D1, 28 January 2015, Pages D339–D344, https://doi.org/10.1093/nar/gku1023
Close - Share Icon Share
Abstract
WD40-repeat proteins, as one of the largest protein families, often serve as platforms to assemble functional complexes through the hotspot residues on their domain surfaces, and thus play vital roles in many biological processes. Consequently, it is highly required for researchers who study WD40 proteins and protein–protein interactions to obtain structural information of WD40 domains. Systematic identification of WD40-repeat proteins, including prediction of their secondary structures, tertiary structures and potential hotspot residues responsible for protein–protein interactions, may constitute a valuable resource upon this request. To achieve this goal, we developed a specialized database WDSPdb (http://wu.scbb.pkusz.edu.cn/wdsp/) to provide these details of WD40-repeat proteins based on our recently published method WDSP. The WDSPdb contains 63 211 WD40-repeat proteins identified from 3383 species, including most well-known model organisms. To better serve the community, we implemented a user-friendly interactive web interface to browse, search and download the secondary structures, 3D structure models and potential hotspot residues provided by WDSPdb.
INTRODUCTION
WD40-repeat protein, named for containing one or multiple WD40 domains, is one of the largest protein families. About 1% genes in human genome encode proteins that belong to this family (1). A typical WD40 domain consists of 6–8 structurally conserved WD40 repeats, each of which containing four anti-parallel β-strands, and then folds into a β-propeller conformation exposing three types of surfaces, i.e. top, bottom and side surfaces. Through certain residues on these surfaces (hotspot residues), WD40 proteins extensively take part in protein–protein interactions (PPIs) (2–4), and as a result, they often serve as hubs in cellular networks. They mainly provide platforms to assemble proteins or nucleic acids into functional complexes, which play vital roles in many biological processes, such as DNA replication, transcription, RNA processing, histone modification/recognition and protein degradation. For example, through PPIs provided by WD40 platforms, protein complexes such as E3 ubiquitin ligase (5,6), G protein (7) and MLL1 (8) are formed and then able to perform their bioactivities. Consequently, it is of great importance to predict or annotate the structural information and potential PPI hotspot residues of WD40 domains in order to understand the functionality of them.
To date, protein family (including the WD40-repeat family) annotations are presented in several databases, such as UniProt (9), SMART (10), Pfam (11), Prosite (12) and Superfamily (13). Although these databases provide valuable information about thousands of protein families or subfamilies, sensitively identifying WD40-repeat proteins and deriving their structure information remains a challenge. As a result, the number of WD40 proteins in proteomes is still much underestimated (1). The main difficulty of family annotation for WD40 proteins is that the average pairwise sequence identity for WD40 domain is too low for most regular HMM or sequence pattern recognition models (14). Moreover, for most WD40 proteins, detailed structural information and potential residues for PPIs are still lacking in those general-purpose databases. It would be highly demanding to develop a comprehensive WD40-repeat protein family-specific knowledgebase to provide such important information.
Recently, we reported a method WDSP (WD40 Structure Predictor) to identify WD40 repeats and to predict the secondary structures of WD40 domains based on their primary sequences (14). More practically, it can determine the repeat and beta-strand boundary more accurately based on better-predicted secondary structures (14). We further improved this tool to build the 3D structure models of WD40 domains as well as the potential surface hotspot residues responsible for PPIs. WDSP incorporated both local residue information (amino acid occurrence preference and loop length propensity) and non-local WD40 family specific structural features (conserved hydrogen-bond network (15,16) and β-bulge (17) into the scoring function, and used a genetic algorithm to combine the identified WD40 repeats into domains. As a result, it achieved higher sensitivity than other general-purpose tools such as Pfam, Prosite and SMART, while keeping very low false positive rates (14).
By using WDSP, we scanned all protein sequences in UniProt database and annotated 63 211 WD40 proteins, each of which comprises at least six WD40 repeats. In addition to the predicted secondary structures and potential hotspot residues, the tertiary structure models for these WD40 domains were built and stored in the database, namely WDSPdb. WDSPdb used a user-friendly 3D structure visualization interface and a color-highlighted texting manner to display the aforementioned detailed information. We believe WDSPdb will benefit the WD40 and PPI research community, especially the experimentalists who are not familiar with protein structure modeling tools.
MATERIALS AND METHODS
Data summary
The data source of protein sequences in WDSPdb is from the UniProt knowledgebase (release version 201310). Taken together, 63 211 WD40-repeat proteins with 71 480 WD40 domains and 489 411 WD40 repeats from 3383 species were identified by the WDSP program (Table 1). Among these proteins, 726 685 potential hotspot residues responsible for PPIs on the top surface were predicted. WD40 proteins were known to be abundant in eukaryotes while considered rare in prokaryotes (1). Interestingly, a large number of bacteria WD40 proteins were also hit by WDSP program and were stored in our database. The WDSP program also identified some WD40 proteins in archaea and viruses.
