-
PDF
- Split View
-
Views
-
Cite
Cite
U. C. Lavania, S. Basu, S. Srivastava, Y. Mukai, S. Lavania, In Situ Chromosomal Localization of rDNA Sites in “Safed Musli” Chlorophytum Ker-Gawl and Their Physical Measurement by Fiber FISH, Journal of Heredity, Volume 96, Issue 2, March/April 2005, Pages 155–160, https://doi.org/10.1093/jhered/esi018
Close - Share Icon Share
Abstract
Fluorescence In Situ Hybridization (FISH) technique has been applied on somatic chromosomes and extended DNA fibers in the medicinally important species of Chlorophytum to elucidate physical localization and measurement of the rDNA sites using two rRNA multigene families homologous to 45S and 5S rDNA. The two species of Chlorophytum, namely C. borivillianum and C. comosum, both with 2n = 28, reveal diversity for copy number and localization of rDNA sites. C. borivillianum is comprised of five 45S-rDNA sites:one each in the secondary constriction region of chromosomes 7, 8, 9; one in the subtelomeric region of the short arm of chromosome 2 and the telomeric region of the short arm of chromosome 12; and one 5S-rDNA site in the subtelomeric region of the long arm of chromosome 1. In C. comosum, there are three 45S-rDNA sites (one each in the short arm of chromosomes 12, 13, and 14) and two 5S-rDNA sites (in the secondary constriction regions of chromosomes 2 and 13). Fiber FISH analysis conducted on extended DNA fibers revealed variation in the size of continuous tandem strings for the two r-DNA families. Taking the standard value of native B DNA equivalent to 3.27 kb for 1 μm, it was estimated that the physical size of continuous DNA strings is of the order of ∼90 kb, 180 kb, and 300 kb for 45S-rDNA and of the order of 60 kb, 150 kb for 5S-rDNA in C. comosum, grossly in correspondence to their respective physical sizes at metaphase.
The genus Chlorophytum (family: Liliaceae) is a fairly large genus comprising over 200 species distributed in the tropical and subtropical regions of the world, mostly in Africa and Australia. Seven species commonly occurring in India exhibit tremendous morphological, chromosomal, and cytotypic diversity (Sheriff and Chennaveeraiah 1972; Naik 1976; Patil et al. 1987; Bordia et al. 1995; Geetha and Maiti 2002). Two species, namely C. borivillianum and C. tuberosum, hold an important position in the Indian materia medica as integral components in herbal-drug prescriptions against general debility. However, the horticultural species C. comosum, which produces profuse tubers, may remain an important source of adulteration in the roots as well as a valuable genomic resource for genetic up-gradation of C. borivillianum. Therefore, a need is felt to undertake basic studies on chromosome identification to elucidate chromosome and cytotypic diversity and to complement the genetic improvement program.
Fluorescence in situ hybridization (FISH) is an effective tool for the physical mapping of DNA sequences on chromosomes. Whereas DNA: DNA dot blot hybridization offers opportunities to elucidate sequence homology and affinities, the in situ hybridization techniques hold the potential to delineate their direct physical visualization under the microscope. With the aid of such molecular cytogenetic techniques, it is now possible to accurately define genomes, chromosomes, and parts thereof on the basis of specific molecular reaction and not just subjective identification of chromosome bands (Lavania 1998a; de Jong et al. 1999; Lavania 2001; Lysak et al. 2003; Mukai 2004). A much higher resolution of target sequences by FISH could be achieved with stretched DNA fibers from interphase nuclei (Fransz et al. 1996), active nuclei/mitotic chromosomes (Lavania et al. 2003), and flow-sorted specific chromosomes (Valarik et al. 2004). Such DNA fiber targets have stretching values of 2.5–3.5 kbp μm−1, close to that of Watson-Crick double-strand DNA molecules (Valarik et al. 2004). Realizing the significance of chromosome diversity and chromosome manipulation in genetic improvement of Chlorophytum, the present study was undertaken to elucidate physical localization of repetitive DNA sequences on metaphase chromosomes as well as their microscopic measurement on extended DNA fibers.
Materials and Methods
Plant Materials and Preparation of Cells for Karyotype and FISH Analysis
Primary roots from fast-growing plants of C. borivillianum Santapau and Fernandes (2n=28) and C. comosum Jacques (2n=28) grown in the experimental field at CIMAP, Lucknow, were excised and pretreated with a saturated aqueous solution of paradichlorobenzene for three hours at 14°C, followed by fixation in ethanol : acetic acid (3:1) overnight. For metaphase chromosome analysis, the fixed roots were softened by hydrolysis in 1N.HCl for two minutes at 60°C, followed by staining in 2% aceto-carmine and squashing to obtain good metaphase spreads. At least five well-spread metaphase plates with proper chromosome contraction were analyzed for karyotype study to prepare the standard karyo-idiogram for the two species.
