-
PDF
- Split View
-
Views
-
Cite
Cite
Qi Zhang, Xiaoting Hua, Zhi Ruan, Yunsong Yu, Ye Feng, Revisiting the contribution of gene duplication of blaOXA-23 in carbapenem-resistant Acinetobacter baumannii, Journal of Antimicrobial Chemotherapy, Volume 73, Issue 1, January 2018, Pages 250–252, https://doi.org/10.1093/jac/dkx339
Close - Share Icon Share
Sir,
Gene duplication has been discovered for many antimicrobial resistance genes in bacterial genomes and has been considered a source of elevated antimicrobial resistance.1 The gene blaOXA-23 is a major determinant in the emergence of carbapenem-resistant Acinetobacter baumannii (CRAB).2–4 We have previously reported the widespread duplication of blaOXA-23 by surveying 113 clinical CRAB isolates in China.5 However, in these isolates the blaOXA-23 copy number did not correlate well with the MIC of imipenem. A similar phenomenon was also reported recently by Yoon et al.6 One reasonable explanation is that, in addition to gene duplications, other mechanisms might also impact on the MIC, such as the presence of specific outer membrane proteins and/or the overexpression of resistance–nodulation–division (RND)-type efflux pumps.7 Often, these mechanisms might vary in their performance when in different genomic contexts. Instead of making comparisons between clinical isolates, in this study we cultured A. baumannii under treatment with carbapenem, thus avoiding any interference induced in different genomic contexts. If an increase in the blaOXA-23 copy number or MIC were to occur within the same strain, the contribution of gene duplication to carbapenem resistance would be acknowledged.
Two CRAB strains were employed in this study: XH386 (ST208 by Oxford MLST scheme) and XH731 (ST195). They each carried a single copy of blaOXA-23, which resided in Tn2009 and Tn2006, respectively. Their initial imipenem MIC was 24 mg/L and the strains were cultured for 2 weeks in imipenem-containing broth at a constant concentration of 16 mg/L. We chose this concentration not only because it was the highest concentration in which each strain can survive, but also because it was the peak serum concentration achievable in patients during the standard treatment. Culture in imipenem-free broth was used as a control. The entire experiment was repeated three times, independently, as biological replicates. Materials and methods are available as Supplementary data at JAC Online.
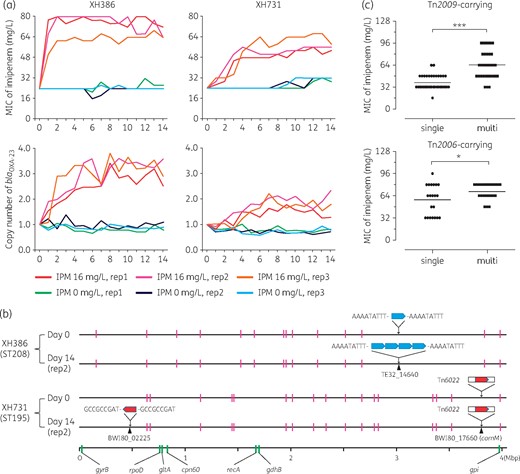
During serial passage, the imipenem MIC and blaOXA-23 copy number increased in both strains (Figure 1a). The three replicates of each strain showed nearly identical patterns, demonstrating that these changes were stable responses to the selection pressure of imipenem. Of note, the changes in the copy number and MIC were not synchronized with one another and the trajectories for XH386 and XH731 also differed. For example, the MIC for XH386 increased sharply immediately after imipenem treatment, but the MIC for XH731 ascended more gradually, and the number of days required for the MIC and copy number to reach their maximum were not identical. These findings suggest that blaOXA-23 duplicates more efficiently in XH386 than in XH731 and that the duplication of blaOXA-23 is not the only determinant of the imipenem MIC.
