-
PDF
- Split View
-
Views
-
Cite
Cite
Nobuyoshi Jinnai, Takuro Sakagami, Takashi Sekigawa, Miho Kakihara, Toshiaki Nakajima, Kenichi Yoshida, Shin Goto, Takashi Hasegawa, Takeshi Koshino, Yoshinori Hasegawa, Hiromasa Inoue, Naohito Suzuki, Yasuyuki Sano, Ituro Inoue, Polymorphisms in the prostaglandin E2 receptor subtype 2 gene confer susceptibility to aspirin-intolerant asthma: a candidate gene approach, Human Molecular Genetics, Volume 13, Issue 24, 15 December 2004, Pages 3203–3217, https://doi.org/10.1093/hmg/ddh332
Close - Share Icon Share
Abstract
Aspirin-intolerant asthma (AIA) is a subtype of bronchial asthma characterized by development of bronchoconstriction evoked by non-steroidal anti-inflammatory drugs (NSAIDs). NSAIDs inhibit the cyclooxygenase pathway, leading to enhancement of the lipoxygenase pathway. We evaluated allelic association of 370 single nucleotide polymorphisms (SNPs) of 63 candidate genes, mostly from the arachidonic acid metabolic cascade, with AIA. After two rounds of screening with 198 AIA patients, multiple SNPs in the prostaglandin E2 receptor subtype 2 (EP2) gene were associated with AIA (P<0.05). Among the 77 SNPs identified in the EP2 gene, we selected 17 SNPs on the basis of linkage disequilibrium and allelic frequencies (minor allele frequency >0.1) for further association study. SNPs in the promoter region of the EP2 gene, uS5, uS5b, and uS7, were significantly associated with AIA (permutation P=0.039–0.001). Analysis of haplotypes constructed according to the LD pattern showed a significant association with AIA (permutation P=0.001). The most significantly associated SNP, uS5, located in the regulatory region of the EP2 gene, was in a STATs-binding consensus sequence [AIA 31.1% versus control 22.1% (permutation P=0.0016) or versus aspirin-tolerant asthma 22.2% (permutation P=0.0017)]. Although STAT1 binding was not observed in gel mobility shift assay with HeLa nuclear extract, an unidentified protein was specifically bound to the allelic sequence. In in vitro reporter assay in HCT116 cells, the site containing the uS5 allele showed reduced transcription activity. Taken together, these results suggest that uS5 allele serves as a target of a transcription repressor protein. A functional SNP of the EP2 gene associated with risk of AIA should decrease the transcription level, resulting in reduction of the PGE2 braking mechanism of inflammation and involvement in the molecular mechanism underlying AIA.
INTRODUCTION
In a subset of asthmatic patients, aspirin and several other non-steroidal anti-inflammatory drugs (NSAIDs) that inhibit cyclooxygenase enzymes (COXs) induce severe asthmatic attack, generally termed aspirin-intolerant asthma (AIA) (1). AIA constitutes 5–15% of asthmatic patients, and is more prevalent in women. AIA is usually more severe, has a later onset than allergic asthma and frequently is associated with nasal polyp or sinusitis. Despite the well-defined pharmacological trigger, the molecular pathogenesis underlying AIA is still obscure. The cyclooxygenase theory is the widely accepted pathogenesis of AIA: a pharmacological action of NSAIDs, inhibition of COXs in the respiratory tract, alters arachidonic acid metabolism in AIA patients (2–4). Thus, aspirin and most other NSAIDs lead to a decrease in the level of PGE2, an anti-inflammatory PG generated as one of the various oxygenated metabolites in the COX pathway, which increases the number of cysteinyl leukotrienes (cys-LTs) that can mediate bronchoconstriction, mucus secretion, vascular permeability, cellular infiltration and eosinophil survival (5–7). An imbalance toward the lipoxygenase (LO) pathway is thought to play a role in accelerating the inflammation reaction in the airway tract. Although AIA is precipitated by inhibition of the COX pathway, it remains unclear why a similar adverse reaction to NSAIDs is not seen in patients with aspirin tolerant asthma (ATA) or in healthy individuals.
There is moderate genetic background in AIA; the European Network on Aspirin-Induced Asthma found that 5.1% of 365 AIA patients had family history of aspirin sensitivity (8). A polymorphism in the promoter of LTC4 synthase, A-444C single nucleotide polymorphism (SNP), has been reported to be associated with AIA in Polish patients (9,10). However, conflicting results were reported in US and Japanese populations (11,12). A recent report showed that a haplotype of the 5-LO gene was weakly associated with AIA in Korean population (13). In the present study, an extensive candidate gene analysis was applied to identify susceptibilities to AIA. On the basis of the well-defined pharmacological actions of NSAIDs, 63 candidate genes for AIA were catalogued and screened for allelic association study in a total of 198 AIA patients. The 833 SNPs of 63 candidate genes were initially identified; 370 SNPs were selected on the basis of linkage disequilibrium and allelic frequency, and evaluated for allelic association with AIA. SNPs in the prostaglandin E2 receptor subtype 2 (EP2) gene were found to be significantly associated with AIA, and the functional impact of a promoter variant was further investigated.
RESULTS
Screening of candidate genes for AIA
On the basis of the pharmacological actions of NSAIDs and the bronchial hyper-responsiveness of AIA, 63 candidate genes were selected for evaluation of association with AIA as follows: 43 genes from the arachidonic acid metabolic cascade, such as LOs, COXs, various leukotriene (LT) and PG synthases and receptors acting on the LO- and COX-pathways, and 20 genes from the immune system and other factors likely to be involved in asthma such as lymphokines, transcription factors of immune cells, matrix metalloproteinases and the platelet activating factor pathway (summarized in Table 1). The 833 SNPs of 63 genes, identified from the public database or by direct sequencing, were genotyped in 96 control subjects; among these, 370 SNPs with minor allele frequency >0.1 or location in the coding region were genotyped for the first screening (Table 1). Distributions of allele frequency of the SNPs were compared in 87 patients with AIA, 192 with ATA [96 atopic asthmatic (AT) and 96 non-atopic asthmatic (NAT) patients] and 96 with non-asthmatic controls (CTR) by a simple chi-square test. Forty-nine SNPs in 15 genes were associated with AIA in comparison with CTR in the first screening (P<0.05). Fifteen SNPs in five genes showed significant differences in allele frequencies in comparisons of both AIA with ATA and AIA with CTR (data not shown).
All the subjects including the individuals in the first screening, 198 AIA, 282 ATA and 274 CTR, were then subjected to genotyping of the 49 SNPs. After increasing the sample size, the association results of most of the SNPs tested by permutation analysis with 10 000 iterations were weakened or disappeared (data not shown), except for the SNPs of EP2 gene. SNPs uS7, S3, S4 and S5 of EP2 gene demonstrated significant associations with AIA in comparison with ATA or with CTR (Table 2). In the second screening, uS7 showed the most significant association in both comparisons of AIA with ATA (permutation P=0.0025) and AIA with CTR (permutation P=0.039).
Linkage disequilibrium mapping of EP2 gene
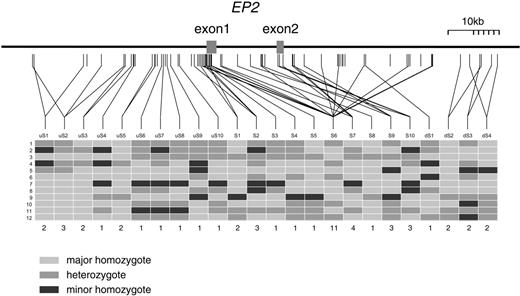
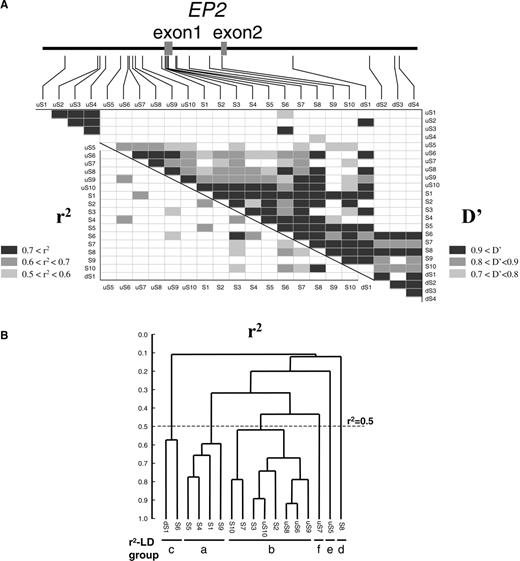
As multiple SNPs of the EP2 gene were associated with AIA, extensive screening of EP2 was undertaken. We identified 77 SNPs covering the entire gene and the regions spanning 35 kb upstream of exon 1 and 43 kb downstream of exon 2 (Fig. 1). The 77 SNPs were categorized into 24 subgroups on the basis of genotype identity after genotyping 12 individuals; SNPs expected to be in tight LD (assuming r2-LD) were grouped together (Fig. 1). Twenty-four representative SNPs of subgroups uS1–uS10, S1–S10 and dS1–dS4 were genotyped in all the patients and controls, and pair-wise linkage disequilibrium was estimated. A highly structured LD pattern, a major LD block structure (|D′|>0.7) covered by uS5–S10 and two minor LD blocks covered by uS1–uS4 and dS1–dS4, were observed (Fig. 2A). SNPs uS5–uS10, S1–S10, and dS1, associated with AIA in the second screening, were located in the major LD block. The 17 SNPs representing the major LD block then were evaluated for association with AIA by genotyping the AIA patients. SNPs in the major LD block were evaluated by r2-statistic, and weak LD was observed (Fig. 2A). In addition to SNPs uS7, S3, S4 and S5, significant associations also were observed for uS5 and uS10 using simple chi-square test and permutation test (Table 2). The minor allele frequency of uS5 was 31.1% for AIA, 22.1% for CTR and 22.2% for ATA, showing significant association (AIA versus CTR: χ2=9.67, P=0.0019, permutation P=0.0016; AIA versus ATA: χ2=9.61, P=0.0019, permutation P=0.0017) (Table 2).
UPGMA-based ‘LD tree’ and haplotype analysis
A UPGMA (unweighted pair-group method using arithmetic averages)-based ‘LD tree’ was developed as a tool for visual inspection of subgroupings of SNPs on the basis of LD structure as described in Materials and Methods (Fig. 2B), and subsequently applied to haplotype analysis. UPGMA is a method for designing a diagram to compare the sequence similarities of a gene between species that is used to evaluate evolutionary processes (14). In the LD tree, (1−r2) value was used to calculate a distance matrix for construction of the tree structure. Seventeen SNPs categorized into six subgroups (r2−a to −f) were in LD estimated by r2-statistic (r2>0.5), as shown in Figure 2B. SNPs uS5, uS7, S1, S3, S6 and S8 represented six subgroups, and were combined to construct haplotypes; the haplotype-based associations were tested with 10 000 iterated permutations. Six major haplotypes (each frequency >5%) were observed in 274 CTR (Table 3). One haplotype, m/m/m/M/M/M at uS5/uS7/S1/S3/S6/S8, showed highly significant difference between AIA and CTR (χ2=11.03, df=1, P=0.0009, permutation P=0.0012) (Table 3). The most common haplotype, M/M/M/m/M/M, and the m/m/m/M/M/M haplotype showed significant differences in frequency between AIA and ATA, further supporting the involvement of EP2 in susceptibility to AIA. The m/m/m/M/M/M haplotype was an at-risk haplotype for AIA, and uS5 a tag-SNP for the haplotype. Similar association evidence with uS5 was observed, indicating that uS5 is likely a causal SNP for AIA.
Transcriptional regulatory motif on uS5 site
The 5′-upstream region of the EP2 gene was surveyed with a TFSEARCH program (15). The sequence surrounding uS5 was predicted to a STATs-binding motif with a possibility to bind subtypes 1, 2, 3, 4 and 6 of the human STAT proteins that play key roles in cytokine signaling (16). Variation of uS5 could affect STAT protein binding affinity, resulting in altered EP2 transcription activity.
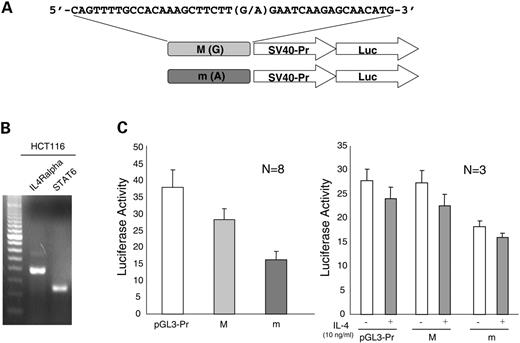
To examine the impact on transcriptional regulation due to the allelic difference of uS5, we cloned the two types of allelic sequence surrounding the uS5 site (41 bp) into upstream of an SV40 promoter-luciferase gene transcriptional unit (Fig. 3A). The effects on transcriptional activity of the 41 bp sequences centering either G allele or A allele of uS5 were investigated in HCT116 cells in which endogenous IL4R alpha and STAT6 gene expressions were confirmed with RT–PCR method (Fig. 3B). Both inserted sequences showed suppressive effects on reporter gene transcription: G allele had 74.6% and A allele had 42.7% activity of control (Fig. 3C, left). If STAT binding was responsible for repressive transcriptional activity, it should be altered by addition of IL-4. However, the reduced luciferase activities due to the inserted sequences were not affected by the addition of IL-4 (Fig. 3C, right). A allele, the susceptibility allele, showed 2.26-fold greater suppressive effect than G allele, suggesting that the uS5 variant affects the rate of transcription due to differences in nuclear factor interaction.
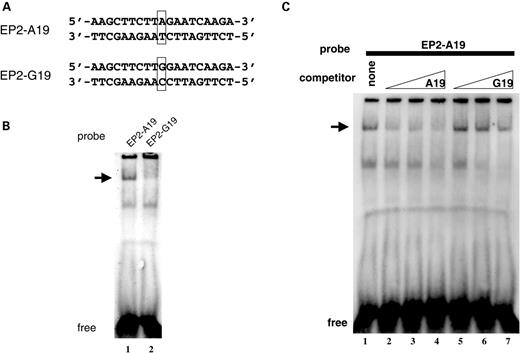
Accordingly, two digoxigenin-labeled double-stranded oligonucleotides (Fig. 4A) containing A allele (EP2-A19) and G allele (EP2-G19) were designed for electrophoretic mobility shift assay (EMSA) using HeLa nuclear extract. DNA–protein binding was observed with higher intensity in EP2-A19 than in EP2-G19 after incubation with the HeLa nuclear extracts (Fig. 4B), and the DNA–protein interaction was more efficiently competed by unlabeled EP2-A19 than by unlabeled EP2-G19 (Fig. 4C). The binding was not observed with purified STAT1 protein (data not shown), and EP2-A19 had a mismatch for STAT consensus motif, suggesting that a nuclear factor other than STAT proteins is involved in the interaction.
DISCUSSION
SNPs are being identified and assembled in large SNP databases at a rapid pace that should facilitate clarification of the genetic basis of complex diseases and drug responses. AIA is a distinct entity of asthma triggered by aspirin or NSAIDs, but little is known of its genetic basis. Identification of genetic susceptibilities to AIA might both clarify its molecular mechanism and reveal promising clinical and therapeutic targets.
Overproduction of cys-LTs in bronchial epithelium and inhibition of prostaglandin synthesis by COX inhibition apparently underlie the pathogenesis AIA (17–19). Cowburn et al. demonstrated that bronchial biopsies from AIA patients exhibited 4-fold increase in eosinophils compared with specimens from ATA patients, and baseline concentrations of cys-LTs measured in BALF of AIA patients correlate well with counts of inflammatory cells having positive immunoreactivity with LTC4 synthase in the bronchial mucosa (6). A similar observation in nasal polyp was reported demonstrating that the number of cells expressing cys-LT1 receptor and the level of cys-LTs in nasal polyp from AIA patients were significantly higher than those from ATA patients, indicating that overexpression of cys-LT1 receptor is involved in the pathogenesis of aspirin sensitivity (20,21). Considering these results together, we speculate that genetic predisposition directed to the regulation of LTC4 synthase expression may play a key role in the pathogenesis of AIA. Sanak et al. reported that a SNP of LTC4 synthase was associated with AIA, and that it functioned to reduce expression of the gene (9,10). The allelic association was not confirmed in Japanese AIA (12), and could not be replicated in the present study with much larger samples. In the current study, we applied an extensive candidate gene approach to identify susceptibility SNPs to AIA. Sixty-three candidate genes were selected on the basis of knowledge of the pharmacological action of aspirin and the plausible factors involving asthma (Table 1). SNPs in the EP2 gene were screened through an extensive two rounds of association studies of the 370 selected SNPs. EP2 is a receptor for PGE2, the well-known inhibitory mediator of inflammation released from mast cells, eosinophils and macrophages that acts as a ‘brake’ on the inflammatory process and thus has a broncho-protective role in the airways, in part by inhibiting the release of chemoattaractants such as LTB4 from alveolar macrophages and airway epithelium and the production of LTC4 in eosinophils (22). PGE2 likely modulates airway tone by inhibiting acetylcholine release of cholinergic nerve endings and mast cell histamine release. More importantly, pre-inhalation of exogenous PGE2 has shown that PGE2 directly suppresses aspirin-induced LT synthesis, most likely from eosinophils and mast cells infiltrating the bronchial mucosa (23). The ability of endogenous and exogenous PGE2 to suppress LTC4 synthesis also has been found in vitro in human eosinophils and other cells (24). Thus, PGE2 may well act to reduce LT synthesis via EP2 receptors on airway leukocytes. At least four subtypes of PGE2 receptor (EPs 1-4), which differ in tissue distribution, ligand-binding affinity and coupling to intracellular signaling pathways, have been cloned to date (25). Although a specific role in blood pressure control was demonstrated in mice lacking EP2 (26,27), involvement in the asthmatic phenotype of EP2 was not known.