For preparation of metaphase plates for in situ hybridization, the fixed root tips were washed in 1 × enzyme buffer at pH 4.6 (prepared by mixing 40 ml of 10 mM citric acid + 60 ml of 10 mM trisodium citrate) to remove the fixative and then transferred to an enzyme solution (mixture of 3% [v/v] pectinase from Aspergillus niger + 2% [w/v] cellulase [1.8% cellulase from Aspergillus niger and 0.2% “Onozuka”RS cellulase]) for 30 min at 37°C to soften the cell wall in order to facilitate squashing. The softened material was again washed in 1 × enzyme buffer, and meristematic cells from the root-tip zone were squeezed out in 45% acetic acid on a clean slide and squashed under a cover glass. The cover glass was removed after freezing the slide on dry ice, and the preparations were allowed to air dry. The air-dried preparations were stored in a moisture-free card box at 37°C for 12 h. The slide preparations were then incubated in 100 μg/ml DNase free RNase in 2 × SSC for 1 h at 37°C, followed by washing in 2 × SSC for 10 min. The slides were then postfixed in 4% (w/v) paraformaldehyde at pH 7.5 in water for 10 min, washed in 2 × SSC for 10 min, dehydrated in a graded ethanol series, and air dried.
Preparation of Extended DNA Fibers for Fiber FISH
A suspension of cell wall free nuclei in 1 × PBS buffer was obtained from the meristematic zone of excised root tips of the target species according to the procedure detailed in Lavania et al. (2003). In brief, the root tips were softened by incubating in enzyme solution (mentioned above) for 30 min at 37°C, followed by squeezing loose cells of the meristematic zone onto a slide in a drop of the enzyme solution. The cell suspension was collected in an Eppendorf tube and sedimented by gentle centrifugation. The cell pellet in the Eppendorf tube was resuspended in about 50 μl of enzyme solution, pipette mixed and incubated for 4–6 h at 37°C to further dissolve the cell wall. The cell suspension was checked intermittently under microscope by staining in a drop of aceto-carmine to ascertain that the cell wall was almost dissolved. Such nuclear suspension was sedimented by gentle centrifugation, the enzyme solution was decanted, and the nuclear pellet was resuspended in 1 × PBS buffer.
Two consecutive drops of 1 μl each of nuclear suspension were applied onto a glass slide toward one end and allowed to air dry for about 10 min. Then 10 μl of STE buffer were applied onto the dried-up nuclear material on the slide and left for ∼5 min. Subsequently, the slide was gently slanted from the farther end to let the buffer solution flow downward. The slides were air dried for 15 min, and the chromatin/DNA fibers on the slides were fixed in freshly prepared acetic-ethanol (1:3) for 3 min., then air dried and stored in a moisture-free slide box for Fiber FISH preparations. Both STE-1 and STE-2 buffers are equally effective to realize EDF in the examined species (STE-1: 0.5% (w/v) SDS, 50 mM EDTA, and 100mM Tris, pH 7.0; STE-2: STE-1 buffer containing 5 mM EDTA; see Fransz et al. 1996).
