-
PDF
- Split View
-
Views
-
Cite
Cite
J. A. W. Morgan, G. D. Bending, P. J. White, Biological costs and benefits to plant–microbe interactions in the rhizosphere, Journal of Experimental Botany, Volume 56, Issue 417, July 2005, Pages 1729–1739, https://doi.org/10.1093/jxb/eri205
Close - Share Icon Share
Abstract
This review looks briefly at plants and their rhizosphere microbes, the chemical communications that exist, and the biological processes they sustain. Primarily it is the loss of carbon compounds from roots that drives the development of enhanced microbial populations in the rhizosphere when compared with the bulk soil, or that sustains specific mycorrhizal or legume associations. The benefits to the plant from this carbon loss are discussed. Overall the general rhizosphere effect could help the plant by maintaining the recycling of nutrients, through the production of hormones, helping to provide resistance to microbial diseases and to aid tolerance to toxic compounds. When plants lack essential mineral elements such as P or N, symbiotic relationships can be beneficial and promote plant growth. However, this benefit may be lost in well-fertilized (agricultural) soils where nutrients are readily available to plants and symbionts reduce growth. Since these rhizosphere associations are commonplace and offer key benefits to plants, these interactions would appear to be essential to their overall success.
Introduction
This paper provides a general overview of the below-ground processes associated with the plant rhizosphere. It covers the interactions between plants and microbes, their chemical communications, the biological processes they sustain, and the costs and benefits to plants associated with these interactions.
Hiltner (1904) first introduced the term rhizosphere, which is derived from the Greek word ‘rhiza’, meaning root, and ‘sphere’, meaning field of influence. He defined the rhizosphere as the zone of soil immediately adjacent to legume roots that supports high levels of bacterial activity. However, more recently the term has been broadened to include both the volume of soil influenced by the root and the root tissues colonized by micro-organisms (reviewed in Pinton et al., 2001). Micro-organisms in the rhizosphere react to the many metabolites released by plant roots. The micro-organisms and their products, also interact with plant roots in a variety of positive, negative, and neutral ways. Such interactions can influence plant growth and development, change nutrient dynamics, and alter a plant's susceptibility to disease and abiotic stress. Many of the obstacles to improving our knowledge of these interactions are methodological (Morgan and Whipps, 2001). Traditional microbial population studies, based on identification and quantification, and the measurement of processes that occur in the rhizosphere, are often difficult or tedious. Similarly, the collection of relevant samples or the simulation of natural conditions in the laboratory can be problematical. However, with the array of molecular techniques that are becoming available, significant improvements in our understanding of rhizosphere microbial communities and processes are expected (Barea et al., 2005; Johnson et al., 2005).
The rhizosphere can be divided into several distinct zones (Lynch, 1987). These include the endorhizosphere (root tissue including the endodermis and cortical layers), the rhizoplane (the root surface with the epidermis and mucilaginous polysaccharide layer), and the ectorhizosphere (the soil immediately adjacent to the root). In addition, plants that are colonized by mycorrhizal fungi have a zone termed the mycorrhizosphere (Lindermann, 1988). As mycorrhizal fungi can extend for some distance out from the plant root, this region can be significant. In soils that are well colonized by plants, the plant roots may affect all the soil present in a particular area, and little non-rhizosphere soil (bulk soil) may be present. Even horticultural plants growing hydroponically possess a rhizosphere, and the layer of solution close to the root surface may differ substantially from the bulk solution (Vanpeer and Schippers, 1989).
Overall, it is the loss of carbon compounds from roots that drives the development of enhanced microbial populations in the rhizosphere when compared with the bulk soil (Grayston et al., 1996). This phenomenon is widespread across all plant species as a general process, although the compounds lost from different plant species, or even cultivars of particular species, can vary markedly in quality and quantity. Conversely, the micro-organisms in the rhizosphere can influence plants in a variety of ways, for example affecting plant growth, nutrition, development, susceptibility to disease, resistance to heavy metals, and the degradation of xenobiotics. As a result these interactions have considerable potential for biotechnological exploitation (Barea et al., 2005).
A living soil
The continued functioning of the soil ecosystem is essential for soil sustainability and productivity in the future (reviewed by van Elsas et al., 1997). Improvements in our understanding of the processes that occur in the soil ecosystem could help us improve management of agricultural practices and conservation methods. In addition, knowledge of the tolerance of soils to changes that may arise from processes like climate change could enable better decision-making for future generations. The capacity of the soil environment to cope with these impacts will not be limitless, and understanding the resistance and resilience of soil to management and perturbation is an area of importance. Suitable levels of food production to maintain our population must be achieved through the development of sustainable agricultural practices. The soil is a living environment that supports extremely diverse communities of micro- and macro-organisms and is often considered a ‘black box’. The soil environment is often overlooked in studies of species diversity and function, as it has little visual appeal in comparison to insects or larger organisms. It is envisaged that the microbially driven nutrient cycling processes which maintain plant growth and productivity will continue no matter what pressures are imposed on them. With changes in the levels of pollution and predicted effects of global warming, many groups of micro-organisms will continue to survive and grow unaffected. However, changes in the relative diversity and functional characteristics of microbial communities may have unpredictable consequences for nutrient cycling processes and the structure and productivity of natural plant communities. Such changes to microbial communities may also impact on the productivity of crop plants necessary for human population growth.