The allele frequency of uS5 of the EP2 gene in AIA patients was quite different from the frequency both in controls and ATA patients [31.3% (AIA) versus 22.3% (CTR) and versus 21.7% (ATA)] (Table 2). The haplotype m/m/m/M/M/M, containing uS5, was most significantly associated with AIA (permutation P=0.0012) (Table 3). uS5 locates in the 5′-upstream of the gene on the STAT-binding motif, the allele likely having impact on transcriptional activity. The 41 bp sequence centered by either G or A allele of uS5 was subcloned into upstream of the SV40 promoter reporter vector. In vitro reporter assay demonstrated stronger transcription repression with A allele of uS5, which is associated with AIA. Despite the site being consensus for STAT binding (G allele of uS5), STAT1 protein did not bind to the sequence; instead, an unknown nuclear factor, presumably a negative regulator, bound particularly to the A allele sequence. Considered together, these data suggest that the low level of EP2 gene expression caused by the uS5 allele could lead to a low response to PGE2, skewed LT activation and bronchoconstriction in response to numerous stimuli, thereby influencing individual susceptibility to AIA. Consistent with the finding that uS5 allele leads to quantitative differences, the homozygous carriers for A allele of uS5 showed the highest odds ratio (OR=3.21, 95% C.I.=1.53–6.75). A modest but persistent failure in the PGE2 braking mechanism together with increased sensitivity to NSAIDs has been postulated to explain why AIA patients overproduce cyc-LTs even without ingestion of NSAIDs or after low doses of NSAIDs. Our hypothesis is consistent with the cyclooxygenase theory in suggesting that a persistent failure of the suppressive activity of PGE2 in AIA patients allows over-activity of the LT and other pathways, both after NSAID exposure and chronically. Thus, the impaired suppressive activity of PGE2 in at least some AIA patients may be related to reduced expression of EP2 receptors due to the polymorphic allele uS5, the transcription of which is reduced by an unidentified repressor protein.
In conclusion, genetic screening of a candidate gene for AIA suggests that variants in the promoter of the EP2 gene are significantly associated with AIA, and are functional by reducing transcriptional activity of the EP2 gene. The functional analysis constitutes only in vitro evidence, so further investigation is required. In addition, this study is preliminary in that it included only Japanese individuals.
MATERIALS AND METHODS
Subjects
Diagnosis of AIA was made on the basis of self-reported history of more than one episode of moderate to severe asthmatic reaction after aspirin or other NSAID ingestion that had been identified by a physician. The oral provocation test was not performed in most patients because of the theoretical risk and the very low likelihood of significant occult disease. (28). ATA was defined as adult asthma diagnosed by expert physicians according to the American Thoracic Society criteria (29) and no history of aspirin or NSAID-induced asthmatic attack. The controls were outpatients with diseases other than asthma and who self-reported no history of aspirin sensitivity. The 198 unrelated individuals with AIA (age: 54.7±13.2 years; 63 males/135 females),282 with ATA (age: 56.0±16.1 years; 132 males/150 females) and 274 non-asthmatic controls (CTR) (age: 50.3±24.6 years; 111 males/163 females) were recruited at Niigata University Hospital, University of Tokyo Hospital, Nagoya University Hospital, Doai Memorial Hospital and Kyushu University Hospital. ATA included 154 AT (age: 48.0±15.6 years; 80 male/74 female) subjects and 128 NAT (age: 65.9±10.0 years; 52 male/76 female) subjects who were genotyped. The patients and controls were all of Japanese ethnicity. Although the Japanese population is considered genetically homogenous, similar numbers of patients and controls from the various locations were used to avoid possible geographical differences in allelic frequencies. For the first screening, 87 randomly selected patients with AIA, 192 patients with ATA and 96 controls were genotyped. All the subjects gave written, informed consent and the study was performed with the approval of the Ethical Committee of Tokyo University. Blood samples of each subject were collected for isolation of genomic DNA.
Selection of SNPs for association study
SNPs were obtained from the two public databases; NCBI dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and IMS-JST JSNP DATABASE (http://snp.ims.u-tokyo.ac.jp/). Additional gene-based SNPs were identified by direct sequencing to cover each gene within a 3 kb SNP-interval. Ninety-six control subjects were genotyped for each SNP and SNPs with minor allele frequencies greater than 0.1 were subjected to further analysis. SNPs in the coding region that might affect gene function were given priority regardless of the allele frequencies. The 833 SNPs of 63 genes were validated and the 370 SNPs then were used for the subsequent study, according to the criteria. Direct sequencing was first performed on the EP2 gene, and 77 SNPs identified were validated; 24 SNPs then were selected and used in the association study.
SNP genotyping
SNPs were genotyped using either the pyrosequencing method on a PSQ96 Instrument (Pyrosequencing AB, Uppsala, Sweden) or direct sequencing using BigDye Terminator cycle sequencing on an ABI PRISM 3700 DNA analyzer (Applied Biosystems, Tokyo, Japan). PCR was performed with a standard protocol except a biotin-labeled primer was used when the pyrosequencing method was applied.
Statistical analysis
Differences in allelic frequencies were evaluated by case–control design with chi-square test. Haplotype frequencies for multiple loci were estimated using the expectation-maximization method with SNPAlyze v3.0 software (DYNACOM, Mobara, Japan). In addition, the permutation test was performed to test deviation of allelic frequencies of SNPs and haplotypes of the EP2 gene (30). Distribution of a test statistic was estimated by evaluating the statistics for a random sampling of 10 000 iterated permutations at fixing the total numbers of both the cases and controls, which is incorporated in SNPAlyze v3.0 software. P-value is estimated by the proportion of permutations for which the permutated data test statistic (Ppermuted) is greater than the initially observed test statistic (Pobserved), so permutation P=P (Pobserved>Ppermuted).
Pair-wise LD was estimated as D=x11−p1q1, where x11 is the frequency of haplotype A1B1, and p1 and q1 are the frequencies of alleles A1 and B1 at loci A and B, respectively. A standardized LD coefficient, r, is given by D/(p1p2q1q2)1/2, where p2 and q2 are the frequencies of the other alleles at loci A and B, respectively (31). Lewontin's coefficient, D′, is given by D′=D/Dmax, where Dmax=min(p1q2, p2q1) when D<0 or Dmax=min(p1q1, p2q2) when D>0 (32).
UPGMA-based LD tree
LD according to r2-statistics was visualized by an ‘LD tree’ constructed on the basis of the UPGMA method of Neighbor program from the PHYLIP package v3.57c, available at web site (http://evolution.gs.washington.edu/phylip.html). (1−r2) was calculated and converted to a distance matrix. Calculated coefficients were within 0–1, and the smaller values represent high LD against uncalculated coefficients.
Transcription regulatory motif
The computer program TFSEARCH based on TRANSFAC databases, available at web site (http://www.cbrc.jp/research/db/TFSEARCH.html), was used to predict potential binding sites of transcription factors in the regulatory region.
Transfection and reporter assays
The complementary oligonucleotide spanning the uS5 promoter allelic sequence, 5′-CCAGTTTTGCCACAAAGCTTCT (G/A)GAATCAAGAGCAACATGC-3′ or 5′-TCGAGCATG TTGCTCTTGATTC(C/T)AAGAAGCTTTGTGGCAAAACT GGGTAC-3′, was annealed and ligated into the KpnI/XhoI-digested pGL3-promoter vector (Promega, Tokyo, Japan), and sequenced. HCT116 cells were cultured in McCoy's 5A medium supplemented with antibiotics and 10% fetal bovine serum. HCT116 cells were transfected with FuGENE6 (Roche Diagnostics, Tokyo, Japan) according to the manufacturer's instructions. Briefly, 200 ng of firefly luciferase reporter plasmid, and 0.6 ng of Renilla luciferase reporter plasmid (pRL-TK, Promega) per 24-well dish were used for each transfection. The cells were harvested 48 h after the transfection, and luciferase assay using the Dual-Luciferase Reporter Assay System was performed in accordance with the manufacturer's protocol (Promega). Experiments were performed at least twice in triplicate, and the relative activities of luciferase were expressed as mean ± S.E, after normalizing with the Renilla luciferase activities.
The total RNA was extracted using Trizol reagent (Invitrogen, Tokyo, Japan) and RT–PCR was performed with the SuperScript One-Step RT–PCR system (Invitrogen) based on the manufacturer's protocol.
Electrophoretic mobility shift assay
EMSA was performed with a DIG Gel Shift Kit (Roche Diagnostics) using digoxigenin (DIG)-labeled double-stranded 19 mer oligonucleotides specific to A allele (EP2-A19) and G allele (EP2-G19) of uS5. DIG-labeled probe was incubated with HeLa nuclear extracts (Promega) for 30 min at 4°C and separated by electrophoresis on a 5% non-denaturing polyacrylamide gel with 0.5× TBE running buffer. DNA–protein complexes were electroblotted onto nylon membrane and the band shift was visualized according to the user's manual for DIG Gel Shift Kit. For the competition assay, we incubated HeLa nuclear extracts with non-labeled competitors for 15 min at 4°C before incubation with labeled EP2-A19.
ACKNOWLEDGEMENTS
We thank DNA donors and supporting medical staff for making this study possible. We are grateful to K. Eguchi, E. Nakamura, Y. Miwa, Y. Sakamoto and Y. Terada for their technical assistance. The critical reading of the manuscript by Dr A. Tajima is gratefully acknowledged. This work was supported by a Research for the Future Program Grant of The Japan Society for the Promotion of Science (II).
Figure 1. SNP location in EP2 gene and subgrouping for screening. Locations of 77 SNPs in the genomic region from 35 kb upstream of exon 1 to 43 kb downstream of exon 2 are depicted. The 77 SNPs were categorized into 12 groups on the basis of the rule of perfect matching of genotypes with 12 individuals (SNP-group is connected by line). SNPs <5% of minor allele frequency were not subgrouped. In the block table, light gray box shows homozygote for major allele, mid-gray box heterozygote and dark gray box homozygote for minor allele. The numbers of SNPs belonging to each group are shown under the block tables.
Figure 2. LD pattern of EP2 gene. Pair-wise LD coefficients, D′ and r2, were determined and expressed as block structure (A) and UPGMA-based tree structure (B). (A) In the schematic block, shaded boxes of dark gray show pair-wise LD of D′>0.9, medium gray 0.8<D′<0.9 and light gray 0.7<D′<0.8. Blank boxes represent D′<0.7. For all the pairs of SNPs in the major D′-LD block (uS5–dS1), pair-wise LD coefficients r2 are also presented. (B) The major LD block (uS5–dS1) was further analyzed with LD tree constructed according to UPGMA method as described in Materials and Methods. For subgrouping of SNPs based on LD structure, r2=0.5 was utilized for cut-off.
Figure 3. uS5 Allele-dependent transcription activity in HCT116 cells. (A) The 41 bp sequences centering on uS5 SNP were subcloned into the reporter vector as described in Materials and Methods. (B) HCT116 cells that endogenously expressed IL4Rα and STAT6 genes were used for the experiments. (C) Each reporter vector was transfected into HCT116 cells, and the firefly luciferase activity was normalized with the Renilla luciferase activity of co-transfected pRL-TK (left). IL-4 stimulation (10 ng/ml) on the effect of transcription was monitored (right).
Figure 4. EMSA with ologonucleotide containing A allele or G allele of uS5. (A) Double-stranded oligonucleotide probes (EP2-A19 and EP2-G19) labeled with digoxigenin for EMSA are shown. (B) Nuclear extract from HeLa cells was incubated with EP2-A19 or EP2-G19 probes. Arrowhead points to specific binding. (C) Specific interaction with EP2-A19 competed with various amounts of non-labeled EP2-A19 or EP2-G19 competitor (6.25-, 12.5- and 25-fold from left to right).