Probe Labeling, FISH, and EDF-FISH
The two rDNA probes (i.e., pTa71 from wheat for 45S rDNA and 5S rDNA probe obtained from onion genomic DNA by PCR) were used. The 45S rDNA was labeled with biotin-16-dUTP (Roche Diagnostics) by nick translation, and 5S rDNA probe was labeled with digoxigenin-11-dUTP (Roche Diagnostics) directly during PCR amplification, according to the manufacturer's instructions. Multicolor FISH was performed essentially in accordance to Mukai et al. (1993), Lavania (2001), and EDF-FISH according to Fransz et al. (1996), with some modifications for signal amplification (Fukui et al. 2001; Yamamoto and Mukai 2005). The DNA probe mixture [0.1 μg of probe DNA, 50% (v/v) deionized formamide, 2 × SSC, 10% (w/v) sodium dextran sulfate, 5 μg of salmon sperm DNA in a final volume of 10 μl] was added to each preparation. Each slide with cover slip was incubated at 80°C for 2 min to denature both the probe DNA and the extended fiber DNA. After incubation at 37°C for 12–16 hours, the slides were rinsed by washing solutions (Fransz et al. 1996; Schwarzacher and Heslop-Harrison 2000; Lavania 2001). All slides for EDF FISH were pretreated by 5% (w/v) nonfat dry milk in 4 × SSC at 37°C and rinsed with 0.05% (v/v) tween-20 in 4 × SSC at room temperature. For conventional FISH, biotin-labeled DNA was detected with 2% fluorescein-conjugated avidin DN (1 mg/ml, Vector Laboratories), and digoxigenin-labeled DNA was detected with 1% rhodamine-conjugated anti-digoxigenin (0.2 mg/ml, Roche Diagnostics). However, for FISH on extended DNA fibers (EDF FISH), the signal detection for biotin-labeled DNA was further amplified with 0.5% biotinylated goat-anti-avidin (0.5 mg/ml, Vector Laboratories) and detected with 2% fluorescein-conjugated avidin DN (1 mg/ml, Vector Laboratories), and digoxigenin-labeled DNA was detected with 1% anti-digoxigenin mouse antibody (0.1 mg/ml, Roche Diagnostics) and 1% Cy3-conjugated anti-mouse IgG (1 mg/ml, Sigma). Each antibody solution was mixed in 4 × SSC containing 1% (w/v) BSA. All further incubation steps were carried out at 37°C for 30 min, each followed by washing steps (4 × SSC, 4 × SSC with 0.1% Triton X-100, 4 × SSC) of 10 min each at room temperature in the dark. Subsequently, the slides were rinsed with 2 × SSC and mounted in DABCO solution (1.25% DABCO in 90% glycerol) with 2.0 ng/μl DAPI as counter-stain. Hybridization signals were observed under the fluorescence microscope. The observations on localization of rDNA sites both at metaphase chromosomes and extended DNA fibers were recorded from at least three preparations each, and the maximum occurrence of in situ hybridization sites was taken into account for final computation.
Results
Karyotypic Distribution of rDNA
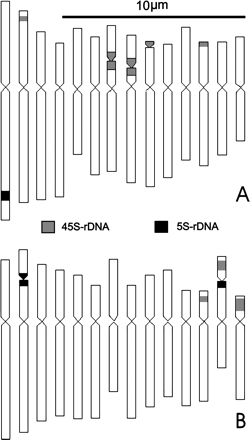
In order to elucidate precisely the chromosomal location of rDNA sites, the standard karyo-idiograms for the two species prepared from aceto-carmine stained metaphase spreads, vis-à-vis their relative comparison drawn from DAPI stained chromosomes, were taken into account. Simultaneous in situ hybridization with biotin-labeled probe pTa71 homologous to 45S rDNA detected as green fluorescing signals with fluorescein-conjugated avidin DN, and digoxigenin-labeled 5S rDNA probe of onion detected as red fluorescing signals with rhodamine-conjugated anti-digoxigenin, facilitated in situ chromosomal localization of the respective DNA sites. It is revealed that there are five pairs of 45S rDNA sites in C. borivillianum and three pairs in C. comosum; there is one pair of 5S rDNA sites in C. borivillianum, and there are two pairs in C. comosum. Taken in conjunction with the karyomorphology, the five 45S rDNA sites in C. borivillianum could be correlated to one site each in the secondary constriction region of chromosomes 7, 8, 9 and in the subtelomeric region of the short arm of chromosome 2 and in the telomeric region of the short arm of chromosome 12. The one 5S rDNA site is localized in the subtelomeric region of the long arm of chromosome 1. The three 45S rDNA sites in C. comosum could be correlated to one site each in the short arm of chromosomes 12, 13 and 14; and the two 5S rDNA sites could be correlated to the secondary constriction of chromosomes 2 and 13. This is idiographically shown in Figure 1a,b, depicting karyomorphological details along with the location of rDNA sites.
Karyo-idiograms of Chlorophytum showing FISH based localization of the two r-DNA sites (45S and 5S rDNA) on somatic chromosomes of: A. C. borivillianum, B. C. comosum.
