-
PDF
- Split View
-
Views
-
Cite
Cite
M. J. López-Piñón, A. Insua, J. Méndez, Chromosome Analysis and Mapping of Ribosomal Genes by One- and Two-Color Fluorescent in situ Hybridization in Hinnites distortus (Bivalvia: Pectinidae), Journal of Heredity, Volume 96, Issue 1, January/February 2005, Pages 52–58, https://doi.org/10.1093/jhered/esi001
Close - Share Icon Share
Abstract
Metaphase chromosomes of the scallop Hinnites distortus were analyzed using Giemsa staining, chromosome measurements, silver staining, one- and two-color fluorescent in situ hybridization (FISH) ribosomal DNA (rDNA) probes, and 4′,6-diamidino-2-phenylindole (DAPI) banding compatible with in situ hybridization. The karyotype (2n = 38) consists of three submetacentric-metacentric, one submetacentric, one subtelocentric-submetacentric, and 14 subtelocentric pairs. The 18S-28S rDNA maps at the centromeric level of two subtelocentric pairs, but not more than two nucleolus organizer region (NOR)-bearing chromosomes were transcriptionally active. The 5S rDNA seems to show a conventional tandem arrangement with a repeat unit of about 450 bp and it maps at the pericentromeric region of the long arm of one subtelocentric pair. Two-color FISH demonstrated that 18S-28S rDNA and 5S rDNA are not syntenic. Sequential FISH/Giemsa staining and subsequent chromosome pairing allow us to propose that pairs 9 and 12 carry the 18S-28S rDNA and pair 13 carries the 5S rDNA. All chromosomes are characterized as containing constitutive heterochromatin at the centromeric region. The data provided are the first contribution toward construction of the molecular karyotype of H. distortus and will be useful in assessing evolutionary relationships within scallops.
In the bivalve family Pectinidae there are some 400 known living species, commonly called scallops, distributed in all the seas of the world (Brand 1991). By virtue of the number of species, their geographical distribution, and the range of habitats they occupy, scallops are a successful group of bivalves and important members of a number of benthic communities (Brand 1991).
Cytogenetic studies on Pectinidae are scarce and have been focused mainly on commercial or potentially commercial species, which represent only a small fraction of the total number of family species. The chromosome number (haploid and/or diploid) was reported in 16 species and the conventional karyotype was described in 10 (review by Beaumont and Zouros 1991; Insua et al. 1998; Thiriot-Quiévreux 2002; von Brand-Skopnik and Ibarra-Humphries 2002). Application of chromosome banding or molecular techniques was carried out in only three species: Pauls and Affonso (2000) report the use of silver staining to detect the nucleolus organizer regions (Ag-NORs) and C-banding in Nodipecten nodosus; Gajardo et al. (2002) applied silver staining, Hoechst 33258/actinomycin D staining, and restriction endonuclease banding in Argopecten purpuratus; and Insua et al. (1998) used fluorescent in situ hybridization (FISH) to map the genes encoding the 18S, 5.8S, and 28S ribosomal RNA (18S-28S rDNA) and genes encoding the 5S rRNA (5S rDNA) in Aequipecten opercularis in addition to silver staining and C-banding.
In this study we undertook a chromosomal analysis in Hinnites distortus (Da Costa, 1778). This species is regarded by Waller (1993) as a junior synonym of Crassodoma pusio (Linnaeus, 1758), but often it is also referred to as Chlamys distorta. It is distributed from Norway to the Azores and the Gulf of Guinea, with southern Spain as the eastern limit, and resembles Chlamys species when not yet attached (Wagner 1991). The data provided here are based on results obtained by conventional Giemsa staining, chromosome measurements, silver staining, and one-color FISH using 18S-28S rDNA and 5S rDNA probes. In addition, two-color FISH was carried out for the first time in bivalves to examine the physical relationships between 18S-28S rDNA and 5S rDNA, as well as 4′,6-diamidino-2-phenylindole (DAPI) banding compatible with in situ hybridization.
Materials and Methods
Samples of H. distortus (1–2 cm) were collected from a wild population in Ría de Arousa (northwest Spain) and fed in the laboratory with the microalgae Isochrysis galbana for 10 days before performing chromosome spreads.