SNPs applied to the first screening
| No. . | Gene . | SNP . | Sequence position . | Variation . | Localization . | rs ID . | Minor allele frequncies (96 CTR) . |
|---|---|---|---|---|---|---|---|
| 1 | ADAM33 | ADAM33S1 | 11 513 | C/A | Intron19 | rs44707 | 0.467 |
| 2 | ADAM33S2 | 12 505 | T/C | Exon20-non-synonymous (M764T) | rs2280091 | 0.098 | |
| 3 | ADAM33S3 | 12 534 | T/C | Exon20-non-synonymous (M774S) | rs2280090 | 0.102 | |
| 4 | ADAM33S4 | 12 612 | T/C | Intron20 | rs2280089 | 0.092 | |
| 5 | ADAM33S5 | 13 018 | G/A | Intron21 | rs628977 | 0.362 | |
| 6 | ADAM33S6 | 13 026 | T/C | Intron21 | rs628965 | 0.362 | |
| 7 | ADAM33S7 | 13 060 | A/C | Intron21 | rs543749 | 0.181 | |
| 8 | ALOX12B | ALOX12BS1 | 1963 | G/A | Intron2 | rs3027303 | 0.189 |
| 9 | ALOX12BS4 | 3276 | G/T | Intron2 | rs3027294 | 0.391 | |
| 10 | ALOX12BS5 | 7053 | G/C | Intron4 | rs2304908 | 0.392 | |
| 11 | ALOX12BS6 | 7223 | C/T | Intron4 | rs2304907 | 0.396 | |
| 12 | ALOX12BS7 | 7341 | C/T | Intron4 | rs2304906 | 0.130 | |
| 13 | ALOX5 | ALOX5S1 | 9223 | A/G | Intron2 | rs4769060 | 0.151 |
| 14 | ALOX5S2 | 26 904 | G/A | Intron3 | New | 0.401 | |
| 15 | ALOX5S3 | 50 760 | G/A | Intron4 | New | 0.198 | |
| 16 | ALOX5S4 | 59 443 | A/G | Intron6 | New | 0.193 | |
| 17 | ALOX5S5 | 69 751 | G/C | Intron7 | New | 0.354 | |
| 18 | CLP | CLPS1 | 1152 | G/A | Intron2 | rs2966305 | 0.401 |
| 19 | CLPS2 | 3607 | C/A | Intron2 | New | 0.032 | |
| 20 | CLPS3 | 3688 | A/G | Intron2 | rs2967871 | 0.315 | |
| 21 | CLPS4 | 9097 | C/T | Intron2 | rs2925050 | 0.495 | |
| 22 | CLPS5 | 9197 | T/G | Intron2 | New | 0.457 | |
| 23 | CLPS6 | 11 744 | T/C | Intron2 | rs1835156 | 0.333 | |
| 24 | CLPS7 | 17 612 | T/C | Intron2 | New | 0.229 | |
| 25 | CLPS8 | 17 865 | G/C | Intron2 | New | 0.356 | |
| 26 | CLPS9 | 17 923 | G/C | Intron2 | rs2967876 | 0.284 | |
| 27 | CLPS10 | 20 308 | C/T | Intron2 | rs2925064 | 0.263 | |
| 28 | CLPS11 | 20 380 | T/C | Intron2 | rs934166 | 0.468 | |
| 29 | CLPS12 | 20 412 | G/A | Intron2 | rs201854 | 0.447 | |
| 30 | CLPS13 | 23 054 | T/C | Intron2 | New | 0.104 | |
| 31 | CLPS17 | 27 698 | A/G | Intron2 | rs2288584 | 0.250 | |
| 32 | CLPS18 | 31 174 | T/C | Intron3 | rs2967855 | 0.283 | |
| 33 | CLPS19 | 31 262 | C/G | Intron3 | New | 0.214 | |
| 34 | CLPS20 | 31 615 | C/T | Intron3 | New | 0.208 | |
| 35 | CLPS21 | 43 186 | A/T | Intron3 | rs2914823 | 0.185 | |
| 36 | CLPS22 | 43 360 | T/G | Intron3 | New | 0.404 | |
| 37 | CLPS23 | 43 394 | A/G | Intron3 | New | 0.250 | |
| 38 | CLPS24 | 47 961 | G/A | Intron3 | New | 0.116 | |
| 39 | CLPS25 | 48 577 | C/T | Intron3 | New | 0.347 | |
| 40 | CLPS26 | 51794 | C/T | Exon4-UTR | rs247862 | 0.297 | |
| 41 | CNOT3 | CNOT3S1 | 328 | C/T | Intron1 | rs42318 | 0.115 |
| 42 | CTSG | CTSGS1 | 308 | G/A | Intron1 | rs2236742 | 0.212 |
| 43 | CTSGS2 | 527 | A/G | Intron1 | New | 0.034 | |
| 44 | CTSGS3 | 726 | A/G | Intron1 | rs1957523 | 0.477 | |
| 45 | CTSGS4 | 1226 | C/T | Intron2 | New | 0.034 | |
| 46 | CTSGS5 | 1349 | G/A | Intron2 | rs2070697 | 0.314 | |
| 47 | CTSGS6 | 1767 | A/G | Exon4-synonymous | New | 0.182 | |
| 48 | CYP4F2 | CYP4F2S1 | 4937 | A/T | Intron1 | New | 0.349 |
| 49 | CYP4F2S4 | 18 733 | T/C | Intron1 | rs2072269 | 0.335 | |
| 50 | CYP4F2S5 | 18 810 | G/C | Intron1 | rs1064796 | 0.495 | |
| 51 | CYP4F2S6 | 30 715 | G/A | Intron1 | rs2018460 | 0.170 | |
| 52 | CYP4F3 | CYP4F3S0 | 3187 | G/A | Intron2 | rs2203998 | 0.328 |
| 53 | CYP4F3S0.5 | 6221 | C/T | Intron4 | rs1290626 | 0.141 | |
| 54 | CYP4F3S1 | 8507 | C/T | Intron6 | rs2283612 | 0.189 | |
| 55 | CYP4F3S2 | 11 871 | A/C | Intron8 | rs2733750 | 0.183 | |
| 56 | CYP4F3S3 | 12 015 | A/G | Exon9-synonymous | New | 0.363 | |
| 57 | CYP4F3S4 | 12 808 | G/A | Intron9 | rs2733752 | 0.136 | |
| 58 | CYP4F3S5 | 12 877 | T/A | Intron9 | New | 0.140 | |
| 59 | CYP4F3S6 | 17 772 | T/C | Intron11 | New | 0.453 | |
| 60 | CYP4F8 | CYP4F8S1 | 3610 | C/T | Intron2 | rs2072599 | 0.168 |
| 61 | CYP4F8S2 | 6771 | G/T | Intron5 | rs2072601 | 0.276 | |
| 62 | CYP4F8S3 | 13 568 | C/T | Intron11 | rs2239366 | 0.276 | |
| 63 | CYSLT1R | CYSLT1RS1 | 927 | C/T | Exon1-synonymous | rs320995 | 0.474 |
| 64 | CYSLT2R | CYSLT2RS1 | 2797 | A/G | Exon1-UTR | rs912277 | 0.417 |
| 65 | CYSLT2RS2 | 3078 | A/C | Exon1-UTR | rs1323552 | 0.489 | |
| 66 | CYSLT2RS3 | 3105 | A/G | Exon1-UTR | New | 0.116 | |
| 67 | FLAP | FLAPS1 | 162 | C/A | Intron1 | rs4769055 | 0.522 |
| 68 | FLAPS2 | 838 | T/G | Intron1 | rs9579645 | 0.096 | |
| 69 | FLAPS3 | 7272 | A/G | Intron1 | rs9551960 | 0.339 | |
| 70 | FLAPS4 | 8640 | A/C | Intron2 | rs3803277 | 0.406 | |
| 71 | FLAPS5 | 8733 | T/C | Intron2 | rs3803278 | 0.370 | |
| 72 | FLAPS6 | 13 674 | G/A | Intron2 | rs4075692 | 0.286 | |
| 73 | FLAPS7 | 20 616 | G/C | Intron3 | New | 0.286 | |
| 74 | FLAPS8 | 20 648 | C/T | Intron3 | rs4468448 | 0.240 | |
| 75 | FLAPS9 | 23 849 | T/A | Intron3 | rs9551964 | 0.277 | |
| 76 | FLAPS9.5 | 24 348 | G/A | Intron3 | New | 0.006 | |
| 77 | FLAPS10 | 28 209 | A/G | Intron3 | rs4769060 | 0.401 | |
| 78 | HPGD | HPGDS1 | 464 | G/A | Exon2-synonymous | rs1050145 | 0.220 |
| 79 | HPGDS2 | 798 | A/G | Intron2 | rs1365613 | 0.447 | |
| 80 | HPGDS3 | 13 332 | A/C | Intron3 | rs2555629 | 0.405 | |
| 81 | HPGDS4 | 13 430 | A/T | Intron3 | New | 0.253 | |
| 82 | HPGDS5 | 20 442 | A/G | Intron4 | New | 0.446 | |
| 83 | HPGDS6 | 20 516 | G/A | Intron4 | New | 0.110 | |
| 84 | IGF1 | IGF1S1 | 3083 | A/G | Intron2 | rs2162679 | 0.368 |
| 85 | IGF1S2 | 9917 | G/T | Intron3 | rs1019731 | 0.021 | |
| 86 | IGF1S3 | 17 640 | G/C | Intron3 | rs2195239 | 0.426 | |
| 87 | IGF1S4 | 17 695 | T/C | Intron3 | rs2195240 | 0.443 | |
| 88 | IGF1S5 | 49 421 | G/A | Intron3 | rs972936 | 0.484 | |
| 89 | IGF1S6 | 60 710 | G/A | Intron3 | rs2072592 | 0.245 | |
| 90 | IGF1S7 | 72 103 | G/A | Intron5 | rs978458 | 0.135 | |
| 91 | IGF1S8 | 84 150 | G/A | Exon6-UTR | rs6219 | 0.271 | |
| 92 | IL13 | IL13S1 | −978 | C/T | Promoter | rs11575055 | 0.179 |
| 93 | IL13S2 | 571 | C/A | Intron1 | rs2066960 | 0.281 | |
| 94 | IL13S3 | 598 | G/C | Intron1 | rs1295987 | 0.120 | |
| 95 | IL13S4 | 805 | C/T | Intron1 | rs2069744 | 0.120 | |
| 96 | IL13S5 | 2100 | G/A | Exon4-non-synonymous (R144Q) | rs20541 | 0.333 | |
| 97 | IL4 | IL4S1 | −219 | T/C | Promoter | rs2243250 | 0.328 |
| 98 | IL4S2 | 3353 | A/C | Intron2 | rs2227284 | 0.255 | |
| 99 | IL4S3 | 3927 | C/G | Intron2 | rs2243263 | 0.073 | |
| 100 | LTA4H | LTA4HS1 | 9153 | A/G | Intron3 | rs763842 | 0.226 |
| 101 | LTA4HS4 | 20 165 | T/C | Intron11 | rs1978331 | 0.335 | |
| 102 | LTB4DH | LTB4DHS1 | 4435 | A/T | Intron2 | New | 0.506 |
| 103 | LTB4DHS2 | 8896 | A/G | Intron4 | New | 0.283 | |
| 104 | LTB4DHS2.5 | 9182 | A/G | Intron4 | New | 0.092 | |
| 105 | LTB4DHS5 | 9502 | C/T | Intron4 | rs1053968 | 0.005 | |
| 106 | LTB4DHS6 | 13 234 | C/G | Intron4 | New | 0.319 | |
| 107 | LTB4DHS6.5 | 13 249 | A/G | Intron4 | New | 0.231 | |
| 108 | LTB4DHS7 | 20 718 | G/A | Intron6 | New | 0.253 | |
| 109 | LTB4DHS8 | 25 971 | A/G | Intron8 | rs1322258 | 0.347 | |
| 110 | LTB4DHS9 | 26 221 | A/G | Intron8 | New | 0.406 | |
| 111 | LTB4DHS10 | 35 589 | A/C | Intron9 | rs2146078 | 0.229 | |
| 112 | LTB4DHS11 | 35 819 | G/A | Intron9 | New | 0.354 | |
| 113 | LTB4R | LTB4RS1 | 1165 | G/C | Exon1-UTR | New | 0.140 |
| 114 | LTC4S | LTC4SS1.5 | −348 | A/C | Promoter | New | 0.208 |
| 115 | LTC4SS2 | 289 | G/C | Intron1 | New | 0.026 | |
| 116 | LTC4SS3 | 1296 | C/T | Intron1 | New | 0.208 | |
| 117 | LTC4SS4 | 2659 | A/G | Intron5 | New | 0.201 | |
| 118 | MGST2 | MGST2S1 | 815 | A/G | Intron1 | rs1000222 | 0.292 |
| 119 | MGST2S2 | 12 971 | C/T | Intron2 | rs795589 | 0.198 | |
| 120 | MGST2S3 | 12 977 | A/C | Intron2 | rs795588 | 0.077 | |
| 121 | MMP1 | MMP1S1 | 193 | G/A | Intron1 | rs470358 | 0.292 |
| 122 | MMP1S2 | 2579 | G/A | Exon5-synonymous | rs470558 | 0.130 | |
| 123 | MMP1S3 | 7300 | T/C | Intron8 | rs470747 | 0.137 | |
| 124 | MMP1S4 | 7815 | C/T | Exon10-UTR | rs2239008 | 0.319 | |
| 125 | MMP1S5 | 7936 | T/C | Exon10-UTR | rs2071230 | 0.298 | |
| 126 | MMP2 | MMP2S1 | 3606 | A/T | Intron1 | rs857403 | 0.214 |
| 127 | MMP2S2 | 3665 | G/A | Intron1 | rs1030868 | 0.229 | |
| 128 | MMP2S3 | 6505 | C/T | Exon5-synonymous | rs1053605 | 0.208 | |
| 129 | MMP2S4 | 6730 | T/G | Intron5 | New | 0.064 | |
| 130 | MMP2S5 | 6958 | A/G | Intron5 | rs866770 | 0.151 | |
| 131 | MMP2S6 | 10 603 | C/T | Exon7-synonymous | rs243849 | 0.151 | |
| 132 | MMP2S7 | 10 896 | T/C | Intron7 | rs243847 | 0.443 | |
| 133 | MMP2S8 | 14 011 | G/A | Exon9-synonymous | rs2287074 | 0.326 | |
| 134 | MMP2S9 | 14 196 | G/A | Intron9 | rs243843 | 0.306 | |
| 135 | MMP2S10 | 14 281 | G/A | Intron9 | New | 0.065 | |
| 136 | MMP2S11 | 14 320 | T/C | Intron9 | rs243842 | 0.364 | |
| 137 | MMP2S12 | 17 660 | G/T | Intron9 | rs171498 | 0.363 | |
| 138 | MMP2S13 | 17 670 | G/T | Intron9 | rs243838 | 0.153 | |
| 139 | MMP2S14 | 17 928 | C/T | Intron10 | New | 0.057 | |
| 140 | MMP2S15 | 20 976 | G/A | Intron11 | New | 0.115 | |
| 141 | MMP2S16 | 21 134 | G/A | Intron11 | rs243836 | 0.386 | |
| 142 | MMP2S17 | 26 345 | C/T | Exon13-UTR | New | 0.021 | |
| 143 | MMP2S18 | 26 512 | A/C | Exon13-UTR | rs7201 | 0.234 | |
| 144 | MMP8 | MMP8S1 | 524 | G/A | Intron1 | rs1939012 | 0.359 |
| 145 | MMP8S2 | 5458 | A/G | Intron4 | rs1940051 | 0.266 | |
| 146 | MMP8S3 | 11 357 | T/A | Intron9 | New | 0.287 | |
| 147 | MMP8S4 | 11 473 | T/C | Exon10-synonymous | New | 0.016 | |
| 148 | MMP9 | MMP9S1 | 2679 | G/A | Exon6-non-synonymous (R279Q) | rs2664538 | 0.382 |
| 149 | MMP9S2 | 3029 | A/C | Intron6 | rs2236416 | 0.179 | |
| 150 | MMP9S3 | 5565 | G/A | Exon12-non-synonymous (R668Q) | rs2274756 | 0.214 | |
| 151 | MMP9S4 | 7419 | G/A | Exon13-synonymous | rs13925 | 0.292 | |
| 152 | P2Y10 | P2Y10S1 | 1192 | A/G | Intron1 | New | 0.016 |
| 153 | P2Y10S2 | 5030 | C/T | Intron1 | rs2858570 | 0.110 | |
| 154 | P2Y10S3 | 5318 | A/T | Intron1 | rs2251477 | 0.184 | |
| 155 | P2Y10S4 | 9417 | T/C | Intron1 | rs2742205 | 0.115 | |
| 156 | PAFAH1B1 | PAFAH1B1S2 | 12 145 | G/T | Intron1 | New | 0.099 |
| 157 | PAFAH1B1S3 | 21 146 | C/G | Intron1 | New | 0.080 | |
| 158 | PAFAH1B1S4 | 37 780 | A/G | Intron1 | rs1266474 | 0.084 | |
| 159 | PAFAH1B1S5 | 38 033 | C/T | Intron1 | New | 0.047 | |
| 160 | PAFAH1B1S6 | 40 522 | G/A | Intron1 | New | 0.130 | |
| 161 | PAFAH1B1S7 | 60 034 | C/T | Intron2 | New | 0.266 | |
| 162 | PAFAH1B1S8 | 68 545 | G/C | Intron2 | New | 0.078 | |
| 163 | PAFAH1B1S9 | 76 403 | T/G | Intron5 | New | 0.032 | |
| 164 | PAFAH1B2 | PAFAH1B2S1 | 3971 | T/C | Intron1 | rs2008908 | 0.446 |
| 165 | PAFAH1B3 | PAFAH1B3S1 | 2538 | C/T | Exon4-synonymous | New | 0.016 |
| 166 | PAFAH2 | PAFAH2S1 | 1279 | G/T | Intron1 | rs3008423 | 0.172 |
| 167 | PAFAH2S2 | 4171 | C/T | Intron1 | New | 0.036 | |
| 168 | PAFAH2S3 | 15456 | G/T | Intron5 | rs1469512 | 0.214 | |
| 169 | PAI1m2 | PAI1m2S1 | 4484 | G/A | Intron1 | rs840088 | 0.401 |
| 170 | PAI1m2S2 | 5094 | A/G | Intron1 | New | 0.089 | |
| 171 | PAI1m2S3 | 19 022 | C/A | Intron1 | rs2099601 | 0.201 | |
| 172 | PAI1m2S4 | 19 047 | A/C | Intron1 | New | 0.201 | |
| 173 | PAI1m2S5 | 19 120 | T/C | Intron1 | rs2083120 | 0.201 | |
| 174 | PAI1m2S6 | 19 348 | A/C | Intron1 | New | 0.393 | |
| 175 | PAI1m2S7 | 62 178 | A/G | Intron8 | New | 0.271 | |
| 176 | PDGFB | PDGFBS1 | 581 | G/A | Intron1 | rs758588 | 0.184 |
| 177 | PDGFBS3 | 20 237 | C/T | Intron3 | rs740750 | 0.310 | |
| 178 | PDGFBS4 | 26 041 | C/T | Intron10 | rs1864972 | 0.198 | |
| 179 | PDGFBS5 | 35 170 | G/A | Intron18 | rs1432878 | 0.108 | |
| 180 | PDGFRL | PDGFRLS1 | 978 | G/C | Intron1 | rs2720576 | 0.365 |
| 181 | PDGFRLS2 | 1187 | C/T | Intron1 | rs2588164 | 0.245 | |
| 182 | PDGFRLS3 | 1416 | T/C | Intron1 | rs2588163 | 0.391 | |
| 183 | PDGFRLS4 | 2963 | G/C | Intron1 | rs2517267 | 0.458 | |
| 184 | PDGFRLS5 | 3794 | C/G | Intron1 | rs2517268 | 0.394 | |
| 185 | PDGFRLS6 | 3882 | G/A | Intron1 | New | 0.216 | |