What does a plant need from below the ground?
The answer to this question is simply that a plant obtains almost everything directly from the soil to support growth, with the notable exceptions of carbon dioxide, oxygen, and light. The soil must have a structure that is physically capable of supporting the above-ground half of the plant through its developing root system as it grows. In addition, the soil needs to be maintained at an appropriate pH, provide protection from toxic substances and pathogens, and contain suitable levels of water. Beyond this, all the essential mineral elements that a plant requires are obtained from the soil. At least 17 elements are essential for plant growth and reproduction (Marschner, 1995). Fourteen of these elements are acquired primarily from the soil solution. These include six macronutrients (N, K, P, S, Mg, and Ca) and eight micronutrients (B, Cl, Cu, Fe, Mn, Mo, Ni, and Zn). In addition, plants will accumulate non-essential and/or toxic mineral elements, such as Cd, Pb, and Na, when these are present in the soil solution. Most of these elements are generally taken up from the soil solution in their ionic form (White, 2003). Plant growth may be limited by the availability of essential elements, as well as by the presence of toxic elements. The interactions between plant roots and organisms within their rhizosphere help them to acquire essential mineral nutrients and prevent the accumulation of toxic elements.
Perhaps the essential mineral element that most frequently limits plant growth is P, which is taken up from the soil solution as phosphate (Pi,
As all the minerals that a plant requires must come from the soil, and as the activity of microbes in the soil are central to the efficient solubilization of these mineral elements, it is not surprising that a series of generalized and specific plant–microbe associations exist to perform this function.
What below-ground plant–microbial associations exist?
The release of carbon compounds from plants into the soil results in greater microbial populations in the rhizosphere relative to the bulk soil, and increased microbial biomass and activity (Lynch, 1987; Bending, 2003). This is considered to be a general effect and to have variable consequences. In addition, plants can have specific mycorrizal- and nodulation-based associations that fulfil unique functions. When considering the rhizosphere effect in general, the rhizosphere/bulk soil (r/s) ratios for bacteria, actinomycetes, and fungi are usually in the ranges 2–20, 5–10, and 10–20, respectively. However, many of the bacteria in the rhizosphere and soil are unable to grow on laboratory media, which makes their study difficult. In young plant roots it is thought that the rhizosphere bacterial communities are dominated by r-strategists, which are species with fast growth rates and capacities to utilize simple substrates (Andrews and Harris, 1986; Brimecombe et al., 2001). As the roots mature, there is a shift in dominance to bacterial communities with relatively slow growth rates and the capacity to degrade more complex substrates (k-strategists). As a rule, although a general increase in micro-organisms in the rhizosphere is always noted, the community structure and functional consequences of this increase are less well understood.
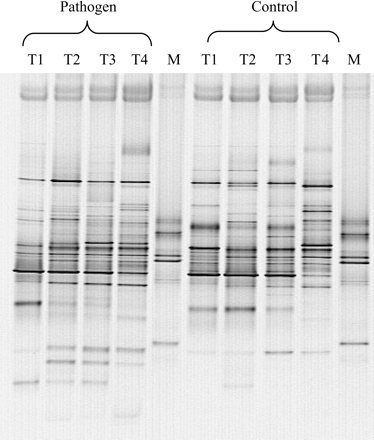
As many of the micro-organisms both in the soil and the rhizosphere are difficult to grow or enumerate using traditional plate count methods (Roszak and Colwell, 1987), a variety of molecular methods have been developed to assay the presence of these micro-organisms in samples. Most recently, the method of choice to determine what micro-organisms are present in environmental samples is to amplify the conserved small subunit rRNA gene (Ford and Olson, 1988). In this process, DNA is isolated from the soil using bead beating, and polymerase chain reaction (PCR) with gene-specific primers is used to amplify the specific gene from the sample. To look at the diversity of small subunit rRNA genes (directly related to the diversity of micro-organisms) present in the sample, the PCR products are either cloned and sequenced, or profiled by gel electrophoresis to allow the analysis of many samples. A variety of techniques are available for microbial community profiling, including denaturant gradient gel electrophoresis (DGGE), temperature gradient gel electrophoresis, single-strand conformation polymorphism, and terminal restriction fragment length polymorphism. DGGE, in particular, has commonly been used to analyse microbial populations in a variety of samples (Calvo-Bado et al., 2003; Singh et al., 2003). The complexity of the banding pattern is used to assess the diversity of micro-organisms present in the sample. Figure 1 shows the DGGE profile of bacterial 16S rRNA gene PCR products generated from tomato roots. Changes in banding caused by experimental variables, such as the age of the plant or growth in the presence of pathogens, can be used to investigate impacts on rhizosphere communities. By investigating the changes in banding patterns on the gel, changes of interest can be targeted, bands isolated from the gel, and sequenced. By comparing the sequence with known sequences in the database, information on the identity and the characteristics of similar species can be obtained. This provides information on specific microbial population changes in the sample. If such populations are universally involved in specific processes such as nutrient cycling or disease suppression, then these processes can be specifically targeted for further study to confirm this.