Chromosome Preparation, Karyotype, and Silver Staining
To obtain metaphases, animals were maintained in 0.005% colchicine in seawater for 7–9 h after several optimization assays. The gills were excised, placed in 50% and 25% seawater solution for 30 min each, and fixed by three incubations of 20 min each in a freshly prepared mixture of absolute ethanol and acetic acid (3:1). Gill cells were dissociated in 50% acetic acid and the suspension obtained was dropped onto slides heated at 42°C (Thiriot-Quiévreux and Ayraud 1982).
Conventional karyotypes were made from slides stained with Giemsa (4%, pH 6.8) for 10 min. Metaphases were photographed with a Nikon photomicroscope using Kodalith film, the chromosomes paired according to their morphology, and then short and long arms measured using the Leica Q-Win 2.2 program (Leica Imaging Systems Ltd). For each chromosome pair, mean and standard deviations of relative length (100 × absolute length/total length of the haploid complement), and centromeric index (100 × length of short arm/total chromosome length) were calculated in a Microsoft Excel spreadsheet. Nomenclature for centromere position follows that of Levan et al. (1964), but when the 95% confidence limits of the centromeric index mean covered two chromosome categories, a binary terminology was adopted.
Silver staining was carried out according to the technique of Howell and Black (1980), modified by Gold and Ellison (1982), on Giemsa-stained slides and discolored with alcohol.
Probe Labeling, DNA Extraction, and Polymerase Chain Reaction Amplification
Recombinant plasmid containing 18S, 5.8S, 28S genes plus intergenic spacers of Drosophila melanogaster was used as a probe to locate the 18S-28S rDNA. Plasmid DNA was purified using the alkaline lysis method (Sambrook et al. 1989) and labeled with digoxigenin-11-dUTP employing a nick translation kit (Roche Diagnostics).
For the 5S rDNA, a specific probe was generated by polymerase chain reaction (PCR). Genomic DNA was extracted from a 30 mg piece of ethanol-preserved adductor muscle according to Winnepenninckx et al. (1993) after two 15 min washes in phosphate buffered saline and sterile deionized water. The 5S rDNA repeat unit was PCR amplified first using a mixture containing 10 ng template DNA/μl, 0.25 mM of each dNTP, 1 μM of each primer (5′-AGCCCGGTTAGTAGTACTTGG-3′ and 5′-CCGACGTTGCTTAACTTCG-3′), buffer (10 mM Tris-HCl, 1.5 mM MgCl2, and 50 mM KCl), and 0.025 U Taq polymerase/μl. Thirty standard PCR amplification cycles were performed (annealing temperature 60°C) and PCR products evaluated by electrophoresis in 2% agarose gels. Labeling was obtained by PCR according to the described procedure, but using 35 μM digoxigenin-11-dUTP or 35 μM tetramethyl-rhodamine-6-dUTP, 160 μM dTTP, 100 μM dATP, 100 μM dCTP, and 100 μM dGTP in the reaction mixture.
FISH
Both probes, initially labeled with digoxigenin-11-dUTP, were used on separate in situ hybridization assays and detected on propidium iodide counterstained chromosomes through fluorescein isothiocyanate (FITC) attached to specific antibodies (one-color FISH) according to Insua et al. (1998).
Two-color FISH was carried out using the 18S-28S rDNA probe labeled with digoxigenin-11-dUTP and the 5S rDNA probe labeled with tetramethyl-rhodamine-6-dUTP. The procedure corresponds to that of Insua et al. (1998), with the following modifications: (1) the hybridization solution consisted of 50% formamide, 10% dextran sulfate, 2× SSC, 250 ng/μl salmon sperm DNA, 0.125% sodium dodecyl sulfate, and 4 ng/μl of each labeled probe; (2) probes were denatured by heat at 75°C for 10 min in the hybridization solution and the chromosomal DNA was denatured at 83°C for 2.5 min in 70% formamide, 2× SSC, and 50 mM sodium phosphate solution; (3) after overnight hybridization, slides were washed twice (7 min each) in 20% formamide, 2× SSC at 42°C, three times (5 min each) in 2× SSC at 42°C, and once (5 min) in 2× SSC at room temperature; (4) for counterstaining, 20 ng/ml DAPI in antifade was used. Images were collected with a Nikon E-800 microscope equipped with a Hamamatsu CCD camera.