| 186 | PDGFRLS7 | 3999 | A/G | Intron1 | rs2517269 | 0.220 | |
| 187 | PDGFRLS8 | 8893 | G/A | Intron1 | New | 0.226 | |
| 188 | PDGFRLS9 | 9922 | G/A | Intron1 | New | 0.232 | |
| 189 | PDGFRLS10 | 14 053 | G/C | Intron2 | rs2720583 | 0.167 | |
| 190 | PDGFRLS11 | 14 194 | A/C | Intron2 | New | 0.396 | |
| 191 | PDGFRLS12 | 18 220 | T/C | Intron2 | rs2427709 | 0.140 | |
| 192 | PDGFRLS13 | 18 473 | G/A | Intron2 | rs2588144 | 0.068 | |
| 193 | PDGFRLS15 | 19 592 | T/G | Intron2 | rs2246488 | 0.339 | |
| 194 | PDGFRLS16 | 28 052 | G/A | Intron2 | rs2517187 | 0.104 | |
| 195 | PDGFRLS17 | 28 449 | C/T | Intron2 | rs2237823 | 0.104 | |
| 196 | PDGFRLS18 | 31 111 | T/C | Intron2 | rs2517198 | 0.094 | |
| 197 | PDGFRLS19 | 31 230 | A/T | Intron2 | New | 0.260 | |
| 198 | PDGFRLS20 | 31 258 | A/G | Intron2 | New | 0.031 | |
| 199 | PDGFRLS21 | 31 499 | T/C | Intron2 | rs2427715 | 0.094 | |
| 200 | PDGFRLS22 | 31 600 | T/C | Intron2 | New | 0.167 | |
| 201 | PDGFRLS23 | 33 975 | G/C | Intron2 | rs2517208 | 0.037 | |
| 202 | PDGFRLS24 | 41 991 | A/G | Intron2 | rs2237831 | 0.189 | |
| 203 | PDGFRLS25 | 42 131 | G/A | Intron2 | rs2237831 | 0.205 | |
| 204 | PDGFRLS26 | 43 810 | G/T | Intron2 | New | 0.073 | |
| 205 | PDGFRLS27 | 46 759 | T/C | Intron3 | New | 0.120 | |
| 206 | PDGFRLS28 | 46 888 | G/A | Intron3 | rs2237835 | 0.226 | |
| 207 | PDGFRLS29 | 46 936 | C/G | Intron3 | New | 0.074 | |
| 208 | PDGFRLS30 | 47 097 | C/G | Intron3 | New | 0.197 | |
| 209 | PDGFRLS31 | 49 411 | A/C | Intron3 | rs2237836 | 0.548 | |
| 210 | PDGFRLS32 | 49 937 | G/T | Intron3 | rs2237837 | 0.103 | |
| 211 | PDGFRLS33 | 53 887 | T/C | Intron4 | rs2237842 | 0.188 | |
| 212 | PDGFRLS34 | 56 021 | C/T | Intron4 | rs2427719 | 0.063 | |
| 213 | PDGFRLS35 | 56 410 | T/C | Intron4 | rs2237845 | 0.240 | |
| 214 | PDGFRLS36 | 65 408 | C/T | Intron5 | New | 0.083 | |
| 215 | PDGFRLS37 | 65 512 | T/C | Exon6-synonymous | rs4705 | 0.521 | |
| 216 | PGDS | PGDSS1 | 5446 | T/C | Intron1 | rs2129595 | 0.191 |
| 217 | PGIS | PGISS1 | 4650 | T/C | Intron1 | rs477627 | 0.063 |
| 218 | PGISS2 | 6734 | A/C | Intron1 | rs927068 | 0.226 | |
| 219 | PGISS3 | 6840 | T/C | Intron1 | New | 0.226 | |
| 220 | PGISS4 | 6870 | G/C | Intron1 | rs498646 | 0.276 | |
| 221 | PGISS4.5 | 6924 | T/C | Intron1 | rs476496 | 0.280 | |
| 222 | PGISS6 | 28 941 | A/G | Intron5 | rs501908 | 0.033 | |
| 223 | PGISS7 | 41 990 | A/G | Intron5 | New | 0.093 | |
| 224 | PGISS8 | 44 026 | G/A | Exon6-synonymous | rs5628 | 0.036 | |
| 225 | PGISS9 | 50 926 | C/T | Intron6 | New | 0.104 | |
| 226 | PGISS10 | 55 002 | C/A | Intron8 | rs5629 | 0.281 | |
| 227 | PGISS11 | 57 532 | A/G | Intron9 | rs729824 | 0.226 | |
| 228 | PGISS12 | 62 730 | T/C | Exon10-UTR | rs5602 | 0.438 | |
| 229 | PLA2G7 | PLA2G7S1 | 2007 | G/T | Intron1 | New | 0.214 |
| 230 | PLA2G7S2 | 2338 | G/A | Intron1 | rs1421369 | 0.468 | |
| 231 | PLA2G7S3 | 6203 | G/A | Intron1 | New | 0.141 | |
| 232 | PLA2G7S4 | 20 965 | G/T | Intron5 | rs1362931 | 0.042 | |
| 233 | PLA2G7S5 | 23 741 | T/C | Exon7-non-synonymous (I198T) | rs1805018 | 0.281 | |
| 234 | PLA2G7S6 | 27 025 | C/G | Intron9 | rs2216465 | 0.542 | |
| 235 | PLA2G7S7 | 30 101 | C/T | Exon11-non-synonymous (A379V) | rs1051931 | 0.042 | |
| 236 | PTAFR | PTAFRS3 | 36 031 | C/A | Exon2-non-synonymous (A224D) | rs5938 | 0.068 |
| 237 | PTGDR | PTGDRS1 | 408 | G/A | Intron1 | New | 0.021 |
| 238 | PTGDRS2 | 3290 | A/G | Intron1 | rs1254609 | 0.194 | |
| 239 | PTGDRS3 | 5754 | T/C | Intron1 | New | 0.075 | |
| 240 | PTGDRS4 | 5793 | A/G | Intron1 | rs708486 | 0.081 | |
| 241 | PTGDS | PTGDSS1 | 4186 | C/A | Exon7-UTR | rs6926 | 0.198 |
| 242 | EP1 | PTGER1S1 | −267 | G/C | Promoter | New | 0.109 |
| 243 | EP2 | PTGER2uS1 | −26 643 | T/C | 5′-Upstream | rs988209 | 0.232 |
| 244 | PTGER2uS2 | −18 461 | T/G | 5′-Upstream | rs1390375 | 0.077 | |
| 245 | PTGER2uS3 | −18 360 | G/A | 5′-Upstream | rs1390374 | 0.022 | |
| 246 | PTGER2uS4 | −15 332 | A/T | 5′-Upstream | rs708490 | 0.444 | |
| 247 | PTGER2uS5 | −12 813 | G/A | 5′-Upstream | New | 0.223 | |
| 248 | PTGER2uS6 | −10 918 | A/G | 5′-Upstream | New | 0.449 | |
| 249 | PTGER2uS7 | −10 814 | T/A | 5′-upstream | New | 0.427 | |
| 250 | PTGER2uS8 | −10 250 | A/G | 5′-upstream | rs714366 | 0.433 | |
| 251 | PTGER2uS9 | −7075 | A/G | 5′-upstream | New | 0.452 | |
| 252 | PTGER2uS10 | −6179 | A/G | 5′-upstream | New | 0.479 | |
| 253 | PTGER2S1 | −609 | G/A | Promoter | rs1254601 | 0.288 | |
| 254 | PTGER2S2 | 300 | G/A | Exon1-UTR | rs1254600 | 0.462 | |
| 255 | PTGER2S3 | 498 | C/G | Exon1-UTR | rs2075797 | 0.441 | |
| 256 | PTGER2S4 | 948 | A/G | Exon1-UTR | rs1353411 | 0.417 | |
| 257 | PTGER2S5 | 1042 | G/A | Exon1-UTR | rs1254598 | 0.350 | |
| 258 | PTGER2S6 | 2803 | G/A | Intron1 | New | 0.164 | |
| 259 | PTGER2S7 | 2988 | C/T | Intron1 | New | 0.339 | |
| 260 | PTGER2S8 | 6063 | C/T | Intron1 | New | 0.116 | |
| 261 | PTGER2S9 | 10 927 | T/G | Intron1 | rs1254585 | 0.446 | |
| 262 | PTGER2S10 | 14 081 | C/T | Exon2-UTR | rs708502 | 0.398 | |
| 263 | PTGER2dS1 | 29 784 | C/T | 3′-Downstream | rs708511 | 0.219 | |
| 264 | PTGER2dS2 | 47 461 | A/G | 3′-Downstream | New | 0.226 | |
| 265 | PTGER2dS3 | 57 931 | C/T | 3′-Downstream | rs708531 | 0.750 | |
| 266 | PTGER2dS4 | 58 051 | C/T | 3′-Downstream | rs708532 | 0.329 | |
| 267 | EP3 | PTGER3S1 | 27 837 | G/A | Intron1 | rs1008484 | 0.058 |
| 268 | PTGER3S2 | 28 078 | G/A | Intron1 | rs1569593 | 0.258 | |
| 269 | PTGER3S5 | 36 177 | A/T | Intron2 | rs5680 | 0.313 | |
| 270 | PTGER3S7 | 44 999 | T/G | Intron2 | rs1983588 | 0.268 | |
| 271 | PTGER3S8 | 45 040 | A/C | Intron2 | rs1983587 | 0.059 | |
| 272 | PTGER3S9 | 54 444 | G/C | Intron2 | rs1883461 | 0.054 | |
| 273 | PTGER3S10 | 54 634 | T/A | Intron2 | rs1883460 | 0.094 | |
| 274 | PTGER3S11 | 58 525 | C/T | Intron2 | New | 0.271 | |
| 275 | PTGER3S12 | 65 699 | G/C | Intron2 | rs647921 | 0.048 | |
| 276 | PTGER3S13 | 65 973 | A/T | Intron2 | rs646621 | 0.443 | |
| 277 | PTGER3S14 | 70 342 | G/A | Intron2 | rs909842 | 0.442 | |
| 278 | PTGER3S15 | 70 357 | A/C | Intron2 | New | 0.300 | |
| 279 | PTGER3S17 | 70 409 | G/A | Intron2 | New | 0.442 | |
| 280 | PTGER3S20 | 70 755 | A/T | Intron2 | rs484675 | 0.441 | |
| 281 | PTGER3S24 | 86 923 | A/G | Intron2 | rs573688 | 0.082 | |
| 282 | PTGER3S25 | 95 146 | A/C | Intron3 | New | 0.037 | |
| 283 | PTGER3S27 | 1 00 052 | T/C | Intron3 | rs1409984 | 0.255 | |
| 284 | PTGER3S29 | 1 14 078 | G/A | Intron3 | rs625617 | 0.371 | |
| 285 | PTGER3S30 | 1 21 798 | C/A | Intron3 | New | 0.328 | |
| 286 | PTGER3S31 | 1 21 846 | G/A | Intron3 | rs602383 | 0.297 | |
| 287 | PTGER3S32 | 1 49 402 | C/T | Intron3 | rs1409165 | 0.370 | |
| 288 | PTGER3S33 | 1 49 626 | G/A | Intron3 | rs1409166 | 0.398 | |
| 289 | PTGER3S35 | 1 60 372 | A/G | Intron3 | New | 0.214 | |
| 290 | PTGER3S36 | 1 60 403 | A/G | Intron3 | rs1409978 | 0.319 | |
| 291 | PTGER3S37 | 1 60 432 | C/G | Intron3 | New | 0.214 | |
| 292 | EP4 | PTGER4S1 | 5748 | C/T | Intron2 | New | 0.389 |
| 293 | PTGER4S2 | 7984 | A/G | Intron2 | New | 0.198 | |
| 294 | PTGER4S3 | 8012 | G/A | Intron2 | New | 0.385 | |
| 295 | PTGES | PTGESS1 | 214 | A/G | Intron1 | rs2241271 | 0.135 |
| 296 | PTGESS2 | 406 | G/A | Intron1 | rs2241270 | 0.120 | |
| 297 | PTGESS3 | 9636 | T/C | Intron2 | New | 0.326 | |
| 298 | PTGESS4 | 9669 | G/A | Intron2 | New | 0.152 | |
| 299 | PTGFR | PTGFRS1 | 714 | A/C | Intron1 | rs3766355 | 0.425 |
| 300 | PTGFRS2 | 823 | G/A | Intron1 | rs3766354 | 0.204 | |
| 301 | PTGFRS3 | 1132 | G/T | Intron1 | rs3766353 | 0.242 | |
| 302 | PTGFRS4 | 3617 | G/A | Intron2 | rs1830763 | 0.489 | |
| 303 | PTGFRS5 | 6578 | A/C | Intron2 | rs1322935 | 0.031 | |
| 304 | PTGFRS6 | 8734 | A/G | Intron2 | rs2057423 | 0.452 | |
| 305 | PTGFRS10 | 24 552 | G/A | Intron2 | rs3766346 | 0.036 | |
| 306 | PTGFRS12 | 33 123 | A/C | Intron2 | rs520171 | 0.276 | |
| 307 | PTGFRS13 | 33 548 | G/C | Intron2 | rs3766345 | 0.516 | |
| 308 | PTGFRS14 | 38 195 | T/C | Intron2 | rs3766338 | 0.266 | |
| 309 | PTGFRS15 | 38 202 | C/T | Intron2 | rs590309 | 0.511 | |
| 310 | PTGFRS16 | 40 734 | G/C | Intron2 | rs622346 | 0.202 | |
| 311 | PTGFRS19 | 46 572 | A/G | Exon3-UTR | rs3766331 | 0.115 | |
| 312 | PTGFRS20 | 46 628 | A/G | Exon3-UTR | rs899 | 0.005 | |
| 313 | PTGS1 | PTGS1S1 | 2850 | C/T | Intron2 | rs1213264 | 0.063 |
| 314 | PTGS1S2 | 3219 | T/C | Intron2 | rs1213265 | 0.067 | |
| 315 | PTGS1S2.5 | 7468 | C/G | Intron3 | rs2282169 | 0.079 | |
| 316 | PTGS1S4 | 23 970 | C/A | Exon11-UTR | rs10306194 | 0.028 | |
| 317 | SCYA5 | SCYA5S1 | 328 | T/C | Intron1 | rs2280789 | 0.349 |
| 318 | SCYB14 | SCYB14S1 | 313 | T/C | Intron1 | rs2072347 | 0.311 |
| 319 | SCYB14S2 | 2099 | G/C | Intron2 | rs2237062 | 0.307 | |
| 320 | SCYB14S3 | 6261 | C/T | Intron3 | rs1016666 | 0.370 | |
| 321 | SLC21A9 | SLC21A9S1 | 196 | T/C | Exon1-UTR | New | 0.158 |
| 322 | SLC21A9S2 | 231 | G/A | Exon1-UTR | rs1944612 | 0.055 | |
| 323 | SLC21A9S3 | 4589 | C/T | Intron1 | New | 0.443 | |
| 324 | SLC21A9S5 | 10 724 | A/G | Intron1 | New | 0.374 | |
| 325 | SLC21A9S6 | 13 387 | G/C | Intron3 | New | 0.453 | |
| 326 | SLC21A9S7 | 15 413 | G/A | Intron4 | rs949069 | 0.391 | |
| 327 | SLC21A9S8 | 15 916 | A/G | Intron4 | New | 0.161 | |
| 328 | SLC21A9S9 | 17 493 | G/A | Intron4 | rs1676878 | 0.136 | |
| 329 | SLC21A9S10 | 24 698 | A/G | Intron7 | rs1676881 | 0.226 | |
| 330 | SLC21A9S10.5 | 24 880 | C/T | Intron7 | rs1612859 | 0.458 | |
| 331 | SLC21A9S11 | 25 005 | T/A | Intron7 | rs1789693 | 0.401 | |
| 332 | SLC21A9S13 | 26 261 | T/G | Intron7 | New | 0.376 | |
| 333 | SLC21A9S14 | 39 441 | G/A | Intron8 | rs1789692 | 0.479 | |
| 334 | SLC21A9S15 | 43 575 | C/T | Intron9 | New | 0.430 | |
| 335 | SLC21A9S16 | 45 422 | C/T | Exon10 | New | 0.432 | |
| 336 | STAT2 | STAT2S1 | 10 525 | A/G | Intron14 | rs2020854 | 0.021 |
| 337 | STAT4 | STAT4S1 | 5564 | C/A | Intron3 | rs1031509 | 0.396 |
| 338 | STAT4S2 | 19 235 | G/A | Intron3 | rs1551443 | 0.214 | |
| 339 | STAT4S3 | 91 147 | A/T | Intron10 | New | 0.516 | |
| 340 | STAT4S4 | 100 038 | G/C | Intron14 | rs1400655 | 0.095 | |
| 341 | STAT4S5 | 118 213 | A/G | Intron21 | rs925847 | 0.553 | |
| 342 | STAT4S6 | 119 708 | A/C | Intron22 | rs1517351 | 0.495 | |
| 343 | STAT4S7 | 119 711 | G/A | Intron22 | New | 0.165 | |
| 344 | TBX21 | TBX21S1 | 8941 | T/C | Intron1 | rs2158079 | 0.201 |
| 345 | TBXA2R | TBXA2RS1 | 10937 | T/C | Exon3-synonymous | rs4523 | 0.184 |
| 346 | TXAs | TXAsS2 | 17778 | A/C | Intron1 | rs41708 | 0.302 |
| 347 | TXAsS3 | 24573 | T/C | Intron1 | rs41706 | 0.355 | |
| 348 | TXAsS4 | 29218 | A/T | Intron1 | rs194150 | 0.443 | |
| 349 | TXAsS5 | 57546 | T/G | Intron3 | rs1015571 | 0.447 | |
| 350 | TXAsS7 | 63115 | C/A | Intron3 | rs2013219 | 0.426 | |
| 351 | TXAsS9 | 69322 | G/A | Intron3 | rs757762 | 0.447 | |
| 352 | TXAsS11 | 80249 | C/T | Intron3 | rs1978180 | 0.028 | |
| 353 | TXAsS13 | 1 06 385 | T/C | Intron4 | rs41733 | 0.117 | |
| 354 | TXAsS14 | 1 06 401 | G/A | Intron4 | rs41732 | 0.117 | |
| 355 | TXAsS15 | 1 12 521 | T/C | Intron5 | New | 0.247 | |
| 356 | TXAsS16 | 1 12 694 | C/T | Intron5 | rs42335 | 0.134 | |
| 357 | TXAsS17 | 1 15 453 | G/A | Intron5 | New | 0.011 | |
| 358 | TXAsS21 | 1 28 260 | G/T | Intron7 | New | 0.165 | |
| 359 | TXAsS22 | 1 39 370 | T/C | Intron9 | rs41718 | 0.146 | |
| 360 | TXAsS23 | 1 46 442 | T/C | Intron9 | rs740150 | 0.389 | |
| 361 | TXAsS24 | 1 52 303 | A/G | Intron9 | rs193949 | 0.177 | |
| 362 | TXAsS25 | 1 52 455 | C/T | Intron9 | New | 0.308 | |
| 363 | TXAsS29 | 1 78 072 | G/A | Intron10 | rs740204 | 0.234 | |
| 364 | TXAsS30 | 1 86 274 | G/A | Intron10 | New | 0.479 | |
| 365 | UPAR | UPARS1 | 11 475 | A/C | Intron3 | New | 0.019 |
| 366 | UPARS2 | 11 639 | G/A | Intron3 | rs2283628 | 0.335 | |
| 367 | UPARS3 | 11 667 | A/C | Intron3 | rs2239373 | 0.385 | |
| 368 | UPARS4 | 11 746 | C/T | Intron3 | rs2239372 | 0.375 | |
| 369 | UPARS5 | 18 100 | C/T | Intron5 | rs2302525 | 0.074 | |
| 370 | UPARS6 | 18 228 | A/G | Exon6-non-synonymous (K220R) | rs2302524 | 0.114 |
| No. . | Gene . | SNP . | Sequence position . | Variation . | Localization . | rs ID . | Minor allele frequncies (96 CTR) . |