Statistics of WDSPdb. The numbers of identified WD40 proteins (with ≥6 repeats), WD40 domains, WD40 repeats, and potential hotspots in total, different taxa and several model organisms
| Category . | WD40 proteins . | WD40 domains . | WD40 repeats . | Potential hotspots . | Species . |
|---|---|---|---|---|---|
| Total | 63 211 | 71 480 | 489 411 | 726 685 | 3383 |
| Eukaryota | 58 284 | 65 311 | 447 323 | 662 833 | 860 |
| Bacteria | 4832 | 6065 | 41 358 | 62 704 | 2476 |
| Archaea | 50 | 59 | 419 | 637 | 34 |
| Virus | 45 | 45 | 311 | 511 | 13 |
| Homo sapiens | 610 | 708 | 4837 | 7084 | |
| Mus musculus | 562 | 659 | 4508 | 6530 | |
| Danio rerio | 407 | 467 | 3242 | 4788 | |
| Drosophila melanogaster | 299 | 319 | 2193 | 3159 | |
| Caenorhabditis elegans | 142 | 157 | 1076 | 1562 | |
| Arabidopsis thaliana | 358 | 384 | 2635 | 3866 | |
| Oryza sativa | 16 | 18 | 123 | 178 | |
| Saccharomyces cerevisiae | 83 | 92 | 635 | 969 | |
| Schizosaccharomyces pombe | 104 | 115 | 787 | 1,147 |
| Category . | WD40 proteins . | WD40 domains . | WD40 repeats . | Potential hotspots . | Species . |
|---|---|---|---|---|---|
| Total | 63 211 | 71 480 | 489 411 | 726 685 | 3383 |
| Eukaryota | 58 284 | 65 311 | 447 323 | 662 833 | 860 |
| Bacteria | 4832 | 6065 | 41 358 | 62 704 | 2476 |
| Archaea | 50 | 59 | 419 | 637 | 34 |
| Virus | 45 | 45 | 311 | 511 | 13 |
| Homo sapiens | 610 | 708 | 4837 | 7084 | |
| Mus musculus | 562 | 659 | 4508 | 6530 | |
| Danio rerio | 407 | 467 | 3242 | 4788 | |
| Drosophila melanogaster | 299 | 319 | 2193 | 3159 | |
| Caenorhabditis elegans | 142 | 157 | 1076 | 1562 | |
| Arabidopsis thaliana | 358 | 384 | 2635 | 3866 | |
| Oryza sativa | 16 | 18 | 123 | 178 | |
| Saccharomyces cerevisiae | 83 | 92 | 635 | 969 | |
| Schizosaccharomyces pombe | 104 | 115 | 787 | 1,147 |
| Category . | WD40 proteins . | WD40 domains . | WD40 repeats . | Potential hotspots . | Species . |
|---|---|---|---|---|---|
| Total | 63 211 | 71 480 | 489 411 | 726 685 | 3383 |
| Eukaryota | 58 284 | 65 311 | 447 323 | 662 833 | 860 |
| Bacteria | 4832 | 6065 | 41 358 | 62 704 | 2476 |
| Archaea | 50 | 59 | 419 | 637 | 34 |
| Virus | 45 | 45 | 311 | 511 | 13 |
| Homo sapiens | 610 | 708 | 4837 | 7084 | |
| Mus musculus | 562 | 659 | 4508 | 6530 | |
| Danio rerio | 407 | 467 | 3242 | 4788 | |
| Drosophila melanogaster | 299 | 319 | 2193 | 3159 | |
| Caenorhabditis elegans | 142 | 157 | 1076 | 1562 | |
| Arabidopsis thaliana | 358 | 384 | 2635 | 3866 | |
| Oryza sativa | 16 | 18 | 123 | 178 | |
| Saccharomyces cerevisiae | 83 | 92 | 635 | 969 | |
| Schizosaccharomyces pombe | 104 | 115 | 787 | 1,147 |
| Category . | WD40 proteins . | WD40 domains . | WD40 repeats . | Potential hotspots . | Species . |
|---|---|---|---|---|---|
| Total | 63 211 | 71 480 | 489 411 | 726 685 | 3383 |
| Eukaryota | 58 284 | 65 311 | 447 323 | 662 833 | 860 |
| Bacteria | 4832 | 6065 | 41 358 | 62 704 | 2476 |
| Archaea | 50 | 59 | 419 | 637 | 34 |
| Virus | 45 | 45 | 311 | 511 | 13 |
| Homo sapiens | 610 | 708 | 4837 | 7084 | |
| Mus musculus | 562 | 659 | 4508 | 6530 | |
| Danio rerio | 407 | 467 | 3242 | 4788 | |
| Drosophila melanogaster | 299 | 319 | 2193 | 3159 | |
| Caenorhabditis elegans | 142 | 157 | 1076 | 1562 | |
| Arabidopsis thaliana | 358 | 384 | 2635 | 3866 | |
| Oryza sativa | 16 | 18 | 123 | 178 | |