Results
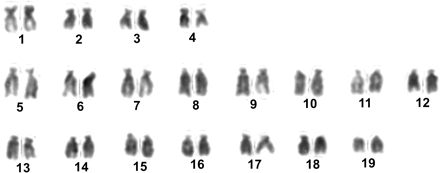
The total number of mitotic metaphases examined in this work are summarized in Table 1. The chromosome number was counted in Giemsa-stained metaphases and most (67%) showed 2n = 38. Aneuploid metaphases (33%) were recorded in most of the individuals examined (17 of 21): 2n = 36 and 2n = 37 were more frequently observed (21.6%), 2n = 39 was found once (0.6%), and 2n < 36 was found in the remaining cases (10.8%). Chromosomes from nine metaphase spreads were measured and their relative lengths and centromeric indexes were calculated (Table 2). According to the centromeric position, the chromosome pairs 1, 2, 3, and 4 were classified as submetacentric and the other pairs as subtelocentric, although some pairs, such as pairs 2, 3, and 4, were borderline to submetacentric-metacentric chromosomes and pair 7 was borderline to subtelocentric-submetacentric chromosomes when 95% confidence limits of the centromeric index means were considered. Chromosome relative length ranged between 6.34% and 3.72% and the 19 chromosome pairs showed decreasing size, maintaining small differences between successive pairs (less than 0.63%), without distinguishable size classes. A representative karyotype is shown in Figure 1.
Summary of metaphases examined
Staining/FISH . | No. of individuals . | No. of metaphases . |
|---|---|---|
| Giemsa staining | 21 | 176 |
| Silver staining | 4 | 14 |
| One-color FISH | ||
| 18S-28S rDNA probe | 8 | 74 |
| 5S rDNA probe | 9 | 97 |
| Two-color FISH | 2 | 14 |
| FISH/DAPI | 5 | 166 |
Staining/FISH . | No. of individuals . | No. of metaphases . |
|---|---|---|
| Giemsa staining | 21 | 176 |
| Silver staining | 4 | 14 |
| One-color FISH | ||
| 18S-28S rDNA probe | 8 | 74 |
| 5S rDNA probe | 9 | 97 |
| Two-color FISH | 2 | 14 |
| FISH/DAPI | 5 | 166 |
Summary of metaphases examined
Staining/FISH . | No. of individuals . | No. of metaphases . |
|---|---|---|
| Giemsa staining | 21 | 176 |
| Silver staining | 4 | 14 |
| One-color FISH | ||
| 18S-28S rDNA probe | 8 | 74 |
| 5S rDNA probe | 9 | 97 |
| Two-color FISH | 2 | 14 |
| FISH/DAPI | 5 | 166 |
Staining/FISH . | No. of individuals . | No. of metaphases . |
|---|---|---|
| Giemsa staining | 21 | 176 |
| Silver staining | 4 | 14 |