|---|---|---|---|---|---|---|---|
| 1 | ADAM33 | ADAM33S1 | 11 513 | C/A | Intron19 | rs44707 | 0.467 |
| 2 | ADAM33S2 | 12 505 | T/C | Exon20-non-synonymous (M764T) | rs2280091 | 0.098 | |
| 3 | ADAM33S3 | 12 534 | T/C | Exon20-non-synonymous (M774S) | rs2280090 | 0.102 | |
| 4 | ADAM33S4 | 12 612 | T/C | Intron20 | rs2280089 | 0.092 | |
| 5 | ADAM33S5 | 13 018 | G/A | Intron21 | rs628977 | 0.362 | |
| 6 | ADAM33S6 | 13 026 | T/C | Intron21 | rs628965 | 0.362 | |
| 7 | ADAM33S7 | 13 060 | A/C | Intron21 | rs543749 | 0.181 | |
| 8 | ALOX12B | ALOX12BS1 | 1963 | G/A | Intron2 | rs3027303 | 0.189 |
| 9 | ALOX12BS4 | 3276 | G/T | Intron2 | rs3027294 | 0.391 | |
| 10 | ALOX12BS5 | 7053 | G/C | Intron4 | rs2304908 | 0.392 | |
| 11 | ALOX12BS6 | 7223 | C/T | Intron4 | rs2304907 | 0.396 | |
| 12 | ALOX12BS7 | 7341 | C/T | Intron4 | rs2304906 | 0.130 | |
| 13 | ALOX5 | ALOX5S1 | 9223 | A/G | Intron2 | rs4769060 | 0.151 |
| 14 | ALOX5S2 | 26 904 | G/A | Intron3 | New | 0.401 | |
| 15 | ALOX5S3 | 50 760 | G/A | Intron4 | New | 0.198 | |
| 16 | ALOX5S4 | 59 443 | A/G | Intron6 | New | 0.193 | |
| 17 | ALOX5S5 | 69 751 | G/C | Intron7 | New | 0.354 | |
| 18 | CLP | CLPS1 | 1152 | G/A | Intron2 | rs2966305 | 0.401 |
| 19 | CLPS2 | 3607 | C/A | Intron2 | New | 0.032 | |
| 20 | CLPS3 | 3688 | A/G | Intron2 | rs2967871 | 0.315 | |
| 21 | CLPS4 | 9097 | C/T | Intron2 | rs2925050 | 0.495 | |
| 22 | CLPS5 | 9197 | T/G | Intron2 | New | 0.457 | |
| 23 | CLPS6 | 11 744 | T/C | Intron2 | rs1835156 | 0.333 | |
| 24 | CLPS7 | 17 612 | T/C | Intron2 | New | 0.229 | |
| 25 | CLPS8 | 17 865 | G/C | Intron2 | New | 0.356 | |
| 26 | CLPS9 | 17 923 | G/C | Intron2 | rs2967876 | 0.284 | |
| 27 | CLPS10 | 20 308 | C/T | Intron2 | rs2925064 | 0.263 | |
| 28 | CLPS11 | 20 380 | T/C | Intron2 | rs934166 | 0.468 | |
| 29 | CLPS12 | 20 412 | G/A | Intron2 | rs201854 | 0.447 | |
| 30 | CLPS13 | 23 054 | T/C | Intron2 | New | 0.104 | |
| 31 | CLPS17 | 27 698 | A/G | Intron2 | rs2288584 | 0.250 | |
| 32 | CLPS18 | 31 174 | T/C | Intron3 | rs2967855 | 0.283 | |
| 33 | CLPS19 | 31 262 | C/G | Intron3 | New | 0.214 | |
| 34 | CLPS20 | 31 615 | C/T | Intron3 | New | 0.208 | |
| 35 | CLPS21 | 43 186 | A/T | Intron3 | rs2914823 | 0.185 | |
| 36 | CLPS22 | 43 360 | T/G | Intron3 | New | 0.404 | |
| 37 | CLPS23 | 43 394 | A/G | Intron3 | New | 0.250 | |
| 38 | CLPS24 | 47 961 | G/A | Intron3 | New | 0.116 | |
| 39 | CLPS25 | 48 577 | C/T | Intron3 | New | 0.347 | |
| 40 | CLPS26 | 51794 | C/T | Exon4-UTR | rs247862 | 0.297 | |
| 41 | CNOT3 | CNOT3S1 | 328 | C/T | Intron1 | rs42318 | 0.115 |
| 42 | CTSG | CTSGS1 | 308 | G/A | Intron1 | rs2236742 | 0.212 |
| 43 | CTSGS2 | 527 | A/G | Intron1 | New | 0.034 | |
| 44 | CTSGS3 | 726 | A/G | Intron1 | rs1957523 | 0.477 | |
| 45 | CTSGS4 | 1226 | C/T | Intron2 | New | 0.034 | |
| 46 | CTSGS5 | 1349 | G/A | Intron2 | rs2070697 | 0.314 | |
| 47 | CTSGS6 | 1767 | A/G | Exon4-synonymous | New | 0.182 | |
| 48 | CYP4F2 | CYP4F2S1 | 4937 | A/T | Intron1 | New | 0.349 |
| 49 | CYP4F2S4 | 18 733 | T/C | Intron1 | rs2072269 | 0.335 | |
| 50 | CYP4F2S5 | 18 810 | G/C | Intron1 | rs1064796 | 0.495 | |
| 51 | CYP4F2S6 | 30 715 | G/A | Intron1 | rs2018460 | 0.170 | |
| 52 | CYP4F3 | CYP4F3S0 | 3187 | G/A | Intron2 | rs2203998 | 0.328 |
| 53 | CYP4F3S0.5 | 6221 | C/T | Intron4 | rs1290626 | 0.141 | |
| 54 | CYP4F3S1 | 8507 | C/T | Intron6 | rs2283612 | 0.189 | |
| 55 | CYP4F3S2 | 11 871 | A/C | Intron8 | rs2733750 | 0.183 | |
| 56 | CYP4F3S3 | 12 015 | A/G | Exon9-synonymous | New | 0.363 | |
| 57 | CYP4F3S4 | 12 808 | G/A | Intron9 | rs2733752 | 0.136 | |
| 58 | CYP4F3S5 | 12 877 | T/A | Intron9 | New | 0.140 | |
| 59 | CYP4F3S6 | 17 772 | T/C | Intron11 | New | 0.453 | |
| 60 | CYP4F8 | CYP4F8S1 | 3610 | C/T | Intron2 | rs2072599 | 0.168 |
| 61 | CYP4F8S2 | 6771 | G/T | Intron5 | rs2072601 | 0.276 | |
| 62 | CYP4F8S3 | 13 568 | C/T | Intron11 | rs2239366 | 0.276 | |
| 63 | CYSLT1R | CYSLT1RS1 | 927 | C/T | Exon1-synonymous | rs320995 | 0.474 |
| 64 | CYSLT2R | CYSLT2RS1 | 2797 | A/G | Exon1-UTR | rs912277 | 0.417 |
| 65 | CYSLT2RS2 | 3078 | A/C | Exon1-UTR | rs1323552 | 0.489 | |
| 66 | CYSLT2RS3 | 3105 | A/G | Exon1-UTR | New | 0.116 | |
| 67 | FLAP | FLAPS1 | 162 | C/A | Intron1 | rs4769055 | 0.522 |
| 68 | FLAPS2 | 838 | T/G | Intron1 | rs9579645 | 0.096 | |
| 69 | FLAPS3 | 7272 | A/G | Intron1 | rs9551960 | 0.339 | |
| 70 | FLAPS4 | 8640 | A/C | Intron2 | rs3803277 | 0.406 | |
| 71 | FLAPS5 | 8733 | T/C | Intron2 | rs3803278 | 0.370 | |
| 72 | FLAPS6 | 13 674 | G/A | Intron2 | rs4075692 | 0.286 | |
| 73 | FLAPS7 | 20 616 | G/C | Intron3 | New | 0.286 | |
| 74 | FLAPS8 | 20 648 | C/T | Intron3 | rs4468448 | 0.240 | |
| 75 | FLAPS9 | 23 849 | T/A | Intron3 | rs9551964 | 0.277 | |
| 76 | FLAPS9.5 | 24 348 | G/A | Intron3 | New | 0.006 | |
| 77 | FLAPS10 | 28 209 | A/G | Intron3 | rs4769060 | 0.401 | |
| 78 | HPGD | HPGDS1 | 464 | G/A | Exon2-synonymous | rs1050145 | 0.220 |
| 79 | HPGDS2 | 798 | A/G | Intron2 | rs1365613 | 0.447 | |
| 80 | HPGDS3 | 13 332 | A/C | Intron3 | rs2555629 | 0.405 | |
| 81 | HPGDS4 | 13 430 | A/T | Intron3 | New | 0.253 | |
| 82 | HPGDS5 | 20 442 | A/G | Intron4 | New | 0.446 | |
| 83 | HPGDS6 | 20 516 | G/A | Intron4 | New | 0.110 | |
| 84 | IGF1 | IGF1S1 | 3083 | A/G | Intron2 | rs2162679 | 0.368 |
| 85 | IGF1S2 | 9917 | G/T | Intron3 | rs1019731 | 0.021 | |
| 86 | IGF1S3 | 17 640 | G/C | Intron3 | rs2195239 | 0.426 | |
| 87 | IGF1S4 | 17 695 | T/C | Intron3 | rs2195240 | 0.443 | |
| 88 | IGF1S5 | 49 421 | G/A | Intron3 | rs972936 | 0.484 | |
| 89 | IGF1S6 | 60 710 | G/A | Intron3 | rs2072592 | 0.245 | |
| 90 | IGF1S7 | 72 103 | G/A | Intron5 | rs978458 | 0.135 | |
| 91 | IGF1S8 | 84 150 | G/A | Exon6-UTR | rs6219 | 0.271 | |
| 92 | IL13 | IL13S1 | −978 | C/T | Promoter | rs11575055 | 0.179 |
| 93 | IL13S2 | 571 | C/A | Intron1 | rs2066960 | 0.281 | |
| 94 | IL13S3 | 598 | G/C | Intron1 | rs1295987 | 0.120 | |
| 95 | IL13S4 | 805 | C/T | Intron1 | rs2069744 | 0.120 | |
| 96 | IL13S5 | 2100 | G/A | Exon4-non-synonymous (R144Q) | rs20541 | 0.333 | |
| 97 | IL4 | IL4S1 | −219 | T/C | Promoter | rs2243250 | 0.328 |
| 98 | IL4S2 | 3353 | A/C | Intron2 | rs2227284 | 0.255 | |
| 99 | IL4S3 | 3927 | C/G | Intron2 | rs2243263 | 0.073 | |
| 100 | LTA4H | LTA4HS1 | 9153 | A/G | Intron3 | rs763842 | 0.226 |
| 101 | LTA4HS4 | 20 165 | T/C | Intron11 | rs1978331 | 0.335 | |
| 102 | LTB4DH | LTB4DHS1 | 4435 | A/T | Intron2 | New | 0.506 |
| 103 | LTB4DHS2 | 8896 | A/G | Intron4 | New | 0.283 | |
| 104 | LTB4DHS2.5 | 9182 | A/G | Intron4 | New | 0.092 | |
| 105 | LTB4DHS5 | 9502 | C/T | Intron4 | rs1053968 | 0.005 | |
| 106 | LTB4DHS6 | 13 234 | C/G | Intron4 | New | 0.319 | |
| 107 | LTB4DHS6.5 | 13 249 | A/G | Intron4 | New | 0.231 | |
| 108 | LTB4DHS7 | 20 718 | G/A | Intron6 | New | 0.253 | |
| 109 | LTB4DHS8 | 25 971 | A/G | Intron8 | rs1322258 | 0.347 | |
| 110 | LTB4DHS9 | 26 221 | A/G | Intron8 | New | 0.406 | |
| 111 | LTB4DHS10 | 35 589 | A/C | Intron9 | rs2146078 | 0.229 | |
| 112 | LTB4DHS11 | 35 819 | G/A | Intron9 | New | 0.354 | |
| 113 | LTB4R | LTB4RS1 | 1165 | G/C | Exon1-UTR | New | 0.140 |
| 114 | LTC4S | LTC4SS1.5 | −348 | A/C | Promoter | New | 0.208 |
| 115 | LTC4SS2 | 289 | G/C | Intron1 | New | 0.026 | |
| 116 | LTC4SS3 | 1296 | C/T | Intron1 | New | 0.208 | |
| 117 | LTC4SS4 | 2659 | A/G | Intron5 | New | 0.201 | |
| 118 | MGST2 | MGST2S1 | 815 | A/G | Intron1 | rs1000222 | 0.292 |
| 119 | MGST2S2 | 12 971 | C/T | Intron2 | rs795589 | 0.198 | |
| 120 | MGST2S3 | 12 977 | A/C | Intron2 | rs795588 | 0.077 | |
| 121 | MMP1 | MMP1S1 | 193 | G/A | Intron1 | rs470358 | 0.292 |
| 122 | MMP1S2 | 2579 | G/A | Exon5-synonymous | rs470558 | 0.130 | |
| 123 | MMP1S3 | 7300 | T/C | Intron8 | rs470747 | 0.137 | |
| 124 | MMP1S4 | 7815 | C/T | Exon10-UTR | rs2239008 | 0.319 | |
| 125 | MMP1S5 | 7936 | T/C | Exon10-UTR | rs2071230 | 0.298 | |
| 126 | MMP2 | MMP2S1 | 3606 | A/T | Intron1 | rs857403 | 0.214 |
| 127 | MMP2S2 | 3665 | G/A | Intron1 | rs1030868 | 0.229 | |
| 128 | MMP2S3 | 6505 | C/T | Exon5-synonymous | rs1053605 | 0.208 | |
| 129 | MMP2S4 | 6730 | T/G | Intron5 | New | 0.064 | |
| 130 | MMP2S5 | 6958 | A/G | Intron5 | rs866770 | 0.151 | |
| 131 | MMP2S6 | 10 603 | C/T | Exon7-synonymous | rs243849 | 0.151 | |
| 132 | MMP2S7 | 10 896 | T/C | Intron7 | rs243847 | 0.443 | |
| 133 | MMP2S8 | 14 011 | G/A | Exon9-synonymous | rs2287074 | 0.326 | |
| 134 | MMP2S9 | 14 196 | G/A | Intron9 | rs243843 | 0.306 | |
| 135 | MMP2S10 | 14 281 | G/A | Intron9 | New | 0.065 | |
| 136 | MMP2S11 | 14 320 | T/C | Intron9 | rs243842 | 0.364 | |
| 137 | MMP2S12 | 17 660 | G/T | Intron9 | rs171498 | 0.363 | |
| 138 | MMP2S13 | 17 670 | G/T | Intron9 | rs243838 | 0.153 | |
| 139 | MMP2S14 | 17 928 | C/T | Intron10 | New | 0.057 | |
| 140 | MMP2S15 | 20 976 | G/A | Intron11 | New | 0.115 | |
| 141 | MMP2S16 | 21 134 | G/A | Intron11 | rs243836 | 0.386 | |
| 142 | MMP2S17 | 26 345 | C/T | Exon13-UTR | New | 0.021 | |
| 143 | MMP2S18 | 26 512 | A/C | Exon13-UTR | rs7201 | 0.234 | |
| 144 | MMP8 | MMP8S1 | 524 | G/A | Intron1 | rs1939012 | 0.359 |
| 145 | MMP8S2 | 5458 | A/G | Intron4 | rs1940051 | 0.266 | |
| 146 | MMP8S3 | 11 357 | T/A | Intron9 | New | 0.287 | |
| 147 | MMP8S4 | 11 473 | T/C | Exon10-synonymous | New | 0.016 | |
| 148 | MMP9 | MMP9S1 | 2679 | G/A | Exon6-non-synonymous (R279Q) | rs2664538 | 0.382 |
| 149 | MMP9S2 | 3029 | A/C | Intron6 | rs2236416 | 0.179 | |
| 150 | MMP9S3 | 5565 | G/A | Exon12-non-synonymous (R668Q) | rs2274756 | 0.214 | |
| 151 | MMP9S4 | 7419 | G/A | Exon13-synonymous | rs13925 | 0.292 | |
| 152 | P2Y10 | P2Y10S1 | 1192 | A/G | Intron1 | New | 0.016 |
| 153 | P2Y10S2 | 5030 | C/T | Intron1 | rs2858570 | 0.110 | |
| 154 | P2Y10S3 | 5318 | A/T | Intron1 | rs2251477 | 0.184 | |
| 155 | P2Y10S4 | 9417 | T/C | Intron1 | rs2742205 | 0.115 | |
| 156 | PAFAH1B1 | PAFAH1B1S2 | 12 145 | G/T | Intron1 | New | 0.099 |
| 157 | PAFAH1B1S3 | 21 146 | C/G | Intron1 | New | 0.080 | |
| 158 | PAFAH1B1S4 | 37 780 | A/G | Intron1 | rs1266474 | 0.084 | |
| 159 | PAFAH1B1S5 | 38 033 | C/T | Intron1 | New | 0.047 | |
| 160 | PAFAH1B1S6 | 40 522 | G/A | Intron1 | New | 0.130 | |
| 161 | PAFAH1B1S7 | 60 034 | C/T | Intron2 | New | 0.266 | |
| 162 | PAFAH1B1S8 | 68 545 | G/C | Intron2 | New | 0.078 | |
| 163 | PAFAH1B1S9 | 76 403 | T/G | Intron5 | New | 0.032 | |
| 164 | PAFAH1B2 | PAFAH1B2S1 | 3971 | T/C | Intron1 | rs2008908 | 0.446 |
| 165 | PAFAH1B3 | PAFAH1B3S1 | 2538 | C/T | Exon4-synonymous | New | 0.016 |
| 166 | PAFAH2 | PAFAH2S1 | 1279 | G/T | Intron1 | rs3008423 | 0.172 |
| 167 | PAFAH2S2 | 4171 | C/T | Intron1 | New | 0.036 | |
| 168 | PAFAH2S3 | 15456 | G/T | Intron5 | rs1469512 | 0.214 | |
| 169 | PAI1m2 | PAI1m2S1 | 4484 | G/A | Intron1 | rs840088 | 0.401 |
| 170 | PAI1m2S2 | 5094 | A/G | Intron1 | New | 0.089 | |
| 171 | PAI1m2S3 | 19 022 | C/A | Intron1 | rs2099601 | 0.201 | |
| 172 | PAI1m2S4 | 19 047 | A/C | Intron1 | New | 0.201 | |
| 173 | PAI1m2S5 | 19 120 | T/C | Intron1 | rs2083120 | 0.201 | |
| 174 | PAI1m2S6 | 19 348 | A/C | Intron1 | New | 0.393 | |
| 175 | PAI1m2S7 | 62 178 | A/G | Intron8 | New | 0.271 | |
| 176 | PDGFB | PDGFBS1 | 581 | G/A | Intron1 | rs758588 | 0.184 |
| 177 | PDGFBS3 | 20 237 | C/T | Intron3 | rs740750 | 0.310 | |
| 178 | PDGFBS4 | 26 041 | C/T | Intron10 | rs1864972 | 0.198 | |
| 179 | PDGFBS5 | 35 170 | G/A | Intron18 | rs1432878 | 0.108 | |
| 180 | PDGFRL | PDGFRLS1 | 978 | G/C | Intron1 | rs2720576 | 0.365 |
| 181 | PDGFRLS2 | 1187 | C/T | Intron1 | rs2588164 | 0.245 | |
| 182 | PDGFRLS3 | 1416 | T/C | Intron1 | rs2588163 | 0.391 | |
| 183 | PDGFRLS4 | 2963 | G/C | Intron1 | rs2517267 | 0.458 | |
| 184 | PDGFRLS5 | 3794 | C/G | Intron1 | rs2517268 | 0.394 | |
| 185 | PDGFRLS6 | 3882 | G/A | Intron1 | New | 0.216 | |
| 186 | PDGFRLS7 | 3999 | A/G | Intron1 | rs2517269 | 0.220 | |
| 187 | PDGFRLS8 | 8893 | G/A | Intron1 | New | 0.226 | |
| 188 | PDGFRLS9 | 9922 | G/A | Intron1 | New | 0.232 | |
| 189 | PDGFRLS10 | 14 053 | G/C | Intron2 | rs2720583 | 0.167 | |
| 190 | PDGFRLS11 | 14 194 | A/C | Intron2 | New | 0.396 | |
| 191 | PDGFRLS12 | 18 220 | T/C | Intron2 | rs2427709 | 0.140 | |