Denaturing gradient gel electrophoresis profile of 16S rRNA genes present in bacteria on roots of tomato plants. Changes seen with time (T1–T4) and in the presence of the plant pathogen Pythium aphanidermatum. Each band represents a single type of bacterium present. Markers for defined strains are run in lane M.
Using such methods, the structures of rhizosphere microbial communities have been shown to be distinct from those of bulk soil, with lower diversity in the rhizosphere relative to bulk soil (Marilley and Aragno, 1999). The specific structure and diversity of the rhizosphere bacterial community varies between plant species and over time (Smalla et al., 2001), and different root zones on the same plant can support distinct bacterial communities, reflecting qualitative and quantitative differences in root exudation (Yang and Crowley, 2000). In addition, soil type has a key role in determining the specific dominant bacteria colonizing the rhizosphere (Marschner et al., 2001). Hence, rhizosphere microbial populations of the same plant species growing in the same field may show great variation, both spatially and temporally. The structure of rhizosphere bacterial communities can also be influenced by root infection by pathogenic bacteria, which promote greater bacterial community variability compared with healthy roots (Yang et al., 2001).
The rhizosphere supports diverse bacteria that can stimulate growth of plants. Such plant growth-promoting rhizobacteria operate by a wide variety of mechanisms, including N2 fixation, enhanced solubilization of P, and phytohormone production (Vessey, 2003; Barrea et al., 2005). Traditionally, pseudomonads have been considered to be important rhizosphere organisms (Lugtenberg et al., 2001). The term ‘pseudomonads’ has in the past been applied to bacteria now placed in different genera (e.g. Burkholderia and Pseudomonas), let alone of different species, and such conclusions need to be reconsidered. While many studies have shown elevated pseudomonad communities in the rhizosphere (Yang et al., 2001; Marilley and Aragno, 1999) this is not always the case, and in some circumstances other microbial groups, such as Bacillus spp., may dominate (Macrae et al., 2001). While many rhizosphere pseudomonads are plant growth-promoting rhizobacteria, others inhibit plant growth and cause disease. However, it is not clear what makes some pseudomonads beneficial and others pathogenic, especially since they colonize the same ecological niches and possess similar mechanisms for plant colonization (Preston, 2004).
As rhizosphere processes result from the activities of diverse groups of micro-organisms, determining the significance of changes to community structure presents a major challenge for the future. So far, the molecular analysis of bacterial community structure has indicated that rhizosphere soil may be dominated by phylogenetically less diverse strains relative to bulk soil. Although the identity of the micro-organisms can be revealed by these approaches, this does not easily relate to the function of the micro-organisms in the samples. Genes for many key processes of interest are present on mobile elements and the small subunit rRNA gene can only place the backbone conserved nature of chromosomal regions with any certainty. Analysis of groups of micro-organisms, where all members perform the same function, or of functional genes themselves, allows the easiest interpretation of results.
For plants, there are two influential specific plant–microbe associations: mycorrhizas and root nodulation. Mycorrhizas are mutualistic symbioses between plant roots and soil fungi (Smith and Read, 1997). Over 80% of land plants are able to form mycorrhizal associations. In these associations there is a bidirectional flow of nutrients. Carbon flows out from the plant host to the fungus, and mineral nutrients flow from the fungus to the plant. It is estimated that between 4% and 20% of net photosynthate can be transferred from the plant to its fungal partner. In return, the mycorrhiza can become the primary organ acquiring mineral nutrients. Nevertheless, there are a number of plant families that are predominantly non-mycorrhizal, including the Brassicaceae (Brassicales), Caryophyllaceae, and Chenopodiaceae (Caryophyllales), Lupinus and Kennedia (Fabales), Cyperaceae and Juncaceae (Poales), and Proteaceae (Proteales), which are all renowned for their ability to acquire P from low-P soils, and are characterized by their ability to proliferate lateral or cluster roots locally in patches of soil rich in P and to secrete organic acids, citrate in particular, into the rhizosphere (Dinkelaker et al., 1995; López-Bucio et al., 2000; Dechassa and Schenk, 2004; Miller, 2005). Non-mycorrhizal plants are characteristic of harsh habitats such as saline and arid soils (Brundrett, 2002).
The establishment of the mycorrhizal network offers a number of basic advantages for the acquisition of mineral nutrients: (i) fungal hyphae extend beyond the area of nutrient depletion surrounding the root; (ii) fungal hyphae greatly increase the surface area for the absorption of nutrients relative to non-mycorrhizal roots; (iii) hyphae are able to extend into soil pores that are too small for roots to enter; and (iv) some mycorrhizal fungi can access forms of N and P that are unavailable to non-mycorrhizal plants, particularly organic forms of these nutrients.