| One-color FISH | ||
| 18S-28S rDNA probe | 8 | 74 |
| 5S rDNA probe | 9 | 97 |
| Two-color FISH | 2 | 14 |
| FISH/DAPI | 5 | 166 |
Measurement and classification of H. distortus chromosomes
Chromosome pair . | Relative length . | . | Centromeric index . | . | . | ||
|---|---|---|---|---|---|---|---|
. | M . | SD . | M . | SD . | Classificationa . | ||
| 1 | 6.34 | 0.54 | 35.10 | 2.89 | sm | ||
| 2 | 5.76 | 0.44 | 35.93 | 2.85 | sm-m | ||
| 3 | 5.14 | 0.51 | 36.48 | 3.22 | sm-m | ||
| 4 | 4.77 | 0.49 | 34.89 | 4.76 | sm-m | ||
| 5 | 6.96 | 0.42 | 21.01 | 3.55 | st | ||
| 6 | 6.30 | 0.41 | 18.48 | 4.07 | st | ||
| 7 | 6.24 | 0.72 | 24.84 | 3.07 | st-sm | ||
| 8 | 5.96 | 0.47 | 19.78 | 3.18 | st | ||
| 9 | 5.58 | 0.54 | 17.25 | 3.51 | st | ||
| 10 | 5.58 | 0.25 | 20.21 | 3.71 | st | ||
| 11 | 5.27 | 0.18 | 19.29 | 3.10 | st | ||
| 12 | 5.10 | 0.41 | 17.75 | 4.15 | st | ||
| 13 | 5.05 | 0.41 | 20.73 | 4.28 | st | ||
| 14 | 4.75 | 0.29 | 19.19 | 3.67 | st | ||
| 15 | 4.58 | 0.34 | 19.03 | 3.95 | st | ||
| 16 | 4.47 | 0.41 | 15.21 | 2.69 | st | ||
| 17 | 4.36 | 0.60 | 16.73 | 2.68 | st | ||
| 18 | 4.08 | 0.44 | 20.73 | 3.74 | st | ||
| 19 | 3.72 | 0.63 | 18.33 | 2.70 | st | ||
Chromosome pair . | Relative length . | . | Centromeric index . | . | . | ||
|---|---|---|---|---|---|---|---|
. | M . | SD . | M . | SD . | Classificationa . | ||
| 1 | 6.34 | 0.54 | 35.10 | 2.89 | sm | ||
| 2 | 5.76 | 0.44 | 35.93 | 2.85 | sm-m | ||
| 3 | 5.14 | 0.51 | 36.48 | 3.22 | sm-m | ||
| 4 | 4.77 | 0.49 | 34.89 | 4.76 | sm-m | ||
| 5 | 6.96 | 0.42 | 21.01 | 3.55 | st | ||
| 6 | 6.30 | 0.41 | 18.48 | 4.07 | st | ||
| 7 | 6.24 | 0.72 | 24.84 | 3.07 | st-sm | ||
| 8 | 5.96 | 0.47 | 19.78 | 3.18 | st | ||
| 9 | 5.58 | 0.54 | 17.25 | 3.51 | st | ||
| 10 | 5.58 | 0.25 | 20.21 | 3.71 | st | ||
| 11 | 5.27 | 0.18 | 19.29 | 3.10 | st | ||
| 12 | 5.10 | 0.41 | 17.75 | 4.15 | st | ||
| 13 | 5.05 | 0.41 | 20.73 | 4.28 | st | ||
| 14 | 4.75 | 0.29 | 19.19 | 3.67 | st | ||
| 15 | 4.58 | 0.34 | 19.03 | 3.95 | st | ||
| 16 | 4.47 | 0.41 | 15.21 | 2.69 | st | ||
| 17 | 4.36 | 0.60 | 16.73 | 2.68 | st | ||
| 18 | 4.08 | 0.44 | 20.73 | 3.74 | st | ||
| 19 | 3.72 | 0.63 | 18.33 | 2.70 | st | ||
m, metacentric; sm, submetacentric; st, subtelocentric.