| 192 | PDGFRLS13 | 18 473 | G/A | Intron2 | rs2588144 | 0.068 | |
| 193 | PDGFRLS15 | 19 592 | T/G | Intron2 | rs2246488 | 0.339 | |
| 194 | PDGFRLS16 | 28 052 | G/A | Intron2 | rs2517187 | 0.104 | |
| 195 | PDGFRLS17 | 28 449 | C/T | Intron2 | rs2237823 | 0.104 | |
| 196 | PDGFRLS18 | 31 111 | T/C | Intron2 | rs2517198 | 0.094 | |
| 197 | PDGFRLS19 | 31 230 | A/T | Intron2 | New | 0.260 | |
| 198 | PDGFRLS20 | 31 258 | A/G | Intron2 | New | 0.031 | |
| 199 | PDGFRLS21 | 31 499 | T/C | Intron2 | rs2427715 | 0.094 | |
| 200 | PDGFRLS22 | 31 600 | T/C | Intron2 | New | 0.167 | |
| 201 | PDGFRLS23 | 33 975 | G/C | Intron2 | rs2517208 | 0.037 | |
| 202 | PDGFRLS24 | 41 991 | A/G | Intron2 | rs2237831 | 0.189 | |
| 203 | PDGFRLS25 | 42 131 | G/A | Intron2 | rs2237831 | 0.205 | |
| 204 | PDGFRLS26 | 43 810 | G/T | Intron2 | New | 0.073 | |
| 205 | PDGFRLS27 | 46 759 | T/C | Intron3 | New | 0.120 | |
| 206 | PDGFRLS28 | 46 888 | G/A | Intron3 | rs2237835 | 0.226 | |
| 207 | PDGFRLS29 | 46 936 | C/G | Intron3 | New | 0.074 | |
| 208 | PDGFRLS30 | 47 097 | C/G | Intron3 | New | 0.197 | |
| 209 | PDGFRLS31 | 49 411 | A/C | Intron3 | rs2237836 | 0.548 | |
| 210 | PDGFRLS32 | 49 937 | G/T | Intron3 | rs2237837 | 0.103 | |
| 211 | PDGFRLS33 | 53 887 | T/C | Intron4 | rs2237842 | 0.188 | |
| 212 | PDGFRLS34 | 56 021 | C/T | Intron4 | rs2427719 | 0.063 | |
| 213 | PDGFRLS35 | 56 410 | T/C | Intron4 | rs2237845 | 0.240 | |
| 214 | PDGFRLS36 | 65 408 | C/T | Intron5 | New | 0.083 | |
| 215 | PDGFRLS37 | 65 512 | T/C | Exon6-synonymous | rs4705 | 0.521 | |
| 216 | PGDS | PGDSS1 | 5446 | T/C | Intron1 | rs2129595 | 0.191 |
| 217 | PGIS | PGISS1 | 4650 | T/C | Intron1 | rs477627 | 0.063 |
| 218 | PGISS2 | 6734 | A/C | Intron1 | rs927068 | 0.226 | |
| 219 | PGISS3 | 6840 | T/C | Intron1 | New | 0.226 | |
| 220 | PGISS4 | 6870 | G/C | Intron1 | rs498646 | 0.276 | |
| 221 | PGISS4.5 | 6924 | T/C | Intron1 | rs476496 | 0.280 | |
| 222 | PGISS6 | 28 941 | A/G | Intron5 | rs501908 | 0.033 | |
| 223 | PGISS7 | 41 990 | A/G | Intron5 | New | 0.093 | |
| 224 | PGISS8 | 44 026 | G/A | Exon6-synonymous | rs5628 | 0.036 | |
| 225 | PGISS9 | 50 926 | C/T | Intron6 | New | 0.104 | |
| 226 | PGISS10 | 55 002 | C/A | Intron8 | rs5629 | 0.281 | |
| 227 | PGISS11 | 57 532 | A/G | Intron9 | rs729824 | 0.226 | |
| 228 | PGISS12 | 62 730 | T/C | Exon10-UTR | rs5602 | 0.438 | |
| 229 | PLA2G7 | PLA2G7S1 | 2007 | G/T | Intron1 | New | 0.214 |
| 230 | PLA2G7S2 | 2338 | G/A | Intron1 | rs1421369 | 0.468 | |
| 231 | PLA2G7S3 | 6203 | G/A | Intron1 | New | 0.141 | |
| 232 | PLA2G7S4 | 20 965 | G/T | Intron5 | rs1362931 | 0.042 | |
| 233 | PLA2G7S5 | 23 741 | T/C | Exon7-non-synonymous (I198T) | rs1805018 | 0.281 | |
| 234 | PLA2G7S6 | 27 025 | C/G | Intron9 | rs2216465 | 0.542 | |
| 235 | PLA2G7S7 | 30 101 | C/T | Exon11-non-synonymous (A379V) | rs1051931 | 0.042 | |
| 236 | PTAFR | PTAFRS3 | 36 031 | C/A | Exon2-non-synonymous (A224D) | rs5938 | 0.068 |
| 237 | PTGDR | PTGDRS1 | 408 | G/A | Intron1 | New | 0.021 |
| 238 | PTGDRS2 | 3290 | A/G | Intron1 | rs1254609 | 0.194 | |
| 239 | PTGDRS3 | 5754 | T/C | Intron1 | New | 0.075 | |
| 240 | PTGDRS4 | 5793 | A/G | Intron1 | rs708486 | 0.081 | |
| 241 | PTGDS | PTGDSS1 | 4186 | C/A | Exon7-UTR | rs6926 | 0.198 |
| 242 | EP1 | PTGER1S1 | −267 | G/C | Promoter | New | 0.109 |
| 243 | EP2 | PTGER2uS1 | −26 643 | T/C | 5′-Upstream | rs988209 | 0.232 |
| 244 | PTGER2uS2 | −18 461 | T/G | 5′-Upstream | rs1390375 | 0.077 | |
| 245 | PTGER2uS3 | −18 360 | G/A | 5′-Upstream | rs1390374 | 0.022 | |
| 246 | PTGER2uS4 | −15 332 | A/T | 5′-Upstream | rs708490 | 0.444 | |
| 247 | PTGER2uS5 | −12 813 | G/A | 5′-Upstream | New | 0.223 | |
| 248 | PTGER2uS6 | −10 918 | A/G | 5′-Upstream | New | 0.449 | |
| 249 | PTGER2uS7 | −10 814 | T/A | 5′-upstream | New | 0.427 | |
| 250 | PTGER2uS8 | −10 250 | A/G | 5′-upstream | rs714366 | 0.433 | |
| 251 | PTGER2uS9 | −7075 | A/G | 5′-upstream | New | 0.452 | |
| 252 | PTGER2uS10 | −6179 | A/G | 5′-upstream | New | 0.479 | |
| 253 | PTGER2S1 | −609 | G/A | Promoter | rs1254601 | 0.288 | |
| 254 | PTGER2S2 | 300 | G/A | Exon1-UTR | rs1254600 | 0.462 | |
| 255 | PTGER2S3 | 498 | C/G | Exon1-UTR | rs2075797 | 0.441 | |
| 256 | PTGER2S4 | 948 | A/G | Exon1-UTR | rs1353411 | 0.417 | |
| 257 | PTGER2S5 | 1042 | G/A | Exon1-UTR | rs1254598 | 0.350 | |
| 258 | PTGER2S6 | 2803 | G/A | Intron1 | New | 0.164 | |
| 259 | PTGER2S7 | 2988 | C/T | Intron1 | New | 0.339 | |
| 260 | PTGER2S8 | 6063 | C/T | Intron1 | New | 0.116 | |
| 261 | PTGER2S9 | 10 927 | T/G | Intron1 | rs1254585 | 0.446 | |
| 262 | PTGER2S10 | 14 081 | C/T | Exon2-UTR | rs708502 | 0.398 | |
| 263 | PTGER2dS1 | 29 784 | C/T | 3′-Downstream | rs708511 | 0.219 | |
| 264 | PTGER2dS2 | 47 461 | A/G | 3′-Downstream | New | 0.226 | |
| 265 | PTGER2dS3 | 57 931 | C/T | 3′-Downstream | rs708531 | 0.750 | |
| 266 | PTGER2dS4 | 58 051 | C/T | 3′-Downstream | rs708532 | 0.329 | |
| 267 | EP3 | PTGER3S1 | 27 837 | G/A | Intron1 | rs1008484 | 0.058 |
| 268 | PTGER3S2 | 28 078 | G/A | Intron1 | rs1569593 | 0.258 | |
| 269 | PTGER3S5 | 36 177 | A/T | Intron2 | rs5680 | 0.313 | |
| 270 | PTGER3S7 | 44 999 | T/G | Intron2 | rs1983588 | 0.268 | |
| 271 | PTGER3S8 | 45 040 | A/C | Intron2 | rs1983587 | 0.059 | |
| 272 | PTGER3S9 | 54 444 | G/C | Intron2 | rs1883461 | 0.054 | |
| 273 | PTGER3S10 | 54 634 | T/A | Intron2 | rs1883460 | 0.094 | |
| 274 | PTGER3S11 | 58 525 | C/T | Intron2 | New | 0.271 | |
| 275 | PTGER3S12 | 65 699 | G/C | Intron2 | rs647921 | 0.048 | |
| 276 | PTGER3S13 | 65 973 | A/T | Intron2 | rs646621 | 0.443 | |
| 277 | PTGER3S14 | 70 342 | G/A | Intron2 | rs909842 | 0.442 | |
| 278 | PTGER3S15 | 70 357 | A/C | Intron2 | New | 0.300 | |
| 279 | PTGER3S17 | 70 409 | G/A | Intron2 | New | 0.442 | |
| 280 | PTGER3S20 | 70 755 | A/T | Intron2 | rs484675 | 0.441 | |
| 281 | PTGER3S24 | 86 923 | A/G | Intron2 | rs573688 | 0.082 | |
| 282 | PTGER3S25 | 95 146 | A/C | Intron3 | New | 0.037 | |
| 283 | PTGER3S27 | 1 00 052 | T/C | Intron3 | rs1409984 | 0.255 | |
| 284 | PTGER3S29 | 1 14 078 | G/A | Intron3 | rs625617 | 0.371 | |
| 285 | PTGER3S30 | 1 21 798 | C/A | Intron3 | New | 0.328 | |
| 286 | PTGER3S31 | 1 21 846 | G/A | Intron3 | rs602383 | 0.297 | |
| 287 | PTGER3S32 | 1 49 402 | C/T | Intron3 | rs1409165 | 0.370 | |
| 288 | PTGER3S33 | 1 49 626 | G/A | Intron3 | rs1409166 | 0.398 | |
| 289 | PTGER3S35 | 1 60 372 | A/G | Intron3 | New | 0.214 | |
| 290 | PTGER3S36 | 1 60 403 | A/G | Intron3 | rs1409978 | 0.319 | |
| 291 | PTGER3S37 | 1 60 432 | C/G | Intron3 | New | 0.214 | |
| 292 | EP4 | PTGER4S1 | 5748 | C/T | Intron2 | New | 0.389 |
| 293 | PTGER4S2 | 7984 | A/G | Intron2 | New | 0.198 | |
| 294 | PTGER4S3 | 8012 | G/A | Intron2 | New | 0.385 | |
| 295 | PTGES | PTGESS1 | 214 | A/G | Intron1 | rs2241271 | 0.135 |
| 296 | PTGESS2 | 406 | G/A | Intron1 | rs2241270 | 0.120 | |
| 297 | PTGESS3 | 9636 | T/C | Intron2 | New | 0.326 | |
| 298 | PTGESS4 | 9669 | G/A | Intron2 | New | 0.152 | |
| 299 | PTGFR | PTGFRS1 | 714 | A/C | Intron1 | rs3766355 | 0.425 |
| 300 | PTGFRS2 | 823 | G/A | Intron1 | rs3766354 | 0.204 | |
| 301 | PTGFRS3 | 1132 | G/T | Intron1 | rs3766353 | 0.242 | |
| 302 | PTGFRS4 | 3617 | G/A | Intron2 | rs1830763 | 0.489 | |
| 303 | PTGFRS5 | 6578 | A/C | Intron2 | rs1322935 | 0.031 | |
| 304 | PTGFRS6 | 8734 | A/G | Intron2 | rs2057423 | 0.452 | |
| 305 | PTGFRS10 | 24 552 | G/A | Intron2 | rs3766346 | 0.036 | |
| 306 | PTGFRS12 | 33 123 | A/C | Intron2 | rs520171 | 0.276 | |
| 307 | PTGFRS13 | 33 548 | G/C | Intron2 | rs3766345 | 0.516 | |
| 308 | PTGFRS14 | 38 195 | T/C | Intron2 | rs3766338 | 0.266 | |
| 309 | PTGFRS15 | 38 202 | C/T | Intron2 | rs590309 | 0.511 | |
| 310 | PTGFRS16 | 40 734 | G/C | Intron2 | rs622346 | 0.202 | |
| 311 | PTGFRS19 | 46 572 | A/G | Exon3-UTR | rs3766331 | 0.115 | |
| 312 | PTGFRS20 | 46 628 | A/G | Exon3-UTR | rs899 | 0.005 | |
| 313 | PTGS1 | PTGS1S1 | 2850 | C/T | Intron2 | rs1213264 | 0.063 |
| 314 | PTGS1S2 | 3219 | T/C | Intron2 | rs1213265 | 0.067 | |
| 315 | PTGS1S2.5 | 7468 | C/G | Intron3 | rs2282169 | 0.079 | |
| 316 | PTGS1S4 | 23 970 | C/A | Exon11-UTR | rs10306194 | 0.028 | |
| 317 | SCYA5 | SCYA5S1 | 328 | T/C | Intron1 | rs2280789 | 0.349 |
| 318 | SCYB14 | SCYB14S1 | 313 | T/C | Intron1 | rs2072347 | 0.311 |
| 319 | SCYB14S2 | 2099 | G/C | Intron2 | rs2237062 | 0.307 | |
| 320 | SCYB14S3 | 6261 | C/T | Intron3 | rs1016666 | 0.370 | |
| 321 | SLC21A9 | SLC21A9S1 | 196 | T/C | Exon1-UTR | New | 0.158 |
| 322 | SLC21A9S2 | 231 | G/A | Exon1-UTR | rs1944612 | 0.055 | |
| 323 | SLC21A9S3 | 4589 | C/T | Intron1 | New | 0.443 | |
| 324 | SLC21A9S5 | 10 724 | A/G | Intron1 | New | 0.374 | |
| 325 | SLC21A9S6 | 13 387 | G/C | Intron3 | New | 0.453 | |
| 326 | SLC21A9S7 | 15 413 | G/A | Intron4 | rs949069 | 0.391 | |
| 327 | SLC21A9S8 | 15 916 | A/G | Intron4 | New | 0.161 | |
| 328 | SLC21A9S9 | 17 493 | G/A | Intron4 | rs1676878 | 0.136 | |
| 329 | SLC21A9S10 | 24 698 | A/G | Intron7 | rs1676881 | 0.226 | |
| 330 | SLC21A9S10.5 | 24 880 | C/T | Intron7 | rs1612859 | 0.458 | |
| 331 | SLC21A9S11 | 25 005 | T/A | Intron7 | rs1789693 | 0.401 | |
| 332 | SLC21A9S13 | 26 261 | T/G | Intron7 | New | 0.376 | |
| 333 | SLC21A9S14 | 39 441 | G/A | Intron8 | rs1789692 | 0.479 | |
| 334 | SLC21A9S15 | 43 575 | C/T | Intron9 | New | 0.430 | |
| 335 | SLC21A9S16 | 45 422 | C/T | Exon10 | New | 0.432 | |
| 336 | STAT2 | STAT2S1 | 10 525 | A/G | Intron14 | rs2020854 | 0.021 |
| 337 | STAT4 | STAT4S1 | 5564 | C/A | Intron3 | rs1031509 | 0.396 |
| 338 | STAT4S2 | 19 235 | G/A | Intron3 | rs1551443 | 0.214 | |
| 339 | STAT4S3 | 91 147 | A/T | Intron10 | New | 0.516 | |
| 340 | STAT4S4 | 100 038 | G/C | Intron14 | rs1400655 | 0.095 | |
| 341 | STAT4S5 | 118 213 | A/G | Intron21 | rs925847 | 0.553 | |
| 342 | STAT4S6 | 119 708 | A/C | Intron22 | rs1517351 | 0.495 | |
| 343 | STAT4S7 | 119 711 | G/A | Intron22 | New | 0.165 | |
| 344 | TBX21 | TBX21S1 | 8941 | T/C | Intron1 | rs2158079 | 0.201 |
| 345 | TBXA2R | TBXA2RS1 | 10937 | T/C | Exon3-synonymous | rs4523 | 0.184 |
| 346 | TXAs | TXAsS2 | 17778 | A/C | Intron1 | rs41708 | 0.302 |
| 347 | TXAsS3 | 24573 | T/C | Intron1 | rs41706 | 0.355 | |
| 348 | TXAsS4 | 29218 | A/T | Intron1 | rs194150 | 0.443 | |
| 349 | TXAsS5 | 57546 | T/G | Intron3 | rs1015571 | 0.447 | |
| 350 | TXAsS7 | 63115 | C/A | Intron3 | rs2013219 | 0.426 | |
| 351 | TXAsS9 | 69322 | G/A | Intron3 | rs757762 | 0.447 | |
| 352 | TXAsS11 | 80249 | C/T | Intron3 | rs1978180 | 0.028 | |
| 353 | TXAsS13 | 1 06 385 | T/C | Intron4 | rs41733 | 0.117 | |
| 354 | TXAsS14 | 1 06 401 | G/A | Intron4 | rs41732 | 0.117 | |
| 355 | TXAsS15 | 1 12 521 | T/C | Intron5 | New | 0.247 | |
| 356 | TXAsS16 | 1 12 694 | C/T | Intron5 | rs42335 | 0.134 | |
| 357 | TXAsS17 | 1 15 453 | G/A | Intron5 | New | 0.011 | |
| 358 | TXAsS21 | 1 28 260 | G/T | Intron7 | New | 0.165 | |
| 359 | TXAsS22 | 1 39 370 | T/C | Intron9 | rs41718 | 0.146 | |
| 360 | TXAsS23 | 1 46 442 | T/C | Intron9 | rs740150 | 0.389 | |
| 361 | TXAsS24 | 1 52 303 | A/G | Intron9 | rs193949 | 0.177 | |
| 362 | TXAsS25 | 1 52 455 | C/T | Intron9 | New | 0.308 | |
| 363 | TXAsS29 | 1 78 072 | G/A | Intron10 | rs740204 | 0.234 | |
| 364 | TXAsS30 | 1 86 274 | G/A | Intron10 | New | 0.479 | |
| 365 | UPAR | UPARS1 | 11 475 | A/C | Intron3 | New | 0.019 |
| 366 | UPARS2 | 11 639 | G/A | Intron3 | rs2283628 | 0.335 | |
| 367 | UPARS3 | 11 667 | A/C | Intron3 | rs2239373 | 0.385 | |
| 368 | UPARS4 | 11 746 | C/T | Intron3 | rs2239372 | 0.375 | |
| 369 | UPARS5 | 18 100 | C/T | Intron5 | rs2302525 | 0.074 | |
| 370 | UPARS6 | 18 228 | A/G | Exon6-non-synonymous (K220R) | rs2302524 | 0.114 |
The SNPs applied to the first screening are listed. The sequence position indicates the location of the SNP relative to the transcription initiation site of exon1of each gene. The variation of allele and the localization in gene structure are shown. Each given SNP is referenced an accession number in dbSNP created by NCBI, rs ID (http://www.ncbi.nlm.nih.gov/SNP/). The New findings are mentioned as ‘new’. The minor allele frequency of each SNP typed in the control sample of 96 individuals is shown.