In addition to this, mycorrhizal fungi are able to provide protection to the host plant against root and shoot pathogens (Whipps, 2004). They might do this in a number of ways, including antibiotic production, induced resistance, competition for root infection sites, and by providing a physical barrier to infection. The significance and function of plant–mycorrhizal associations, while not as diverse as the plant–rhizosphere association, can vary greatly (Smith and Read, 1997). A single plant root may be colonized by many different mycorrhizal fungi, and mycorrhizal fungi often have low specificity, and are able to colonize a variety of different plant species. There is also great variation between fungal species in the benefits they provide to their host. The mycelial network can link plants of the same and/or different species. Several studies have shown a transfer of C between individual plants of the same and different plant species through the mycelial network (Francis and Read, 1984; Fitter et al., 1998), but other studies have shown no such transfer (Pfeffer et al., 2004). Clearly a range of factors could influence the direction and rates of such C translocation, including the age and nutrition of donor and recipient plants, and the functional characteristics of the individual fungal and plant species involved.
In the same way as the rhizosphere effect is seen for plant roots, a mycorrhizosphere effect can be seen where the soil surrounding fungal hyphae supports distinct bacterial communities compared with the bulk soil (Lindermann, 1988). Mycorrhizosphere inhabitants can include intra-hyphal bacteria in ectomycorrhizal fungi (Bertaux et al., 2003), and intra-spore bacteria in some arbuscular fungi (Bianciotto et al., 1996). It has been shown that some mycorrhizosphere bacteria can promote mycorrhiza formation, with a variety of Gram-positive and -negative strains involved (Garbaye, 1994), although the precise mechanisms involved are unclear. There are several categories of mycorrhiza, currently defined on the basis of structure and morphology.
Arbuscular mycorrhizas (AM) are the commonest mycorrhizal group, and are found on angiosperms, gymnosperms, pteridophytes, and bryophytes. The association is very close and the fungal hyphae penetrate root cortical cells to form arbuscules to exchange nutrients and carbon. About 150–200 obligate biotrophic AM fungi have been described so far, which all belong to the Glomeromycota (Schussler et al., 2001). Indeed, the roots of the earliest land plants contained arbuscular mycorrhizal structures, and the symbiosis of ancestral plants with the Glomeromycota may have enabled plants to colonize the land (Taylor et al., 2004). The arbuscular fungi spread into the soil to form the extramatrical mycelium, the size of which, and the relative proportion of mycelium within the root and in the soil, varies greatly between different AM species (Hart and Reader, 2002). The uptake and translocation to the host of ions with low diffusion coefficients that are relatively immobile in the soil solution, particularly Pi, but also Zn2+ and Cu2+, can be very important (Smith and Read, 1997). However, AM fungi may also have a role in providing mineral forms of N, K, and other nutrients to the host. There is evidence that AM fungi could play a role in the tolerance of some plants to heavy-metal contamination, with the development of metal tolerance by the fungi, and binding of metals to polyphosphate within fungal hyphae implicated (Barea et al., 2005).
It is thought that any AM-fungus/host-plant combination is possible, but that the colonization rates and types of mycorrhizal association formed differ between the individual combinations and are controlled by the interplay of fungal and plant genomes (Graham, 2000; Lerat et al., 2003). Thus, the dependence of fungal colonization, C-transfers, and P-transfers on soil P availability differs between individual AM-fungus/host-plant symbioses (Graham, 2000; Lerat et al., 2003; Dennison and Kiers, 2005).
In natural environments the diversity of AM fungi is a key contributor to the diversity and productivity of plant communities (van der Heijden et al., 1998). Different plant species within natural communities have different AM-fungus assemblages on their roots, suggesting that there are different AM-fungus/host-plant preferences (Vandenkoornhuyse et al., 2003). Furthermore, the composition of root-inhabiting AM communities within individual host plants shows seasonal variation (Heinemeyer, 2004) and can change as plants mature (Husband, 2002). Further changes in the AM community colonizing plant roots can be induced by shading and pesticide treatment (Vandenkoornhuyse et al., 2003). It is acknowledged that there is high functional diversity among AM fungus species in terms of improving P uptake by the host (Munkvold et al., 2004). However, the implications of changes in the AM community structure for the functional interactions between the symbionts and the C cost of the symbiosis are unknown.
Many farming practices, including fertilizer application, cultivation, and fumigation can have deleterious impacts on communities of AM fungi (Kurle and Pfleger, 1994), which are known to be less diverse and abundant in conventional agricultural systems relative to organically managed and semi-natural areas (Bending et al., 2004; Oehl et al., 2004). Furthermore, there is evidence that the AM-fungus communities selected by intensive conventional practices are relatively less beneficial for crop yields than those from organic practices (Scullion et al., 1998; Eason et al., 1999). Commercial inocula of AM fungi are available for use in degraded habitats and agricultural systems, although application of these products has been relatively limited to date (Gianinazzi and Vosatka, 2004).