Measurement and classification of H. distortus chromosomes
Chromosome pair . | Relative length . | . | Centromeric index . | . | . | ||
|---|---|---|---|---|---|---|---|
. | M . | SD . | M . | SD . | Classificationa . | ||
| 1 | 6.34 | 0.54 | 35.10 | 2.89 | sm | ||
| 2 | 5.76 | 0.44 | 35.93 | 2.85 | sm-m | ||
| 3 | 5.14 | 0.51 | 36.48 | 3.22 | sm-m | ||
| 4 | 4.77 | 0.49 | 34.89 | 4.76 | sm-m | ||
| 5 | 6.96 | 0.42 | 21.01 | 3.55 | st | ||
| 6 | 6.30 | 0.41 | 18.48 | 4.07 | st | ||
| 7 | 6.24 | 0.72 | 24.84 | 3.07 | st-sm | ||
| 8 | 5.96 | 0.47 | 19.78 | 3.18 | st | ||
| 9 | 5.58 | 0.54 | 17.25 | 3.51 | st | ||
| 10 | 5.58 | 0.25 | 20.21 | 3.71 | st | ||
| 11 | 5.27 | 0.18 | 19.29 | 3.10 | st | ||
| 12 | 5.10 | 0.41 | 17.75 | 4.15 | st | ||
| 13 | 5.05 | 0.41 | 20.73 | 4.28 | st | ||
| 14 | 4.75 | 0.29 | 19.19 | 3.67 | st | ||
| 15 | 4.58 | 0.34 | 19.03 | 3.95 | st | ||
| 16 | 4.47 | 0.41 | 15.21 | 2.69 | st | ||
| 17 | 4.36 | 0.60 | 16.73 | 2.68 | st | ||
| 18 | 4.08 | 0.44 | 20.73 | 3.74 | st | ||
| 19 | 3.72 | 0.63 | 18.33 | 2.70 | st | ||
Chromosome pair . | Relative length . | . | Centromeric index . | . | . | ||
|---|---|---|---|---|---|---|---|
. | M . | SD . | M . | SD . | Classificationa . | ||
| 1 | 6.34 | 0.54 | 35.10 | 2.89 | sm | ||
| 2 | 5.76 | 0.44 | 35.93 | 2.85 | sm-m | ||
| 3 | 5.14 | 0.51 | 36.48 | 3.22 | sm-m | ||
| 4 | 4.77 | 0.49 | 34.89 | 4.76 | sm-m | ||
| 5 | 6.96 | 0.42 | 21.01 | 3.55 | st | ||
| 6 | 6.30 | 0.41 | 18.48 | 4.07 | st | ||
| 7 | 6.24 | 0.72 | 24.84 | 3.07 | st-sm | ||
| 8 | 5.96 | 0.47 | 19.78 | 3.18 | st | ||
| 9 | 5.58 | 0.54 | 17.25 | 3.51 | st | ||
| 10 | 5.58 | 0.25 | 20.21 | 3.71 | st | ||
| 11 | 5.27 | 0.18 | 19.29 | 3.10 | st | ||
| 12 | 5.10 | 0.41 | 17.75 | 4.15 | st | ||
| 13 | 5.05 | 0.41 | 20.73 | 4.28 | st | ||
| 14 | 4.75 | 0.29 | 19.19 | 3.67 | st | ||
| 15 | 4.58 | 0.34 | 19.03 | 3.95 | st | ||
| 16 | 4.47 | 0.41 | 15.21 | 2.69 | st | ||
| 17 | 4.36 | 0.60 | 16.73 | 2.68 | st | ||
| 18 | 4.08 | 0.44 | 20.73 | 3.74 | st | ||
| 19 | 3.72 | 0.63 | 18.33 | 2.70 | st | ||
m, metacentric; sm, submetacentric; st, subtelocentric.
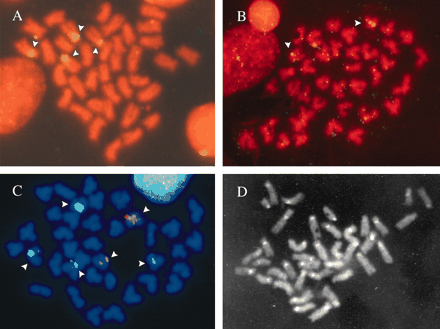
In situ hybridization assays using the 18S-28S rDNA probe alone showed four hybridization signals located at the centromeric level of two subtelocentric pairs (Figure 2a). Depending on the metaphase spreads, hybridization signals were spread along the centromeric region or limited to one of the pericentromeric regions.
Metaphase chromosomes of H. distortus. Propidium iodide-counterstained chromosomes after one-color FISH with (a) 18S-28S rDNA probe and (b) 5S rDNA probe. (c) DAPI-counterstained chromosomes after two-color FISH with 18S-28S rDNA probe (green signal) and 5S rDNA probe (red signal). (d) C-banded metaphase obtained after FISH and DAPI counterstaining. Arrows indicate the hybridization sites.