SNPs applied to the first screening
| No. . | Gene . | SNP . | Sequence position . | Variation . | Localization . | rs ID . | Minor allele frequncies (96 CTR) . |
|---|---|---|---|---|---|---|---|
| 1 | ADAM33 | ADAM33S1 | 11 513 | C/A | Intron19 | rs44707 | 0.467 |
| 2 | ADAM33S2 | 12 505 | T/C | Exon20-non-synonymous (M764T) | rs2280091 | 0.098 | |
| 3 | ADAM33S3 | 12 534 | T/C | Exon20-non-synonymous (M774S) | rs2280090 | 0.102 | |
| 4 | ADAM33S4 | 12 612 | T/C | Intron20 | rs2280089 | 0.092 | |
| 5 | ADAM33S5 | 13 018 | G/A | Intron21 | rs628977 | 0.362 | |
| 6 | ADAM33S6 | 13 026 | T/C | Intron21 | rs628965 | 0.362 | |
| 7 | ADAM33S7 | 13 060 | A/C | Intron21 | rs543749 | 0.181 | |
| 8 | ALOX12B | ALOX12BS1 | 1963 | G/A | Intron2 | rs3027303 | 0.189 |
| 9 | ALOX12BS4 | 3276 | G/T | Intron2 | rs3027294 | 0.391 | |
| 10 | ALOX12BS5 | 7053 | G/C | Intron4 | rs2304908 | 0.392 | |
| 11 | ALOX12BS6 | 7223 | C/T | Intron4 | rs2304907 | 0.396 | |
| 12 | ALOX12BS7 | 7341 | C/T | Intron4 | rs2304906 | 0.130 | |
| 13 | ALOX5 | ALOX5S1 | 9223 | A/G | Intron2 | rs4769060 | 0.151 |
| 14 | ALOX5S2 | 26 904 | G/A | Intron3 | New | 0.401 | |
| 15 | ALOX5S3 | 50 760 | G/A | Intron4 | New | 0.198 | |
| 16 | ALOX5S4 | 59 443 | A/G | Intron6 | New | 0.193 | |
| 17 | ALOX5S5 | 69 751 | G/C | Intron7 | New | 0.354 | |
| 18 | CLP | CLPS1 | 1152 | G/A | Intron2 | rs2966305 | 0.401 |
| 19 | CLPS2 | 3607 | C/A | Intron2 | New | 0.032 | |
| 20 | CLPS3 | 3688 | A/G | Intron2 | rs2967871 | 0.315 | |
| 21 | CLPS4 | 9097 | C/T | Intron2 | rs2925050 | 0.495 | |
| 22 | CLPS5 | 9197 | T/G | Intron2 | New | 0.457 | |
| 23 | CLPS6 | 11 744 | T/C | Intron2 | rs1835156 | 0.333 | |
| 24 | CLPS7 | 17 612 | T/C | Intron2 | New | 0.229 | |
| 25 | CLPS8 | 17 865 | G/C | Intron2 | New | 0.356 | |
| 26 | CLPS9 | 17 923 | G/C | Intron2 | rs2967876 | 0.284 | |
| 27 | CLPS10 | 20 308 | C/T | Intron2 | rs2925064 | 0.263 | |
| 28 | CLPS11 | 20 380 | T/C | Intron2 | rs934166 | 0.468 | |
| 29 | CLPS12 | 20 412 | G/A | Intron2 | rs201854 | 0.447 | |
| 30 | CLPS13 | 23 054 | T/C | Intron2 | New | 0.104 | |
| 31 | CLPS17 | 27 698 | A/G | Intron2 | rs2288584 | 0.250 | |
| 32 | CLPS18 | 31 174 | T/C | Intron3 | rs2967855 | 0.283 | |
| 33 | CLPS19 | 31 262 | C/G | Intron3 | New | 0.214 | |
| 34 | CLPS20 | 31 615 | C/T | Intron3 | New | 0.208 | |
| 35 | CLPS21 | 43 186 | A/T | Intron3 | rs2914823 | 0.185 | |
| 36 | CLPS22 | 43 360 | T/G | Intron3 | New | 0.404 | |
| 37 | CLPS23 | 43 394 | A/G | Intron3 | New | 0.250 | |
| 38 | CLPS24 | 47 961 | G/A | Intron3 | New | 0.116 | |
| 39 | CLPS25 | 48 577 | C/T | Intron3 | New | 0.347 | |
| 40 | CLPS26 | 51794 | C/T | Exon4-UTR | rs247862 | 0.297 | |
| 41 | CNOT3 | CNOT3S1 | 328 | C/T | Intron1 | rs42318 | 0.115 |
| 42 | CTSG | CTSGS1 | 308 | G/A | Intron1 | rs2236742 | 0.212 |
| 43 | CTSGS2 | 527 | A/G | Intron1 | New | 0.034 | |
| 44 | CTSGS3 | 726 | A/G | Intron1 | rs1957523 | 0.477 | |
| 45 | CTSGS4 | 1226 | C/T | Intron2 | New | 0.034 | |
| 46 | CTSGS5 | 1349 | G/A | Intron2 | rs2070697 | 0.314 | |
| 47 | CTSGS6 | 1767 | A/G | Exon4-synonymous | New | 0.182 | |
| 48 | CYP4F2 | CYP4F2S1 | 4937 | A/T | Intron1 | New | 0.349 |
| 49 | CYP4F2S4 | 18 733 | T/C | Intron1 | rs2072269 | 0.335 | |
| 50 | CYP4F2S5 | 18 810 | G/C | Intron1 | rs1064796 | 0.495 | |
| 51 | CYP4F2S6 | 30 715 | G/A | Intron1 | rs2018460 | 0.170 | |
| 52 | CYP4F3 | CYP4F3S0 | 3187 | G/A | Intron2 | rs2203998 | 0.328 |
| 53 | CYP4F3S0.5 | 6221 | C/T | Intron4 | rs1290626 | 0.141 | |
| 54 | CYP4F3S1 | 8507 | C/T | Intron6 | rs2283612 | 0.189 | |
| 55 | CYP4F3S2 | 11 871 | A/C | Intron8 | rs2733750 | 0.183 | |
| 56 | CYP4F3S3 | 12 015 | A/G | Exon9-synonymous | New | 0.363 | |
| 57 | CYP4F3S4 | 12 808 | G/A | Intron9 | rs2733752 | 0.136 | |
| 58 | CYP4F3S5 | 12 877 | T/A | Intron9 | New | 0.140 | |
| 59 | CYP4F3S6 | 17 772 | T/C | Intron11 | New | 0.453 | |
| 60 | CYP4F8 | CYP4F8S1 | 3610 | C/T | Intron2 | rs2072599 | 0.168 |
| 61 | CYP4F8S2 | 6771 | G/T | Intron5 | rs2072601 | 0.276 | |
| 62 | CYP4F8S3 | 13 568 | C/T | Intron11 | rs2239366 | 0.276 | |
| 63 | CYSLT1R | CYSLT1RS1 | 927 | C/T | Exon1-synonymous | rs320995 | 0.474 |
| 64 | CYSLT2R | CYSLT2RS1 | 2797 | A/G | Exon1-UTR | rs912277 | 0.417 |
| 65 | CYSLT2RS2 | 3078 | A/C | Exon1-UTR | rs1323552 | 0.489 | |
| 66 | CYSLT2RS3 | 3105 | A/G | Exon1-UTR | New | 0.116 | |
| 67 | FLAP | FLAPS1 | 162 | C/A | Intron1 | rs4769055 | 0.522 |
| 68 | FLAPS2 | 838 | T/G | Intron1 | rs9579645 | 0.096 | |
| 69 | FLAPS3 | 7272 | A/G | Intron1 | rs9551960 | 0.339 | |
| 70 | FLAPS4 | 8640 | A/C | Intron2 | rs3803277 | 0.406 | |
| 71 | FLAPS5 | 8733 | T/C | Intron2 | rs3803278 | 0.370 | |
| 72 | FLAPS6 | 13 674 | G/A | Intron2 | rs4075692 | 0.286 | |
| 73 | FLAPS7 | 20 616 | G/C | Intron3 | New | 0.286 | |
| 74 | FLAPS8 | 20 648 | C/T | Intron3 | rs4468448 | 0.240 | |
| 75 | FLAPS9 | 23 849 | T/A | Intron3 | rs9551964 | 0.277 | |
| 76 | FLAPS9.5 | 24 348 | G/A | Intron3 | New | 0.006 | |
| 77 | FLAPS10 | 28 209 | A/G | Intron3 | rs4769060 | 0.401 | |
| 78 | HPGD | HPGDS1 | 464 | G/A | Exon2-synonymous | rs1050145 | 0.220 |
| 79 | HPGDS2 | 798 | A/G | Intron2 | rs1365613 | 0.447 | |
| 80 | HPGDS3 | 13 332 | A/C | Intron3 | rs2555629 | 0.405 | |
| 81 | HPGDS4 | 13 430 | A/T | Intron3 | New | 0.253 | |
| 82 | HPGDS5 | 20 442 | A/G | Intron4 | New | 0.446 | |
| 83 | HPGDS6 | 20 516 | G/A | Intron4 | New | 0.110 | |
| 84 | IGF1 | IGF1S1 | 3083 | A/G | Intron2 | rs2162679 | 0.368 |
| 85 | IGF1S2 | 9917 | G/T | Intron3 | rs1019731 | 0.021 | |
| 86 | IGF1S3 | 17 640 | G/C | Intron3 | rs2195239 | 0.426 | |
| 87 | IGF1S4 | 17 695 | T/C | Intron3 | rs2195240 | 0.443 | |
| 88 | IGF1S5 | 49 421 | G/A | Intron3 | rs972936 | 0.484 | |
| 89 | IGF1S6 | 60 710 | G/A | Intron3 | rs2072592 | 0.245 | |
| 90 | IGF1S7 | 72 103 | G/A | Intron5 | rs978458 | 0.135 | |
| 91 | IGF1S8 | 84 150 | G/A | Exon6-UTR | rs6219 | 0.271 | |
| 92 | IL13 | IL13S1 | −978 | C/T | Promoter | rs11575055 | 0.179 |
| 93 | IL13S2 | 571 | C/A | Intron1 | rs2066960 | 0.281 | |
| 94 | IL13S3 | 598 | G/C | Intron1 | rs1295987 | 0.120 | |
| 95 | IL13S4 | 805 | C/T | Intron1 | rs2069744 | 0.120 | |
| 96 | IL13S5 | 2100 | G/A | Exon4-non-synonymous (R144Q) | rs20541 | 0.333 | |
| 97 | IL4 | IL4S1 | −219 | T/C | Promoter | rs2243250 | 0.328 |
| 98 | IL4S2 | 3353 | A/C | Intron2 | rs2227284 | 0.255 | |
| 99 | IL4S3 | 3927 | C/G | Intron2 | rs2243263 | 0.073 | |
| 100 | LTA4H | LTA4HS1 | 9153 | A/G | Intron3 | rs763842 | 0.226 |
| 101 | LTA4HS4 | 20 165 | T/C | Intron11 | rs1978331 | 0.335 | |
| 102 | LTB4DH | LTB4DHS1 | 4435 | A/T | Intron2 | New | 0.506 |
| 103 | LTB4DHS2 | 8896 | A/G | Intron4 | New | 0.283 | |
| 104 | LTB4DHS2.5 | 9182 | A/G | Intron4 | New | 0.092 | |
| 105 | LTB4DHS5 | 9502 | C/T | Intron4 | rs1053968 | 0.005 | |
| 106 | LTB4DHS6 | 13 234 | C/G | Intron4 | New | 0.319 | |
| 107 | LTB4DHS6.5 | 13 249 | A/G | Intron4 | New | 0.231 | |
| 108 | LTB4DHS7 | 20 718 | G/A | Intron6 | New | 0.253 | |
| 109 | LTB4DHS8 | 25 971 | A/G | Intron8 | rs1322258 | 0.347 | |
| 110 | LTB4DHS9 | 26 221 | A/G | Intron8 | New | 0.406 | |
| 111 | LTB4DHS10 | 35 589 | A/C | Intron9 | rs2146078 | 0.229 | |
| 112 | LTB4DHS11 | 35 819 | G/A | Intron9 | New | 0.354 | |
| 113 | LTB4R | LTB4RS1 | 1165 | G/C | Exon1-UTR | New | 0.140 |
| 114 | LTC4S | LTC4SS1.5 | −348 | A/C | Promoter | New | 0.208 |
| 115 | LTC4SS2 | 289 | G/C | Intron1 | New | 0.026 | |
| 116 | LTC4SS3 | 1296 | C/T | Intron1 | New | 0.208 | |
| 117 | LTC4SS4 | 2659 | A/G | Intron5 | New | 0.201 | |
| 118 | MGST2 | MGST2S1 | 815 | A/G | Intron1 | rs1000222 | 0.292 |
| 119 | MGST2S2 | 12 971 | C/T | Intron2 | rs795589 | 0.198 | |
| 120 | MGST2S3 | 12 977 | A/C | Intron2 | rs795588 | 0.077 | |
| 121 | MMP1 | MMP1S1 | 193 | G/A | Intron1 | rs470358 | 0.292 |
| 122 | MMP1S2 | 2579 | G/A | Exon5-synonymous | rs470558 | 0.130 | |
| 123 | MMP1S3 | 7300 | T/C | Intron8 | rs470747 | 0.137 | |
| 124 | MMP1S4 | 7815 | C/T | Exon10-UTR | rs2239008 | 0.319 | |
| 125 | MMP1S5 | 7936 | T/C | Exon10-UTR | rs2071230 | 0.298 | |
| 126 | MMP2 | MMP2S1 | 3606 | A/T | Intron1 | rs857403 | 0.214 |
| 127 | MMP2S2 | 3665 | G/A | Intron1 | rs1030868 | 0.229 | |
| 128 | MMP2S3 | 6505 | C/T | Exon5-synonymous | rs1053605 | 0.208 | |
| 129 | MMP2S4 | 6730 | T/G | Intron5 | New | 0.064 | |
| 130 | MMP2S5 | 6958 | A/G | Intron5 | rs866770 | 0.151 | |
| 131 | MMP2S6 | 10 603 | C/T | Exon7-synonymous | rs243849 | 0.151 | |
| 132 | MMP2S7 | 10 896 | T/C | Intron7 | rs243847 | 0.443 | |
| 133 | MMP2S8 | 14 011 | G/A | Exon9-synonymous | rs2287074 | 0.326 | |
| 134 | MMP2S9 | 14 196 | G/A | Intron9 | rs243843 | 0.306 | |
| 135 | MMP2S10 | 14 281 | G/A | Intron9 | New | 0.065 | |
| 136 | MMP2S11 | 14 320 | T/C | Intron9 | rs243842 | 0.364 | |
| 137 | MMP2S12 | 17 660 | G/T | Intron9 | rs171498 | 0.363 | |
| 138 | MMP2S13 | 17 670 | G/T | Intron9 | rs243838 | 0.153 | |
| 139 | MMP2S14 | 17 928 | C/T | Intron10 | New | 0.057 | |
| 140 | MMP2S15 | 20 976 | G/A | Intron11 | New | 0.115 | |
| 141 | MMP2S16 | 21 134 | G/A | Intron11 | rs243836 | 0.386 | |
| 142 | MMP2S17 | 26 345 | C/T | Exon13-UTR | New | 0.021 | |
| 143 | MMP2S18 | 26 512 | A/C | Exon13-UTR | rs7201 | 0.234 | |
| 144 | MMP8 | MMP8S1 | 524 | G/A | Intron1 | rs1939012 | 0.359 |
| 145 | MMP8S2 | 5458 | A/G | Intron4 | rs1940051 | 0.266 | |
| 146 | MMP8S3 | 11 357 | T/A | Intron9 | New | 0.287 | |
| 147 | MMP8S4 | 11 473 | T/C | Exon10-synonymous | New | 0.016 | |
| 148 | MMP9 | MMP9S1 | 2679 | G/A | Exon6-non-synonymous (R279Q) | rs2664538 | 0.382 |
| 149 | MMP9S2 | 3029 | A/C | Intron6 | rs2236416 | 0.179 | |
| 150 | MMP9S3 | 5565 | G/A | Exon12-non-synonymous (R668Q) | rs2274756 | 0.214 | |
| 151 | MMP9S4 | 7419 | G/A | Exon13-synonymous | rs13925 | 0.292 | |
| 152 | P2Y10 | P2Y10S1 | 1192 | A/G | Intron1 | New | 0.016 |
| 153 | P2Y10S2 | 5030 | C/T | Intron1 | rs2858570 | 0.110 | |
| 154 | P2Y10S3 | 5318 | A/T | Intron1 | rs2251477 | 0.184 | |
| 155 | P2Y10S4 | 9417 | T/C | Intron1 | rs2742205 | 0.115 | |
| 156 | PAFAH1B1 | PAFAH1B1S2 | 12 145 | G/T | Intron1 | New | 0.099 |
| 157 | PAFAH1B1S3 | 21 146 | C/G | Intron1 | New | 0.080 | |
| 158 | PAFAH1B1S4 | 37 780 | A/G | Intron1 | rs1266474 | 0.084 | |
| 159 | PAFAH1B1S5 | 38 033 | C/T | Intron1 | New | 0.047 | |
| 160 | PAFAH1B1S6 | 40 522 | G/A | Intron1 | New | 0.130 | |
| 161 | PAFAH1B1S7 | 60 034 | C/T | Intron2 | New | 0.266 | |
| 162 | PAFAH1B1S8 | 68 545 | G/C | Intron2 | New | 0.078 | |
| 163 | PAFAH1B1S9 | 76 403 | T/G | Intron5 | New | 0.032 | |
| 164 | PAFAH1B2 | PAFAH1B2S1 | 3971 | T/C | Intron1 | rs2008908 | 0.446 |
| 165 | PAFAH1B3 | PAFAH1B3S1 | 2538 | C/T | Exon4-synonymous | New | 0.016 |