| Saccharomyces cerevisiae | 83 | 92 | 635 | 969 | |
| Schizosaccharomyces pombe | 104 | 115 | 787 | 1,147 |
The framework of WDSPdb
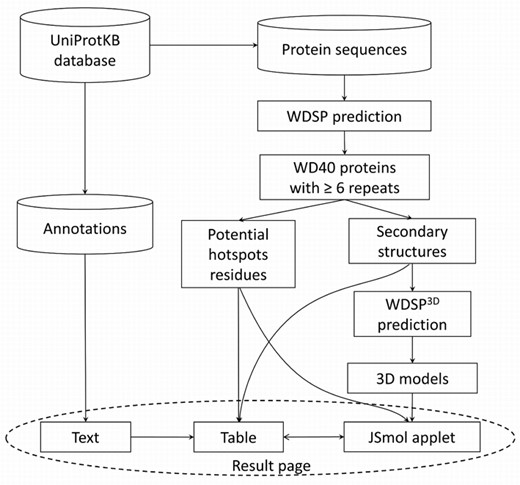
Figure 1 shows the details of identifying and classifying the WD40-repeat proteins in WDSPdb. First, for each protein sequence in the UniProt database, the WDSP program was used to identify WD40 repeats. If no less than six WD40 repeats were identified, the protein was classified as a WD40 protein that contains one or more WD40 domains (if more than eight repeats). Second, for each WD40 domain, the secondary structure of the domain and potential hotspot residues for PPIs were predicted and displayed in a table of the result page (Figure 2A). Third, the predicted 3D structure models were presented in the interactive JSmol applet (http://jsmol.sourceforge.net/). Finally, general annotations extracted from the UniProt database were also shown in the result page.
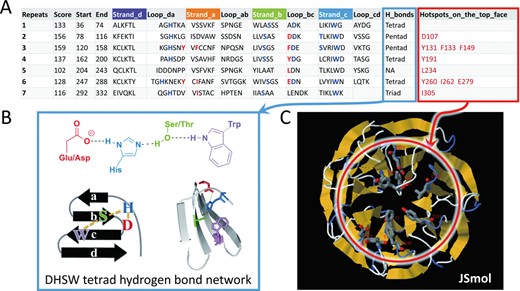
(A) The secondary structure table provided by the output of the WDSP. Each row represents a WD40 repeat sequence. Secondary structure markers are colored in the table heading. Residues shown in blue in each repeat form family-conserved DHSW tetrad hydrogen bond networks for structure stabilization. Residues shown in red are hotspot residues predicted to be responsible for PPI. (B) The structure of DHSW tetrad hydrogen bonds network. (C) The interactive interface implemented by Jsmol applet for viewing and manipulating the 3D structure. When clicking on the potential hotspot residues listed in the table, they will display as sticks with red labels.
DHSW tetrad hydrogen bond networks and 3D structure models generation
In most WD40 proteins, one or more DHSW tetrad (four residues consisting of Asp, His, Ser and Trp) hydrogen bond networks (blue residues in Figure 2A and B) can always be identified. These tetrads are specifically conserved in WD40 protein family and were proved to contribute much to structural stabilization (15,16). In fact, besides tetrads, pentad and triad hydrogen bond networks also exist in WD40 proteins widely. They are all important WD40 structural features, and highlighting them in the result page will help researchers understand the structural stability intuitively. Moreover, identification of these structural features substantially benefits the 3D structure prediction procedure that follows.
The 3D structure models of WD40 domain were generated by an in-house program WDSP3D. It combines Modeller v9.12 homology modeling package (18) and secondary structure-based sequence alignment to obtain more accurate 3D structure predictions. In the Modeller input file, DHSW tetrads identified by WDSP were treated with distance restraints. The PDB structure with the closest sequence identity was used as the template for each annotated WD40 domain, and simulated annealing Molecular Dynamics (MD) refinement process was used for each model. In our test, the backbone structures of predicted models (using different PDB structures as templates) are quite consistent with the original PDB structure, while the long loop structure is more arbitrary. The loops and side-chains structures can further be refined using longer-time MD simulations. The refined structures will be updated to the database in the future.