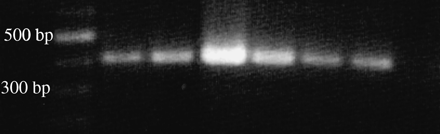
Polymerase chain reaction amplification of the 5S rDNA repeat unit yielded a single band of about 450 bp in length in several individuals (Figure 3). To ensure the specificity of the probe derived from this product, a control in situ hybridization was performed on slides of the bivalve Cerastoderma edule, where location of 5S rDNA is known (Insua et al. 1999). When the 5S rDNA probe was used alone in H. distortus slides it revealed two hybridization signals located at the pericentromeric region of the long arm of one subtelocentric pair (Figure 2b).
PCR product of the 5S rDNA repeat unit of H. distortus. First lane on the left, DNA molecular marker (100 bp ladder); consecutive lanes, PCR product of different individuals.
Because the karyotype displayed mainly subtelocentric chromosomes with small differences in size, a two-color FISH was performed to detect both gene families simultaneously. In this case the 18S-28S rDNA was labeled with digoxigenin-11-dUTP and the 5S rDNA was labeled with tetramethyl-rhodamine-6-dUTP. Figure 2c illustrates the results of this assay, 18S-28S rDNA and 5S rDNA mapped on different chromosome pairs.
After carrying out in situ hybridization assays, some slides were Giemsa stained, several metaphases photographed, and chromosomes paired. This allowed us to assign the 18S-28S rDNA-bearing chromosomes to pairs 9 and 12, and the 5S rDNA-bearing chromosomes to pair 13.
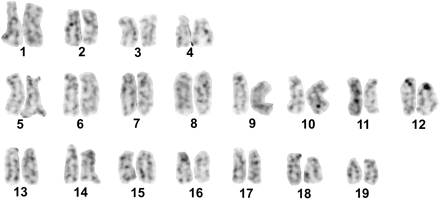
Silver staining was performed on slides stained directly with Giemsa and subsequently discolored. The metaphases revealed not more than one or two Ag-NORs (Figure 4), despite 18S-28S rDNA being found at two chromosome pairs.
Karyotype of H. distortus after silver staining. Note Ag-NORs in chromosome pair 12.
Counterstaining with DAPI after in situ hybridization revealed brightly fluorescent bands at the centromeric region of all the chromosomes (C-band-like pattern). Identical results were obtained when DAPI was added to slides previously counterstained with propidium iodide (Figure 2d).
Discussion
The diploid number of 2n = 38 reported here for H. distortus confirms the results provided by Beaumont and Gruffydd (1974). This chromosome number is the most usual in the Pectinidae family (Beaumont and Zouros 1991; Insua et al. 1998; Thiriot-Quiévreux 2002; von Brand-Skopnik and Ibarra-Humphries 2002) and also in the Bivalvia class (Insua 1993; Thiriot-Quiévreux 2002). However, it should be noted that most of the individuals examined displayed aneuploid metaphases and that these represent one-third of the total cells counted. This may be due to technical factors, but it cannot be ignored that it reveals a chromosomal abnormality. Evidence of a genetic basis for chromosome loss in somatic cells was reported in the oyster Crassostrea gigas (Leitão et al. 2001).
The karyotype of H. distortus (1 sm, 3 sm-m, 1 st-sm, 14 st) is characterized by the absence of clear metacentric and telocentric chromosomes, which is not usual among the pectinids studied so far. In general terms, karyotypes in the family Pectinidae contain from 1 to 6 metacentric pairs and from 1 to 14 telocentric pairs (Insua et al. 1998; Thiriot-Quiévreux 2002). Only the karyotype reported for A. purpuratus (Gajardo et al. 2002) and Placopecten magellanicus (Xiang et al. 1993) lacks metacentric pairs, and that described for one population of N. nodosus (Pauls et al. 1996) lacks telocentric pairs. Nevertheless, the karyotype of H. distortus follows the pectinid tendency to show a higher number of mono-armed (subtelocentric and telocentric) than of bi-armed chromosomes (metacentric and submetacentric).