Ectomycorrhizas are found almost entirely on woody perennials including members of the Pinaceae, Betulaceae, Fagaceae, and Dipterocarpaceae (Smith and Read, 1997). Over 2000 fungus species are known to be capable of forming ectomycorrhizas, with most being basidiomycetes or ascomycetes. Hyphae penetrate into the root cortex where they ramify between cells to form a ‘Hartig net’, through which materials are exchanged. The fungus forms a mantle of hyphae on the outside of the plant root which extends into the surrounding soil. The structure of the ectomycorrhizal fungus extramatrical mycelium varies considerably between fungal species, ranging from a weft of undifferentiated mycelium around the root, to highly differentiated mycelium comprising a foraging fungal front connected to roots via rhizomorphs (Agerer, 2001). These different systems reflect differences in exploration and nutrient mobilization strategies. The primary function of the fungal mycelium is absorption of nutrients from the soil, and the translocation of these materials to the host. Ectomycorrhizal fungi have been shown to translocate mineral forms of N, P, and micronutrients from the soil to the host. Some ectomycorrhizal fungi typical of temperate and boreal forests are able to produce a suite of extracellular enzymes that mobilize organic forms of N and P, which are otherwise unavailable to the host plant (Read and Perez-Moreno, 2003).
As well as the fungus–plant interactions there are bacterium–plant interactions, although these are limited (Squartini, 2003). The symbiotic associations between N2-fixing bacteria and plant roots are the best studied. In many environments N limits plant growth. Plants of the angiosperm Rosid I clade (APG II, 2003) have evolved symbioses with specific bacterial strains to acquire N through the strain's capacity for biological N2 fixation (Gualtieri and Bisseling, 2000). The Rosid I clade includes the Fabaceae and Ulmaceae, which form nodules with rhizobia, and species from eight other families (Betulaceae, Casuarinaceae, Coriariaceae, Datiscacae, Elaeagnaceae, Myricaceae, Rhamnaceae, and Rosaceae), which form nodules with Frankia. Thus, like mycorrhizal associations, the precise benefits to plants of hosting N2-fixing symbionts depends not only upon the N availability in the soil but also differs between individual symbiont/host-plant associations (Dennison and Kiers, 2005).
In terms of agricultural importance, the most significant interactions are the Fabaceae–Rhizobium spp./Bradyrhizobium spp. root nodule symbioses (Squartini, 2003). In the bulk soil, the bacteria persist in a dormant or saprophytic state. In the presence of a suitable host the bacteria infect the plant through the root hair. Legume roots are known to exude various flavonoid and isoflavonoid molecules that induce expression of nod (nodulation) genes by bacteria in rhizobia. This results in the formation by the bacterium of lipo-oligosaccharide Nod factors, the precise structure of which determines the host range and specificity of the association. Once inside the plant, an infolded plasma membrane forms around the bacterium to produce an infection thread. The bacteria multiply and are released into the cytoplasm. Here, root cells are induced to divide, resulting in formation of a root nodule containing enlarged non-motile bacterial cells. Within the nodule the host plant provides the bacteria with the carbohydrates they need. In return, the rhizobial bacteria fix N2 from the atmosphere into
Recent work on root nodule bacteria has demonstrated that the interaction is not restricted to Rhizobium/Bradyrhizobium (Sawada et al., 2003). N2-fixing strains of Ralstonia, Burkholderia, and Methylobacterium have been isolated from the nodules of some tropical Fabaceae. The key characteristic among all these strains appears to be the expression of nod genes (Dakora, 2003). Where these genes have been studied they have been shown to reside on plasmids or mobile regions of the chromosome. In several instances nif (N2 fixing), nod, and other genes involved in nodule formation and functioning are clustered together. The genomes of a number of rhizobial species and the model host Lotus japonicus have been sequenced. These developments should allow progress in understanding the molecular mechanisms underlying the functioning of the root nodule symbiosis (Colebatch et al., 2002).
Costs and gains to the plant
The cost to the plant for interacting with non-symbiotic micro-organisms in the rhizosphere is likely to purchase a variety of benefits. These include the development of defences to prevent infection by pathogens, and enhanced uptake of nutrients and water (Lynch, 1987). However, since rhizodeposition can promote directional growth of some soil-borne pathogens towards the root, the cost of rhizodeposition can be added to by the cost of pathogen infection (Whipps, 2001). These forms of cost and benefit cannot be valued numerically. It may simply be that there is an absolute requirement for the rhizosphere microbial community and the plant could not exist without it. If plants could exist without rhizosphere micro-organisms, then they could simply excrete a general biocide and avoid attack.
Root carbon budgets have been determined for a number of plant species in laboratory investigations using 14C tracer techniques (Whipps, 1987; Grayston et al., 1996). While these methods are effective at quantifying fluxes, they are not able to distinguish root from microbial respiration and, therefore, are unable to quantify absolute amounts of C lost as rhizodeposits. Using tracer techniques, the proportion of net fixed photosynthate translocated to the roots of wheat, maize, tomato, and pea seedlings has been estimated to be between 30% and 60%. When respiration (arising from both plant roots and rhizosphere organisms) is considered, the amount of C lost from the roots is estimated to be between 40% and 80% of photosynthate. Generally between 10% and 30% of the C lost from the roots is recovered as rhizodeposits.