Potential hotspot residues responsible for PPIs
Hotspot residues are the major contributors for a certain PPI. In the initial version of this database, we provided the potential hotspot residues prediction on the top surface. Gaudet et al. and Wu et al. first reported that WD40 protein binds other proteins on the top surface by the 16th, 18th and 34th residues of each WD40 blade (19,20). We found it is actually a common phenomenon in the WD40 protein family (17). These three residues are at the R1, R1–2 and D-1 positions, where the R1 is one of the three residues (R1, R2, X) in a WD40-protein-family-conserved β-bulge, the R1–2 is the position two residues ahead of the R1 and the D-1 refers to the position just ahead of the Asp residue forming the DHSW tetrad. If binding-type residues (Arg, His, Lys, Asp, Glu, Trp, Tyr, Phe, Leu, Ile, Met, Asn, Gln) occur at these R1, R1–2 and D-1 positions, we assign them as potential hotspot residues. Thus, up to 18–24 residues (each WD40 domain has 6–8 WD40 repeats) are possibly assigned as potential hotspot residues (red residues in Figure 2A and C) in a WD40 domain. We believe this information can help not only experimentalists to select mutagenesis residues, but also computational biologists to build protein complex models. We also provided a convenient way to accentuate these residues on 3D structure models, i.e. when clicking on a potential hotspot residue listed in the secondary structure table, this residue will display as sticks in the 3D model panel (Figure 2C).
WEB INTERFACE
Database organization
MySQL was used as the database management system. Two tables were created to store the data. One table stored the general information of proteins, and the other stored the detailed structural information of WD40 domains. UniProt ID of each protein is the main key to organize and link the two tables. We adopted Tomcat as the web server utility, and JSP technology was utilized to display the results from browsing and searching.
Data browse and search
To present the data clearly and nicely, WDSPdb provided two different ways to view the data: (i) Users can browse by species. In the ‘DataBase’ drop-down menu, several well-known species names can be directly selected to display all identified WD40 proteins within this organism. (ii) Users can view the data by searching UniProt ID, gene name, Genbank ID or description. Users can also restrict their search within a specified taxon or organism by a drop-down menu.
The result page
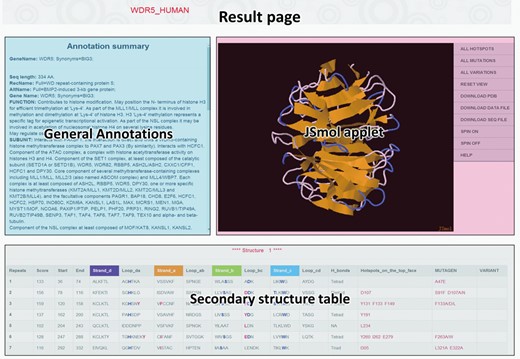
The result page (Figure 3) from the database browsing or searching is composed of three parts, that is, the general annotations extracted from UniProt database, the interactive JSmol applet to present the 3D structure model and a table to show the detailed secondary structure, specific hydrogen bond network for structure stabilization and hotspot residues for PPIs. Within the structure panel of the JSmol applet, users are not only able to rotate, zoom and move the structure to get better visual angles, but also can do more sophisticated operations with the imbedded JSmol console. All of these data, including 3D structures, can be downloaded for further investigation.
The result page for each identified WD40-repeat protein, which comprises general annotation from UniProt database, JSmol applet presenting the predicted 3D structure and the detailed secondary structure table.
The WDSP predictor page
We reserved the WDSP program page to allow the users to input their own protein sequences and to judge whether the query sequence contains WD40 domains. If a WD40 domain exists, the secondary structure and hotspot residues table will be shown in the result page.
DISCUSSION
Comparison with other databases
We compared WD40 proteins in our database with those in other widely used protein family databases: UniProt, SMART, Pfam and Prosite (Table 2). By WDSP, we have identified 99 262 proteins with at least one WD40 repeat and 63 211 proteins with at least six WD40 repeats. Compared with other four databases, WDSPdb contains many more predicted WD40 proteins especially for those with at least six WD40 repeats. These WD40 repeats could form single or multiple complete WD40 domains potentially. Using human protein WDR46 as an example, which is a well-known WD40 protein, only WDSPdb identified seven WD40 repeats and a regular WD40 domain can thus be inferred. However, none of the other databases annotated them completely. Moreover, WDSPdb is superior to other databases in the record number of multiple-WD40-domain proteins (with more than eight WD40 repeats). Taken together, WDSPdb stored 7444 proteins with multiple WD40 domains, and we found that they are more likely to appear in bacteria than in eukaryota.