Fluorescent in situ hybridization with an 18S-28S rDNA probe revealed that H. distortus contains this type of sequence on two subtelocentric pairs. The chromosomal position of 18S-28S rDNA was found at the centromeric level, but the variation detected among metaphases (centromeric or pericentromeric region) precludes giving a more precise description. Differences in the hybridization process caused by an unequal condensation state of chromosomes could explain the position variation detected. Further analysis on less contracted chromosomes, often obtained from early embryos, will provide a better resolution. FISH results contrast with those obtained by silver staining, since only one or two Ag-NORs were detected. Intra- and interindividual variation in the number of Ag-NORs is common in bivalves (e.g., Cross et al. 2003; Insua and Méndez 1998; Leitão et al. 1999; Torreiro et al. 1999), however, the maximum number of Ag-NORs usually corresponds to the 18S-28S sites revealed by FISH. Since silver staining only detects the transcriptionally active NORs at the preceding interphase (Howell 1977; Miller et al. 1976a,b), the low number of Ag-NORs seen in H. distortus indicates that the transcription process does not occur, or occurs rarely, at the same time at the four sites of 18S-28S rDNA. Similar results found in fish (Fujiwara et al. 1998) suggest that an interlocus inactivation could be operating.
The distribution of 18S-28S rDNA in H. distortus is very different from that reported for A. opercularis, where it is located at the telomere of the long arm of a telocentric pair (Insua et al. 1998). But coincidences could occur with A. purpuratus. For this species, Gajardo et al. (2002) described terminal Ag-NORs on the short arm of two pairs and pericentric Ag-NORs on another pair. Available data on bivalves indicate that the most common situation is the occurrence of one or two chromosome pairs with NORs located terminally on one arm (Insua et al. 2001a).
The 5S rDNA of H. distortus seems to exhibit, at least in part, the conventional tandem arrangement, seeing that successful amplification was obtained using contiguous primers with opposite orientation. The size of the repeat unit (∼450 pb) was close to that of A. opercularis (∼460 pb) (Insua et al. 1998), shorter than that of the cockle Ceratoderma edule (∼550 bp) (Insua et al. 1999), and intermediate between two types of units occurring in mussels (∼250 bp and ∼760 bp) (Insua et al. 2001b). The chromosomal location of the 5S rDNA at the pericentromeric region of the subtelocentric pair is similar to that of A. opercularis, Mytilus edulis, and Mytilus galloprovincialis (Insua et al. 1998, 2001b) in the sense that these also show at least a 5S rDNA site near the centromere, but in these species the number of sites (two or four per haploid complement) and bearing-chromosome morphology (metacentric) differ.
The two-color FISH used in this work for the first time on bivalve chromosomes is usually employed in vertebrates, but its use in invertebrates is still very restricted. The results provided here by this technique encourage future research into the construction of molecular karyotypes in bivalve species. Simultaneous detection of the rDNA in H. distortus demonstrated unambiguously that 18S-28S rDNA and 5S rDNA are not syntenic. In addition, sequential FISH/Giemsa staining and subsequent chromosome pairing allow us to propose that pairs 9 and 12 carry 18S-28S rDNA and pair 13 carries 5S rDNA. The location of the 18S-28S rDNA and 5S rDNA on different chromosomes was also observed in the bivalves investigated (Insua et al. 1998, 1999, 2001b), which is very common in high eukaryotes (e.g., Kost et al. 1995; Martins and Galetti 2001; Matsuda et al. 1994; Suzuki et al. 1996; Wimber and Steffensen 1970). Although in some invertebrate species the 5S rDNA is linked to the repeat units of other tandemly repeated gene families such as trans-spliced leader genes, histone genes (Drouin and Moniz de Sá 1995), and U1 small nuclear RNA genes (Pelliccia et al. 2001), it seems unlikely for this situation to occur in H. distortus. The size of the repeat unit (∼450 bp) suggests that other sequences besides the coding and spacer region of the 5S rDNA could not be included.