Contribution of gene duplication of blaOXA-23 to carbapenem resistance. (a) Response of two A. baumannii strains to imipenem treatment. XH386 and XH731 were passaged for 14 days at two concentrations: 0 and 16 mg/L. Each strain had three biological replicates. The imipenem MIC was determined using an agar dilution method and the copy number of blaOXA-23 was measured by quantitative RT–PCR. Both the copy number and MIC were measured three times for each biological replicate and the mean values are displayed. (b) How duplication occurred in the two strains. Days 0 and 14 (replicate two for both XH386 and XH731) represent the strains before and following serial passage. Blue and red arrows represent the single-copy Tn2009 and Tn2006, respectively. The sequences surrounding the arrows are the direct repeats generated by the transposition event. Tn2006 itself was carried within Tn6022 in strain XH731 prior to imipenem treatment. The distribution of ISAba1 is indicated by the magenta bars. The scale at the bottom indicates relative position according to the XH386 reference genome. The positions of the seven genes employed by the Oxford MLST scheme are indicated by the green bars. (c) Reanalysis of the data of Hua et al.5 The 69 Tn2009-carrying and 44 Tn2006-carrying CRAB isolates were further divided into two groups according to their copy number of blaOXA-23, i.e. the single-copy group (copy number <1.5) and the multi-copy group (copy number >1.5). Student’s t-test was performed for comparison: *P < 0.05; ***P < 0.001. IPM, imipenem. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
By employing the PacBio RS II sequencer, we sequenced the complete genomes of replicate strains for both XH386 and XH731 on Day 14 and compared these with the genomes prior to imipenem treatment. The blaOXA-23 copy number revealed by genome sequencing was basically consistent with that measured by quantitative RT–PCR. For XH386, the single-copy Tn2009 increased to four copies via tandem duplication. For XH731, Tn2006 duplicated at a novel genomic integration site (Figure 1b). Other genomic variations occurring during serial passage, which might also contribute to elevated imipenem resistance, are listed in Tables S1 and S2 (available as Supplementary data at JAC Online).
The in vivo situation within patients is assumed to be much more complicated than the in vitro passage experiment. The host immune response as well as the use of non-carbapenem antimicrobial products via empirical therapy alters bacterial gene expression and is therefore predicted to affect carbapenem resistance. Furthermore, the genomic contexts of the clinical isolates are not exactly identical to one another. All of these factors may participate in concealing the dosage effect of gene amplification on the resistance level, which may explain the low Pearson correlation coefficient between the MIC and copy number.5 When we revisited our clinical CRAB isolates by simply grouping them according to the blaOXA-23 copy number, the multi-copy group displayed a significantly higher MIC than the single-copy group (Figure 1c). This is possibly because this simple grouping approach is less affected by the other factors and therefore highlights the contribution of gene amplification. Interestingly, the differences between the effects of the single- and multi-grouping of the Tn2009-carrying isolates seem greater than that of the Tn2006-carrying ones. Recently, Yoon et al.6 published a related finding that Tn2009-carrying strains are much more prone to producing mutants with higher imipenem resistance than Tn2006 strains. These two findings, together with the discovery in this study that the blaOXA-23 copy number increases more rapidly under imipenem treatment in XH386 than in XH731, point to the same premise that Tn2009 duplicates more efficiently than Tn2006, although the exact mechanism awaits future research.
Taken together, the above findings suggest that carbapenem resistance in CRAB is multifactorial and can indeed increase via the duplication of blaOXA-23. Similar issues may also occur with other antimicrobial resistance genes and the contribution of their amplification to antimicrobial resistance should likewise be carefully evaluated.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81401698, 81230039), the Natural Science Foundation of Zhejiang Province (LY15H190004, LY15H190005) and the Zhejiang Provincial Medical Scientific Research Foundation (2015RCA017, 2016KYA108, 2016DTA003).
Transparency declarations
None to declare.
Supplementary data
Materials and methods and Tables S1 and S2 are available as Supplementary data at JAC Online.
References
Author notes
Qi Zhang and Xiaoting Hua authors contributed equally to this work.