Comparison of WD40 proteins among WDSPdb, SMART, Pfam, Prosite, UniProt databases and the union set of SMART+Pfam+Prosite+UniProt database
| . | Protein (repeat≥1) . | 1Protein (repeat≥6) . | 2Mutil-WD40-domain proteins . |
|---|---|---|---|
| WDSPdb | 99 262 | 63 211 | 7444 |
| SMART | 83 877 | 39 378 | 6511 |
| Pfam | 73 298 | 15 018 | 2256 |
| Prosite | 68 376 | 9750 | 1749 |
| UniProt | 3196 | 2033 | 198 |
| SMART+Pfam+Prosite+UniProt | 84 912 | 39 883 | 6610 |
| . | Protein (repeat≥1) . | 1Protein (repeat≥6) . | 2Mutil-WD40-domain proteins . |
|---|---|---|---|
| WDSPdb | 99 262 | 63 211 | 7444 |
| SMART | 83 877 | 39 378 | 6511 |
| Pfam | 73 298 | 15 018 | 2256 |
| Prosite | 68 376 | 9750 | 1749 |
| UniProt | 3196 | 2033 | 198 |
| SMART+Pfam+Prosite+UniProt | 84 912 | 39 883 | 6610 |
1Only proteins with at least six WD40 repeats are stored in WDSPdb, since a WD40 domain requires at least six WD40 repeats to form a complete structure.
2Proteins with more than eight WD40 repeats.
| . | Protein (repeat≥1) . | 1Protein (repeat≥6) . | 2Mutil-WD40-domain proteins . |
|---|---|---|---|
| WDSPdb | 99 262 | 63 211 | 7444 |
| SMART | 83 877 | 39 378 | 6511 |
| Pfam | 73 298 | 15 018 | 2256 |
| Prosite | 68 376 | 9750 | 1749 |
| UniProt | 3196 | 2033 | 198 |
| SMART+Pfam+Prosite+UniProt | 84 912 | 39 883 | 6610 |
| . | Protein (repeat≥1) . | 1Protein (repeat≥6) . | 2Mutil-WD40-domain proteins . |
|---|---|---|---|
| WDSPdb | 99 262 | 63 211 | 7444 |
| SMART | 83 877 | 39 378 | 6511 |
| Pfam | 73 298 | 15 018 | 2256 |
| Prosite | 68 376 | 9750 | 1749 |
| UniProt | 3196 | 2033 | 198 |
| SMART+Pfam+Prosite+UniProt | 84 912 | 39 883 | 6610 |
1Only proteins with at least six WD40 repeats are stored in WDSPdb, since a WD40 domain requires at least six WD40 repeats to form a complete structure.
2Proteins with more than eight WD40 repeats.
Conclusion and future perspectives
WDSPdb is a specialized WD40-repeat protein structures and potential PPI hotspot residues database. It contains the most comprehensive list of WD40 proteins while keeping low false positive rate. WDSPdb will be a powerful tool for scientists who are studying WD40 proteins or WD40 interacting proteins. From the structural point of view, to visualize potential hotspot residues and variants in the 3D structures is very helpful for understanding why some variants are disease-causing but others are not. Actually, our result from WDSPdb was successfully applied to interpret several recently discovered disease-causing mutations (21). We believe that WDSPdb will be utilized in a broader spectrum of circumstances for comprehending the structural basis of many biological processes.
We will continue to add comprehensive annotations and links from the literature and other databases to WDSPdb. We will also improve the underlying algorithm of WDSP to get more accurate results for WDSPdb. It is worth pointing out that the top surface of WD40 domain is the most active surface for PPIs, while the side surface and bottom surface could also participate in binding. We will gradually include potential PPI or protein–DNA interaction hotspot residues on these surfaces into our database. Moreover, our recently developed RSFF1 force field (22) will be used to refine our 3D structure models, especially for those with long loops.
AVAILABILITY
WDSPdb is freely available at http://wu.scbb.pkusz.edu.cn/wdsp/.
The authors would like to thank Dr Hui Zhang, Dr Xin-Hao Zhang, Dr Fan Jiang and Dr Fei Lu for valuable suggestions and discussions. They acknowledge Mr Jun Lu for assistance in the website development.
FUNDING
National Natural Science Foundation of China [21133002, 31471243]; National Basic Research Program of China [2013CB911501]; Shenzhen Peacock Program [KQTD201103]. Funding for open access charge: Shenzhen Peacock Program [KQTD201103].
Conflict of interest statement. None declared.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.




Comments