Despite the frequent use of the C-band protocol of Sumner (1972) to detect constitutive heterochromatin, several other methods have been developed (e.g., Arrighi and Hsu 1971; Dev et al. 1972; Fernández et al. 2002). Heng and Tsui (1993) demonstrated that adjusting the denaturation time in the presence of formamide during the in situ hybridization protocol made it possible to obtain a C-banding pattern in human chromosomes stained with DAPI. On the other hand, Fernandez et al. (2002) obtained a reliable pattern of C bands in mammalian species using a method based on two steps comprised in the in situ hybridization protocol: heat denaturation of chromosomal DNA in the presence of formamide and incubation in 2× SSC at room temperature. Thus it seems very likely that the C-band-like pattern obtained here in H. distortus reflects real constitutive heterochromatin regions. The occurrence of consistent heterochromatic blocks in the centromeric region of all chromosomes in H. distortus contrasts with the distribution in A. opercularis, where the main heterochromatin blocks are in an intercalary or subterminal position in four chromosome pairs (Insua et al. 1998), and also with that of A. purpuratus, showing centromeric heterochromatin in only 11 pairs (Gajardo et al. 2002). Considering other bivalve species, the existence of constitutive heterochromatin at the centromeres is not common in mussels (Martínez-Lage et al. 1994, 1995), but is in oysters (Leitão et al. 2002; Li and Havenhand 1997).
In summary, this is the most exhaustive cytogenetic study carried out so far on scallops. It describes the chromosome number and morphology, the chromosome mapping of ribosomal genes by one- and two-color FISH, the identification of transcriptionally active NORs, and the distribution of constitutive heterochromatin. The data provided contribute to the construction of the molecular karyotype of H. distortus and will be useful in assessing the evolutionary relationships within scallops.
Corresponding Editor: Stephen O'Brien
We thank Dr. Guillermo Román for supplying the samples and Ms. Rosa García Díaz for her technical assistance in the laboratory. This work was funded by Xunta de Galicia through project XUGA10302B97.
References
Arrighi FE and Hsu TE,
Beaumont AR and Gruffydd LLD,
Beaumont AR and Zouros E,
Brand AR,
Cross I, Vega L, and Rebordinos L,
Dev VG, Miller DA, Allder-Dice PW, and Miller OJ,
Drouin G and Moniz de Sá M,
Fernández R, Barragán MJL, Bullejos M, Marchal JA, Díaz de la Guardia R, and Sánchez A,
Fujiwara A, Abe S, Yamaha E, Yamazaki F, and Yoshida MC,
Gajardo G, Parraguez M, and Colihueque N,
Gold JR and Ellison JR,
Heng HH and Tsui LC,
Howell WM,
Howell WM and Black DA,
Insua A, Freire R, and Méndez J,
Insua A, Freire R, and Ríos J,
Insua A, Freire R, Ríos J, and Méndez J,
Insua A, López-Piñón MJ, and Méndez J,
Insua A and Mendez J,
Kost MV, Alimov AA, Sarafanov AG, Tikchomirova TP, Gumeniuk RR, Timofeeva MJ, and Zelenin AV,
Leitão A, Boudry P, Labat JP, and Thiriot-Quiévreux C,
Leitão A, Boudry P, McCombie H, Gérard A, and Thiriot-Quiévreux C,
Leitão A, Chaves R, Santos S, Boudry P, Guedes-Pinto H, and Thiriot-Quiévreux,
Levan A, Fredga K, and Sandberg AA,
Li XX and Havenhand JN,
Martínez-Lage A, González-Tizón A, and Méndez J,
Martínez-Lage A, González-Tizón A, and Méndez J,
Martins C and Galetti PM Jr,
Matsuda Y, Moriwaki K, Chapman VW, Hoi-Sen Y, Akbarzadeh J, and Suzuki H,
Miller DA, Dev VG, Tantravahi R, and Miller OJ,
Miller DA, Miller OJ, Dev VG, Tantravahi R, and Croce CM,
Pauls E and Affonso PRAM,
Pauls E, Pacheco ML, and Cabeza MP,
Pelliccia F, Barzotti R, Bucciarelli E, and Rocchi A,
Sambrook J, Fritsch EF, and Maniatis T,
Sumner AT,
Suzuki H, Sakurai S, and Matsuda Y,
Thiriot-Quiévreux C,
Thiriot-Quiévreux C and Ayraud N,
Torreiro A, Martínez-Expósito MJ, Trucco MI, and Pasantes JJ,
von Brand-Skopnik E and Ibarra-Humphries AM,
Waller TR,
Wimber DE and Steffensen DM,
Winnepenninckx B, Backeljau T, and De Wachter R,