Much less is known about rhizodeposition from mature plants. In long-term experiments with wheat and white mustard, the amounts of C recovered in the root biomass at crop harvest were between 20% and 35% of that which had been translocated to the root. Assuming that root respiration consumed 30% of the C translocated to the root, the amount of material lost via rhizodeposition was estimated to be 0.5 and 2.8 t C ha−1 year−1 for wheat and mustard, respectively. In field experiments with wheat, the proportion of fixed C transferred to the root declined from 50% after 42 d to 2% at 154 d. The total C lost via rhizodeposition, including soil respiration and root residues, was estimated to be between 1.2 and 1.9 t C ha−1 year−1, representing 15% of the C fixed by this cereal. In the case of forest trees, 40–73% of the net fixed C can be transferred to the roots, and losses of C, including that mediated through mycorrhizal fungi, has been calculated at 5.8–7.5 t C ha−1 year−1. When bark was removed from forest trees to stop the flow of assimilate to roots, it was found that current assimilate provided energy for over 50% of soil respiration (Hogberg et al., 2001), demonstrating the reliance of soil micro-organisms for root-derived C. Significant quantities of N are also lost from the plant as rhizodeposits (Brimecombe et al., 2001). In barley and pea, rhizodeposition in 7–14-week-old plants has been estimated to account for between 32% and 71% and 15% and 48%, respectively, of the below-ground N budget, amounting to 20% and 7%, respectively, of total plant N at maturity.
Advances in reporter gene technology could help to improve our understanding of rhizosphere C fluxes (Kilham and Yeomans, 2001). Bacteria harbouring a marker gene reporting on single C sources have the potential to provide quantitative information on the spatial and temporal dynamics of rhizosphere C flow. However, application of such technology has been limited to date.
Plants secrete a variety of extracellular P-mobilizing enzymes, the induction of which is triggered by P deficiency (Vance et al., 2003; Hammond et al., 2004). Most roots can produce acid phosphatase, which hydrolyses inorganic P from organic phosphomonoesters, which form 30–80% of the soil P reserve in agricultural soils. In lupins, acid phosphatase production can be increased by up to 20 times under P stress. Similarly, many plant species exude organic anions such as citric and malic acids in response to deficiency of several nutrients, including P, K, Fe, and Mn (Jones et al., 2003). These compounds lower rhizosphere pH, increasing the availability of
Organic acids play a role in mediating detoxification of metals by plants (Pinton et al., 2001; Jones et al., 2003). Al toxicity can limit crop production in many areas. In several crop plants including wheat and maize, Al3+ tolerance is the result of increased exudation of malate or citrate in response to the metal. The organic acids complex Al3+ rendering it non-toxic. The exudation of organic acids allows the plant to grow at higher Al3+ concentrations.
Mucilage, consisting of polysaccharides containing hexose and pentose sugars and uronic acids, is secreted by root cells as the root grows through the soil (Czarnes et al., 2000). Additional mucilage is secreted by rhizosphere microbes. On contact with the soil, mucilage forms a gel which has a number of beneficial properties. The gel binds soil particles and microbes together with the root to form a ‘rhizosheath’. As the moisture content of soil falls following plant uptake, the mucilage dries. Since it binds the soil and root together, the gel ensures that gaps do not form as the soil shrinks, so that hydraulic conductivity is maintained. Other possible benefits to the plant of mucilages include aiding lubrication as roots move through the soil, and the absorption of ions, including Fe2+/3+, Ca2+, and
Rhizodeposition also stimulates the germination of pathogen propagules and directed growth towards the root, which can lead to disease (Whipps, 2001). Soil-borne pathogens fall into two broad groupings. Nectrotrophic pathogens, including Fusarium, Verticillium, and Pythium rapidly kill all or part of the host following their entry through plant roots. These pathogens characteristically have wide host ranges and attack young, debilitated, or senescing tissues. For some nectrotrophic fungal pathogens with a broad host range including Pythium and Fusarium, plant exudate components including sugars and amino acids stimulate propagule germination and growth towards the root. Subsequent infection occurs through wounds or breaks in the root surface. For those pathogens with limited host ranges, propagule germination stimulants can be compounds specific to the host family, such as organic S compounds in the case of the interaction of Sclerotium cepivorum with Allium spp. Host cells are rapidly killed by cytolytic enzymes or toxins. Biotrophic pathogens such as Plasmodiophora brassicae have a narrow host range and infect plants directly or through natural openings. Such pathogens initially grow within host tissues without cell death, although they can cause changes to root structure and physiology. Specialized parasitic structures called haustoria are formed within host cells, through which nutrients are drawn from the host.
The rhizosphere also supports populations of bacteria that may have negative effects on plant growth and development without infection of root tissues (Nehl et al., 1996). The mechanisms by which such deleterious rhizosphere bacteria operate include the production of phytotoxins and phytohormones, competition for nutrients, and the inhibition of mycorrhizal fungi.
There are many examples of bacteria that can suppress the growth of pathogenic fungi in the rhizosphere (Whipps, 2001). Effective colonization of the root is a key factor determining the ability of these bacteria to exert biocontrol. A number of these bacteria produce anti-fungal metabolites, including antibiotics, extracellular enzymes, and HCN (Brimecombe et al., 2001). Competition between rhizosphere bacteria and fungal pathogens for nutrients has also been identified as a biocontrol mechanism. For example, the sequestration of Fe3+ by bacterial siderophores and chelators can limit availability of the nutrient to pathogens, restricting their growth through the rhizosphere. Exposure of roots to non-pathogenic rhizosphere bacteria, including strains of Bacillus spp. and Pseudomonas spp., can induce resistance of host plants to some pathogenic fungi. Several mechanisms have been implicated in induced resistance, including enhanced production of phytoalexins, production of stress-related proteins and degradative enzymes, and the strengthening of epidermal cells (van Loon et al., 1998).