Fiber FISH and Measurement of rDNA Sites as Extended DNA Fibers
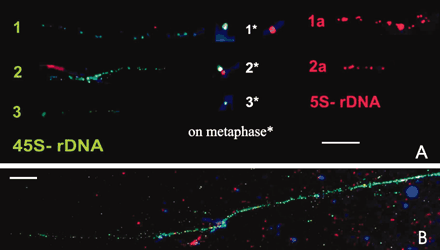
Fiber FISH with the two rDNA probes conducted on extended DNA fibers from C. comosum revealed differences in fiber tandem size hybridizing with the two rDNA families. Representative fiber FISH on rDNA fiber strings, vis-à-vis excised rDNA loci as detected by conventional FISH on metaphase chromosomes, are shown in Figure 2a. Taking the standard value of native B DNA equivalent to 3.27 kb for 1 μm (Fransz et al. 1996), it was estimated that the physical size of continuous DNA strings is of the order of ∼90 kb, 180 kb, and 300 kb for 45S rDNA and of 60 kb and 150 kb for 5S rDNA in C. comosum.
A. FISH based localization of the two rDNA sites on extended DNA fibers obtained from somatic nuclei of C. comosum vis-à-vis their signal intensity as seen on metaphase chromosomes*. Note the variable size of extended DNA fibers/metaphase signals denoted by artificial numbers as 1, 2, 3 for the two rDNA families: 45S rDNA is depicted in green and 5S rDNA is shown in red fluorescent signals. B. 45S rDNA fiber of Allium cepa to depict relatively much longer continuous fiber string. Bar = 10 μm.
Discussion
Ribosomal RNA (rRNA) multigene families consist of the 18S-5.8S-26S (45S) and 5S rRNA genes. In yeast, the 5S and 45S genes are juxtaposed in the same locus; whereas in higher eukaryotes, they are organized as families of tandemly repeated units located at one or a few chromosomal sites (Flavell 1986) and may be unlinked on the same chromosome arm or located on different chromosomes (Mukai et al. 1990). In the last decade, FISH studies have been conducted in numerous plant species to elucidate the number and localization of rDNA sites. It is observed that most of diploid plants have two sites (i.e., a single locus) of both 5S and 45S rDNA (Mishima et al. 2002), although some diploids may have multiple sites (Fukui et al. 1994; Badaeva et al. 1996; de Bustos et al. 1996; Lavania 1998b; Ansari et al. 1999; Raina and Mukai 1999; Zhang and Sang 1999). The derived polyploids expectedly would have the sum of the rDNA sites present in their diploid ancestors, as demonstrated in artificial polyploids of Rubus (Lim et al. 1998). Prokopowich et al. (2003), based on exhaustive analysis on the rDNA copy number and genome size in large number of animal and plant taxa extending across the taxonomic boundaries, have suggested a strong positive correlation between genome size and rDNA copy number, obviating that need for ribosomes would increase as genome size increases if the relative proportion of protein-coding genes remain constant.
The present study revealed several rDNA sites in the two examined species: five pairs of 45S rDNA + one pair of 5S rDNA sites in C. borivillianum; and three pairs of 45S rDNA + two pairs of 5S rDNA sites in C. comosum, both with 2n=28. Whereas most of the 45S rDNA sites are localized in NOR regions in C. borivillianum, in C. comosum these are the 5S rDNA sites that are located in NOR regions. Such an occurrence of multiple rDNA sites localized to specific chromosomes may have value in chromosome identification and elucidation of evolutionary relationships and delineation of possible break point sites.
These two species are supposedly tetraploids from 2n=14 ancestors, with base number x=7 (Sheriff and Chennaveeraiah 1972; Naik 1976). The observations recorded for the number of rDNA sites in these species does not support the contention that there has been a simple duplication of rDNA sites during evolutionary polyploidization. Occurrence of rDNA sites in odd numbers points to the possibility that the two species may have either originated through allopolypoidization combining two different genomes, or there has been elimination of some rDNA sites during speciation, both for 45S rDNA and 5S rDNA sites. Based on exhaustive analysis of site-number change for rDNA loci during polyploid evolution in Sanguisorba (Rosaceae), Mishima et al. (2002) have suggested that the duplicated 45S rDNA sites may remain conserved, but 5S rDNA sites have a tendency toward elimination after polyploidization. As such, the evolutionary dynamics of the rDNA site number still remain unclear, as both rDNA sites are implicated in evolutionary elimination in Chlorophytum. While most natural polyploid plants have multiple (duplicated) sites, sometimes the number of sites detected is different from the number expected based on ploidy level (Adachi et al. 1997; Ansari et al. 1999; Raina and Mukai 1999). These differences indicate that the evolutionary dynamics of rDNA sites in polyploid plants are complex (Shi et al. 1996), although Dvorak (1990) has suggested that rDNA loci show a general tendency to be eliminated if rendered dispensable by polyploid evolution.