Many rhizosphere fungi, including mycorrhizal fungi, are able to suppress soil-borne plant pathogens (Whipps, 2001). Fungi have the advantage over bacterial biocontrol agents in that they are generally more effective at spreading through the soil and rhizosphere. A variety of mechanisms are involved in the control of fungal pathogens by rhizosphere fungi, including competition for nutrients, antibiotic production, and induced resistance. In addition, many fungi are able to parasitize spores, sclerotia, or hyphae of other fungi, resulting in biocontrol. Mycoparasitism is initiated by host sensing, which is generally followed by directed growth towards it, recognition, penetration, and degradation. Production of a number of degradative enzymes, including chitinases, proteases, and glucanases is involved in the biocontrol process.
Although many agrochemicals are available to control root disease, concerns over the environmental and human-health implications of pesticide use, and the desire for sustainable agricultural systems, have driven great interest in developing biological disease control based on rhizosphere antagonists of pathogens (Whipps, 2001). A number of such biocontrol agents are available as commercial formulations for use in agriculture and horticulture.
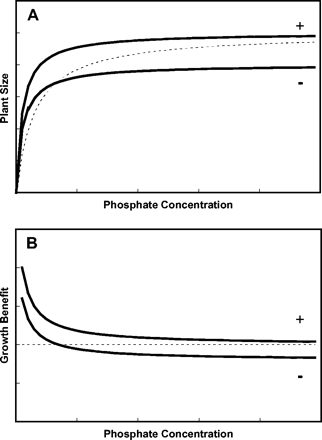
For AM associations a variety of different forms of cost–benefit analysis have been carried out (Fitter, 1991; Tinker et al., 1994; Johnson et al., 1997; Graham, 2000). The main benefit to plants of AM is generally considered to be enhanced P nutrition, and for this reason, cost–benefit analyses of the AM symbiosis consider P delivered to the root by the fungus, and C provided by the plant to the fungus, as the currency driving the symbiosis. For this review, a simple cost–benefit curve for a particular combination of AM fungus and host plant has been generated (Fig. 2). As P concentration increases, plant growth increases, following a hyperbolic relationship. This can be approximated by a Michaelis–Menton equation. The presence of an AM fungus will decrease the Km value (the P concentration at which growth is half maximum). A cost-neutral association will produce equivalent maximal plant yield. A costly AM association will reduce maximal plant yield. But both mycorrhizal associations are beneficial to the plant at low P concentrations. Mycorrhizal fungi may be parasitic on plants when net cost of the symbiosis exceeds benefits. In general, it has been estimated that between 4% and 20% of total photosynthate may be consumed by mycorrhizas (Johnson et al., 1997). It is clear that the C costs of promoting mycorrhizal associations may not always be compensated for by improved P acquisition when Pi is readily available, which may account for the decrease in mycorrhizal colonization of roots as plant P status is improved (Graham, 2000). The relative benefit to plants in terms of P gained per unit of C provided to the AM fungus can range between 0.4 and 10.2, indicating considerable differences between species (Pearson and Jakobsen, 1993).
Schematic cost–benefit curves for combinations of AM-fungus and host-plant symbiosis at different soil P concentrations. (A) Growth of mycorrhizal and non-mycorrhizal plants at different P concentrations in the soil solution. (B) Cost–benefit curve of mycorrhizal associations calculated as the quotient of growth in the presence/growth in the absence of mycorrhiza. Plant lacking mycorrhiza (broken line). Plants possessing cost neutral (+) or costly (−) mycorrhizal associations.
However, it should be recognized that mycorrhizal symbioses also contribute to disease resistance, protection from toxic minerals, and the acquisition of water and nitrogen (Dennison and Kiers, 2005). Quantifying benefits to plants of AM association purely in terms of P gain may therefore be misleading.
It has been speculated that the evolution of mycorrhizal associations laid the foundation for rhizobial nodulation, since there is genetic evidence that they share some common intermediates in their signalling pathways (Ana et al., 2004; Borisov et al., 2004; Levy et al., 2004). Although the costs of hosting a rhizobial symbiont have been estimated as 20–30% of total photosynthate (Provorov and Tikhonovich, 2003), the availability of N for plant growth is paramount on low-N soils. Nevertheless, strains of Rhizobium and Frankia vary greatly in their ability to fix N2.
Benefits to humans from an improved understanding of the rhizosphere
It is difficult to quantify the benefits to humans from an improved understanding of the rhizosphere and its processes. These benefits are qualitative, but possibilities for future improvements exist. In general, plants grow well in natural soils without any interference from humans. Intensive agricultural practices have, on occasions, led to major failings in plant growth. For example, the addition of unsuitable manures containing heavy metals lead to crop losses because Rhizobium species are extremely sensitive to heavy metal toxicity and roots fail to nodulate (McGrath et al., 1995). Therefore, a greater understanding of below-ground processes might help us to foresee the consequences of problems such as climate change, global warming, or intensive agriculture, but the benefit of this knowledge is difficult to assess. Maintaining the microbial diversity of our soils is universally believed to be a good thing. Experiments can be conducted to look at changes in diversity with particular treatments, these results may then offer limits of tolerance of the systems to treatments and enable management strategies to be designed to try and maintain the diversity and productivity of a soil.
We thank Leo Calvo-Bado (University of Warwick) for providing information on DGGE and Fig. 1, and BBSRC and DEFRA (HH3501SFV, HH3506SFV, HH3508SFV, HH3504SPO, HH1751SX, and HH3207HX) for funding elements of the work presented in this review.
References
Agerer R.
APG II.
Ana JM, Kiss GB, Riely BK, et al.
Andrews JH, Harris RF.
Barea J-M, Pozo MJ, Azcón R, Azcón-Aguilar C.
Bending GD.
Bending GD, Turner MK, Rayns F, Marx MC, Wood M.
Bertaux J, Schmid M, Prevost-Boure NC, Churin JL, Hartmann A, Garbaye J, FreyKlett P.
Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy HV, Bonfante P.
Borisov AY, Danilova TN, Koroleva TA, et al.
Brimecombe MJ, De Leij FA, Lynch JM.
Calvo-Bado L, Pettitt T, Parsons N, Petch GM, Morgan JAW, Whipps JM.
Colebatch G, Trevaskis B, Udvardi M.
Czarnes S, Hallett PD, Bengough AG, Young IM.
Dakora FD.
Deaker R, Roughley RJ, Kennedy IR.
Dechassa N, Schenk MK.
Dennison RF, Kiers ET.
Dinkelaker B, Hengeler C, Marschner H.
Eason WR, Scullion J, Scott EP.
Fitter AH.
Fitter AH, Graves JD, Watkins NK, Robinson D, Scrimgeour C.
Ford S, Olsen BH.
Francis R, Read DJ.
Garbaye J.
Gianinazzi S, Vosatka M.
Graham JH.
Grayston SJ, Vaughan D, Jones D.
Hammond JP, Broadley MR, White PJ.
Hart MM, Reader RJ.
Heinemeyer A, Ridgway KP, Edwards EJ, Benham DG, Young JPW, Fitter AH.
Hiltner L.
Hogberg P, Nordgren A, Buchmann N, Taylor AFS, Ekblad A, Hogberg MN, Nyberg G, Ottosson-Lofvenius M, Read DJ.
Husband R, Herre EA, Turner SL, Gallery R, Young JPW.
Johnson D, Ijdo M, Genney DR, Anderson IC, Alexander IJ.
Johnson NC, Graham JH, Smith FA.
Jones DL, Dennis PG, Owen AG, van Hees PAW.
Killham K, Yeomans C.
Kurle JE, Pfleger FL.
Lerat S, Lapointe L, Gutjahr S, Piché Y, Verheilig H.
Levy J, Bres C, Geurts R, et al.
Linderman RG.
López-Bucio J, Nieto-Jacobo MF, Ramírez-Rodríguez V, Herrera-Estrella L.
Lugtenberg BJJ, Dekkers L, Bloemberg GV.
Marilley L, Aragno M.
Macrae A, Lucon CMM, Rimmer DL, O'Donnell AG.
Marschner P, Yang CH, Lieberei R, Crowley DE.
McGrath SP, Chaudri AM, Giller KE.
Miller RM.
Morgan JAW, Whipps JM.
Munkvold L, Kjoller R, Vestberg M, Rosendahl S, Jakobsen I.
Nehl DB, Allen SJ, Brown JF.
Oehl F, Sieverding E, Mader P, Dubois D, Ineichen K, Boller T, Wiemken A.
Pearson JN, Jakobsen I.
Pfeffer PE, Douds DD, Bucking H, Schwartz DP, Shachar-Hill Y.
Pinton R, Varanini Z, Nannipieri P. (eds)
Preston GM.
Provorov NA, Tikhonovich IA.
Raghothama KG.
Read DJ, Perez-Moreno J.
Roszak DB, Colwell RR.
Sawada H, Kuykendall LD, Young JM.
Schussler A, Schwarzott D, Walker C.
Scullion J, Eason WR, Scott EP.
Singh BK, Walker A, Morgan JAW, Wright DJ.
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G.
Squartini A.
Taylor TN, Klavins SD, Krings M, Taylor EL, Kerp H, Hass H.
Tinker PB, Durall DM, Jones MD.
Vance CP, Uhde-Stone C, Allan DL.
Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW.
van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis P, Streitwolf-Engel R, Boller T, Wiemken A, Sanders IR.
van Elsas D, Wellington EMH, Trevors JT. (eds)
van Loon LC, Bakker PAHM, Pieterse CMJ.
Vanpeer R, Schippers B.
Whipps JM.
Whipps JM.
White PJ.
Yang CH, Crowley DE.




Comments