FISH on extended DNA fibers is a valuable tool in physical mapping studies that can facilitate the determination of the physical size of target sequences by direct measurement under the microscope (Mukai 2004). The Chlorophytum species examined here reveal the occurrence of multiple rDNA sites located on several chromosomes. In C. comosum in particular, the two families occupy more than one location with clear-cut physical differences in locus size. Therefore, this species offers a possibility to unravel the possible tandem size differences in the different localization sites. The extended rDNA fibers obtained in this species when localized by in situ hybridization reveal differences in the fiber string size of rDNA repeat units. Based on fiber FISH analysis for known target sequences in Arabidopsis, Fransz et al. (1996) and de Jong et al. (1999) have deduced a correlation between the molecular size of the target DNA sequences and its physical size as extended DNA fiber, suggesting that the 1 μm length of extended DNA fiber is equivalent to ∼3.27 kb, which is in close agreement to the Watson-Crick DNA standard length estimate of 2.97 kb for 1 μm B DNA.
Considering this as a reference for measuring the physical size of DNA as extended DNA fibers, and taking the standard value of native B DNA equivalent to 3.27 kb for 1 μm (Fransz et al. 1996), it was estimated that the physical size of continuous DNA strings in C. comosum is of the order of ∼90 kb, 180 kb, and 300 kb for 45S rDNA and 60 kb and 150 kb for 5S rDNA, respectively. These values are apparently quite low compared to those reported in other plants—for example, 5S rDNA continuous string size reported for tomato is 660 kb (Fransz et al. 1996). Therefore, to ascertain the validity of present observations, a simultaneous test run was conducted on rDNA fibers of Allium cepa (Figure 2b), suggesting that extended DNA fibers are in the expected order. However, there could be some variation to these measurements, depending upon the stretching of chromatin/DNA fibers. Nevertheless, observation on fiber strings of such discrete sizes points to the possibility that such variation in string size is probably locus-specific, corresponding to the given rDNA locus. However, directed efforts on the measurement of DNA fibers from the specific locus of super-stretched flow sorted chromosomes (Valarik et al. 2004) could facilitate to substantiate such an assumption.
The information about discrete variation in rDNA continuous tandem strings is the first report of its kind. Of course, additional data covering a range of species may be required to substantiate the incidence of discrete variation in DNA fiber strings commensurate with locus specific structural organization of DNA repeat families. Possibly discrete variation in rDNA continuous string size is correlated with signal intensity at metaphase (Figure 2b), but this remains to be proven on other species.
Corresponding Editor: Reid G. Palmer
Financial support from Department of Biotechnology, India, and Japan Society for the Promotion of Science to U.C.L., and JRF-NET (Council of Scientific and Industrial Research), New Delhi, to S.B. is gratefully acknowledged. This paper is dedicated to Prof. A.K. Sharma on his 80th birthday.
References
Adachi J, Watnabe K, Fukui K, Ohmido N, and Kosuge K,
Ansari HA, Ellison NW, Reader SM, Badaeva ED, Friebe B, Miller TE, and Williams WM,
Badaeva ED, Friebe B, and Gill BS,
Bordia PC, Joshi A, and Simlot MM,
de Bustos A, Cuadrado A, Soler C, and Jove N,
de Jong JH, Fransz PF, and Zabel P,
Dvorak J,
Flavell RB,
Fransz PF, Alonso-Blanco C, Liharska TB, Peeters AJM, Zabel P, and de Jong JH,
Fukui K, Ohimido N, and Khush GS,
Fukui K-N, Suzuki G, Lagudah ES, Rahman R, Appels R, Yamamoto M, and Mukai Y,
Geetha KA and Maiti S,
Lavania UC,
Lavania UC,
Lavania UC,
Lavania UC, Yamamoto M, and Mukai Y,
Lim KY, Leitch IJ, and Leitch AR,
Lysak MA, Pecinka A, and Schubert I,
Mishima M, Ohmido N, Fukai K, and Yahara T,
Mukai Y,
Mukai Y, Endo TR, and Gill BS,
Mukai Y, Nakahara Y, and Yamamoto M,
Naik VN,
Patil VP, Kumbhojkar MS, and Gandhi SS,
Prokopowich CD, Gregory TR, and Crease TJ,
Raina SN and Mukai Y,
Schwarzacher T and Heslop-Harrison P,
Sheriff A and Chennaveeraiah MS,
Shi L, Zhu T, and Keim P,
Valarik M, Bartos J, Kovarova P, Kubalakova M, de Jong JH, and Dolezel J,
Yamamoto M and Mukai Y,