| 166 | PAFAH2 | PAFAH2S1 | 1279 | G/T | Intron1 | rs3008423 | 0.172 |
| 167 | PAFAH2S2 | 4171 | C/T | Intron1 | New | 0.036 | |
| 168 | PAFAH2S3 | 15456 | G/T | Intron5 | rs1469512 | 0.214 | |
| 169 | PAI1m2 | PAI1m2S1 | 4484 | G/A | Intron1 | rs840088 | 0.401 |
| 170 | PAI1m2S2 | 5094 | A/G | Intron1 | New | 0.089 | |
| 171 | PAI1m2S3 | 19 022 | C/A | Intron1 | rs2099601 | 0.201 | |
| 172 | PAI1m2S4 | 19 047 | A/C | Intron1 | New | 0.201 | |
| 173 | PAI1m2S5 | 19 120 | T/C | Intron1 | rs2083120 | 0.201 | |
| 174 | PAI1m2S6 | 19 348 | A/C | Intron1 | New | 0.393 | |
| 175 | PAI1m2S7 | 62 178 | A/G | Intron8 | New | 0.271 | |
| 176 | PDGFB | PDGFBS1 | 581 | G/A | Intron1 | rs758588 | 0.184 |
| 177 | PDGFBS3 | 20 237 | C/T | Intron3 | rs740750 | 0.310 | |
| 178 | PDGFBS4 | 26 041 | C/T | Intron10 | rs1864972 | 0.198 | |
| 179 | PDGFBS5 | 35 170 | G/A | Intron18 | rs1432878 | 0.108 | |
| 180 | PDGFRL | PDGFRLS1 | 978 | G/C | Intron1 | rs2720576 | 0.365 |
| 181 | PDGFRLS2 | 1187 | C/T | Intron1 | rs2588164 | 0.245 | |
| 182 | PDGFRLS3 | 1416 | T/C | Intron1 | rs2588163 | 0.391 | |
| 183 | PDGFRLS4 | 2963 | G/C | Intron1 | rs2517267 | 0.458 | |
| 184 | PDGFRLS5 | 3794 | C/G | Intron1 | rs2517268 | 0.394 | |
| 185 | PDGFRLS6 | 3882 | G/A | Intron1 | New | 0.216 | |
| 186 | PDGFRLS7 | 3999 | A/G | Intron1 | rs2517269 | 0.220 | |
| 187 | PDGFRLS8 | 8893 | G/A | Intron1 | New | 0.226 | |
| 188 | PDGFRLS9 | 9922 | G/A | Intron1 | New | 0.232 | |
| 189 | PDGFRLS10 | 14 053 | G/C | Intron2 | rs2720583 | 0.167 | |
| 190 | PDGFRLS11 | 14 194 | A/C | Intron2 | New | 0.396 | |
| 191 | PDGFRLS12 | 18 220 | T/C | Intron2 | rs2427709 | 0.140 | |
| 192 | PDGFRLS13 | 18 473 | G/A | Intron2 | rs2588144 | 0.068 | |
| 193 | PDGFRLS15 | 19 592 | T/G | Intron2 | rs2246488 | 0.339 | |
| 194 | PDGFRLS16 | 28 052 | G/A | Intron2 | rs2517187 | 0.104 | |
| 195 | PDGFRLS17 | 28 449 | C/T | Intron2 | rs2237823 | 0.104 | |
| 196 | PDGFRLS18 | 31 111 | T/C | Intron2 | rs2517198 | 0.094 | |
| 197 | PDGFRLS19 | 31 230 | A/T | Intron2 | New | 0.260 | |
| 198 | PDGFRLS20 | 31 258 | A/G | Intron2 | New | 0.031 | |
| 199 | PDGFRLS21 | 31 499 | T/C | Intron2 | rs2427715 | 0.094 | |
| 200 | PDGFRLS22 | 31 600 | T/C | Intron2 | New | 0.167 | |
| 201 | PDGFRLS23 | 33 975 | G/C | Intron2 | rs2517208 | 0.037 | |
| 202 | PDGFRLS24 | 41 991 | A/G | Intron2 | rs2237831 | 0.189 | |
| 203 | PDGFRLS25 | 42 131 | G/A | Intron2 | rs2237831 | 0.205 | |
| 204 | PDGFRLS26 | 43 810 | G/T | Intron2 | New | 0.073 | |
| 205 | PDGFRLS27 | 46 759 | T/C | Intron3 | New | 0.120 | |
| 206 | PDGFRLS28 | 46 888 | G/A | Intron3 | rs2237835 | 0.226 | |
| 207 | PDGFRLS29 | 46 936 | C/G | Intron3 | New | 0.074 | |
| 208 | PDGFRLS30 | 47 097 | C/G | Intron3 | New | 0.197 | |
| 209 | PDGFRLS31 | 49 411 | A/C | Intron3 | rs2237836 | 0.548 | |
| 210 | PDGFRLS32 | 49 937 | G/T | Intron3 | rs2237837 | 0.103 | |
| 211 | PDGFRLS33 | 53 887 | T/C | Intron4 | rs2237842 | 0.188 | |
| 212 | PDGFRLS34 | 56 021 | C/T | Intron4 | rs2427719 | 0.063 | |
| 213 | PDGFRLS35 | 56 410 | T/C | Intron4 | rs2237845 | 0.240 | |
| 214 | PDGFRLS36 | 65 408 | C/T | Intron5 | New | 0.083 | |
| 215 | PDGFRLS37 | 65 512 | T/C | Exon6-synonymous | rs4705 | 0.521 | |
| 216 | PGDS | PGDSS1 | 5446 | T/C | Intron1 | rs2129595 | 0.191 |
| 217 | PGIS | PGISS1 | 4650 | T/C | Intron1 | rs477627 | 0.063 |
| 218 | PGISS2 | 6734 | A/C | Intron1 | rs927068 | 0.226 | |
| 219 | PGISS3 | 6840 | T/C | Intron1 | New | 0.226 | |
| 220 | PGISS4 | 6870 | G/C | Intron1 | rs498646 | 0.276 | |
| 221 | PGISS4.5 | 6924 | T/C | Intron1 | rs476496 | 0.280 | |
| 222 | PGISS6 | 28 941 | A/G | Intron5 | rs501908 | 0.033 | |
| 223 | PGISS7 | 41 990 | A/G | Intron5 | New | 0.093 | |
| 224 | PGISS8 | 44 026 | G/A | Exon6-synonymous | rs5628 | 0.036 | |
| 225 | PGISS9 | 50 926 | C/T | Intron6 | New | 0.104 | |
| 226 | PGISS10 | 55 002 | C/A | Intron8 | rs5629 | 0.281 | |
| 227 | PGISS11 | 57 532 | A/G | Intron9 | rs729824 | 0.226 | |
| 228 | PGISS12 | 62 730 | T/C | Exon10-UTR | rs5602 | 0.438 | |
| 229 | PLA2G7 | PLA2G7S1 | 2007 | G/T | Intron1 | New | 0.214 |
| 230 | PLA2G7S2 | 2338 | G/A | Intron1 | rs1421369 | 0.468 | |
| 231 | PLA2G7S3 | 6203 | G/A | Intron1 | New | 0.141 | |
| 232 | PLA2G7S4 | 20 965 | G/T | Intron5 | rs1362931 | 0.042 | |
| 233 | PLA2G7S5 | 23 741 | T/C | Exon7-non-synonymous (I198T) | rs1805018 | 0.281 | |
| 234 | PLA2G7S6 | 27 025 | C/G | Intron9 | rs2216465 | 0.542 | |
| 235 | PLA2G7S7 | 30 101 | C/T | Exon11-non-synonymous (A379V) | rs1051931 | 0.042 | |
| 236 | PTAFR | PTAFRS3 | 36 031 | C/A | Exon2-non-synonymous (A224D) | rs5938 | 0.068 |
| 237 | PTGDR | PTGDRS1 | 408 | G/A | Intron1 | New | 0.021 |
| 238 | PTGDRS2 | 3290 | A/G | Intron1 | rs1254609 | 0.194 | |
| 239 | PTGDRS3 | 5754 | T/C | Intron1 | New | 0.075 | |
| 240 | PTGDRS4 | 5793 | A/G | Intron1 | rs708486 | 0.081 | |
| 241 | PTGDS | PTGDSS1 | 4186 | C/A | Exon7-UTR | rs6926 | 0.198 |
| 242 | EP1 | PTGER1S1 | −267 | G/C | Promoter | New | 0.109 |
| 243 | EP2 | PTGER2uS1 | −26 643 | T/C | 5′-Upstream | rs988209 | 0.232 |
| 244 | PTGER2uS2 | −18 461 | T/G | 5′-Upstream | rs1390375 | 0.077 | |
| 245 | PTGER2uS3 | −18 360 | G/A | 5′-Upstream | rs1390374 | 0.022 | |
| 246 | PTGER2uS4 | −15 332 | A/T | 5′-Upstream | rs708490 | 0.444 | |
| 247 | PTGER2uS5 | −12 813 | G/A | 5′-Upstream | New | 0.223 | |
| 248 | PTGER2uS6 | −10 918 | A/G | 5′-Upstream | New | 0.449 | |
| 249 | PTGER2uS7 | −10 814 | T/A | 5′-upstream | New | 0.427 | |
| 250 | PTGER2uS8 | −10 250 | A/G | 5′-upstream | rs714366 | 0.433 | |
| 251 | PTGER2uS9 | −7075 | A/G | 5′-upstream | New | 0.452 | |
| 252 | PTGER2uS10 | −6179 | A/G | 5′-upstream | New | 0.479 | |
| 253 | PTGER2S1 | −609 | G/A | Promoter | rs1254601 | 0.288 | |
| 254 | PTGER2S2 | 300 | G/A | Exon1-UTR | rs1254600 | 0.462 | |
| 255 | PTGER2S3 | 498 | C/G | Exon1-UTR | rs2075797 | 0.441 | |
| 256 | PTGER2S4 | 948 | A/G | Exon1-UTR | rs1353411 | 0.417 | |
| 257 | PTGER2S5 | 1042 | G/A | Exon1-UTR | rs1254598 | 0.350 | |
| 258 | PTGER2S6 | 2803 | G/A | Intron1 | New | 0.164 | |
| 259 | PTGER2S7 | 2988 | C/T | Intron1 | New | 0.339 | |
| 260 | PTGER2S8 | 6063 | C/T | Intron1 | New | 0.116 | |
| 261 | PTGER2S9 | 10 927 | T/G | Intron1 | rs1254585 | 0.446 | |
| 262 | PTGER2S10 | 14 081 | C/T | Exon2-UTR | rs708502 | 0.398 | |
| 263 | PTGER2dS1 | 29 784 | C/T | 3′-Downstream | rs708511 | 0.219 | |
| 264 | PTGER2dS2 | 47 461 | A/G | 3′-Downstream | New | 0.226 | |
| 265 | PTGER2dS3 | 57 931 | C/T | 3′-Downstream | rs708531 | 0.750 | |
| 266 | PTGER2dS4 | 58 051 | C/T | 3′-Downstream | rs708532 | 0.329 | |
| 267 | EP3 | PTGER3S1 | 27 837 | G/A | Intron1 | rs1008484 | 0.058 |
| 268 | PTGER3S2 | 28 078 | G/A | Intron1 | rs1569593 | 0.258 | |
| 269 | PTGER3S5 | 36 177 | A/T | Intron2 | rs5680 | 0.313 | |
| 270 | PTGER3S7 | 44 999 | T/G | Intron2 | rs1983588 | 0.268 | |
| 271 | PTGER3S8 | 45 040 | A/C | Intron2 | rs1983587 | 0.059 | |
| 272 | PTGER3S9 | 54 444 | G/C | Intron2 | rs1883461 | 0.054 | |
| 273 | PTGER3S10 | 54 634 | T/A | Intron2 | rs1883460 | 0.094 | |
| 274 | PTGER3S11 | 58 525 | C/T | Intron2 | New | 0.271 | |
| 275 | PTGER3S12 | 65 699 | G/C | Intron2 | rs647921 | 0.048 | |
| 276 | PTGER3S13 | 65 973 | A/T | Intron2 | rs646621 | 0.443 | |
| 277 | PTGER3S14 | 70 342 | G/A | Intron2 | rs909842 | 0.442 | |
| 278 | PTGER3S15 | 70 357 | A/C | Intron2 | New | 0.300 | |
| 279 | PTGER3S17 | 70 409 | G/A | Intron2 | New | 0.442 | |
| 280 | PTGER3S20 | 70 755 | A/T | Intron2 | rs484675 | 0.441 | |
| 281 | PTGER3S24 | 86 923 | A/G | Intron2 | rs573688 | 0.082 | |
| 282 | PTGER3S25 | 95 146 | A/C | Intron3 | New | 0.037 | |
| 283 | PTGER3S27 | 1 00 052 | T/C | Intron3 | rs1409984 | 0.255 | |
| 284 | PTGER3S29 | 1 14 078 | G/A | Intron3 | rs625617 | 0.371 | |
| 285 | PTGER3S30 | 1 21 798 | C/A | Intron3 | New | 0.328 | |
| 286 | PTGER3S31 | 1 21 846 | G/A | Intron3 | rs602383 | 0.297 | |
| 287 | PTGER3S32 | 1 49 402 | C/T | Intron3 | rs1409165 | 0.370 | |
| 288 | PTGER3S33 | 1 49 626 | G/A | Intron3 | rs1409166 | 0.398 | |
| 289 | PTGER3S35 | 1 60 372 | A/G | Intron3 | New | 0.214 | |
| 290 | PTGER3S36 | 1 60 403 | A/G | Intron3 | rs1409978 | 0.319 | |
| 291 | PTGER3S37 | 1 60 432 | C/G | Intron3 | New | 0.214 | |
| 292 | EP4 | PTGER4S1 | 5748 | C/T | Intron2 | New | 0.389 |
| 293 | PTGER4S2 | 7984 | A/G | Intron2 | New | 0.198 | |
| 294 | PTGER4S3 | 8012 | G/A | Intron2 | New | 0.385 | |
| 295 | PTGES | PTGESS1 | 214 | A/G | Intron1 | rs2241271 | 0.135 |
| 296 | PTGESS2 | 406 | G/A | Intron1 | rs2241270 | 0.120 | |
| 297 | PTGESS3 | 9636 | T/C | Intron2 | New | 0.326 | |
| 298 | PTGESS4 | 9669 | G/A | Intron2 | New | 0.152 | |
| 299 | PTGFR | PTGFRS1 | 714 | A/C | Intron1 | rs3766355 | 0.425 |
| 300 | PTGFRS2 | 823 | G/A | Intron1 | rs3766354 | 0.204 | |
| 301 | PTGFRS3 | 1132 | G/T | Intron1 | rs3766353 | 0.242 | |
| 302 | PTGFRS4 | 3617 | G/A | Intron2 | rs1830763 | 0.489 | |
| 303 | PTGFRS5 | 6578 | A/C | Intron2 | rs1322935 | 0.031 | |
| 304 | PTGFRS6 | 8734 | A/G | Intron2 | rs2057423 | 0.452 | |
| 305 | PTGFRS10 | 24 552 | G/A | Intron2 | rs3766346 | 0.036 | |
| 306 | PTGFRS12 | 33 123 | A/C | Intron2 | rs520171 | 0.276 | |
| 307 | PTGFRS13 | 33 548 | G/C | Intron2 | rs3766345 | 0.516 | |
| 308 | PTGFRS14 | 38 195 | T/C | Intron2 | rs3766338 | 0.266 | |
| 309 | PTGFRS15 | 38 202 | C/T | Intron2 | rs590309 | 0.511 | |
| 310 | PTGFRS16 | 40 734 | G/C | Intron2 | rs622346 | 0.202 | |
| 311 | PTGFRS19 | 46 572 | A/G | Exon3-UTR | rs3766331 | 0.115 | |
| 312 | PTGFRS20 | 46 628 | A/G | Exon3-UTR | rs899 | 0.005 | |
| 313 | PTGS1 | PTGS1S1 | 2850 | C/T | Intron2 | rs1213264 | 0.063 |
| 314 | PTGS1S2 | 3219 | T/C | Intron2 | rs1213265 | 0.067 | |
| 315 | PTGS1S2.5 | 7468 | C/G | Intron3 | rs2282169 | 0.079 | |
| 316 | PTGS1S4 | 23 970 | C/A | Exon11-UTR | rs10306194 | 0.028 | |
| 317 | SCYA5 | SCYA5S1 | 328 | T/C | Intron1 | rs2280789 | 0.349 |
| 318 | SCYB14 | SCYB14S1 | 313 | T/C | Intron1 | rs2072347 | 0.311 |
| 319 | SCYB14S2 | 2099 | G/C | Intron2 | rs2237062 | 0.307 | |
| 320 | SCYB14S3 | 6261 | C/T | Intron3 | rs1016666 | 0.370 | |
| 321 | SLC21A9 | SLC21A9S1 | 196 | T/C | Exon1-UTR | New | 0.158 |
| 322 | SLC21A9S2 | 231 | G/A | Exon1-UTR | rs1944612 | 0.055 | |
| 323 | SLC21A9S3 | 4589 | C/T | Intron1 | New | 0.443 | |
| 324 | SLC21A9S5 | 10 724 | A/G | Intron1 | New | 0.374 | |
| 325 | SLC21A9S6 | 13 387 | G/C | Intron3 | New | 0.453 | |
| 326 | SLC21A9S7 | 15 413 | G/A | Intron4 | rs949069 | 0.391 | |
| 327 | SLC21A9S8 | 15 916 | A/G | Intron4 | New | 0.161 | |
| 328 | SLC21A9S9 | 17 493 | G/A | Intron4 | rs1676878 | 0.136 | |
| 329 | SLC21A9S10 | 24 698 | A/G | Intron7 | rs1676881 | 0.226 | |
| 330 | SLC21A9S10.5 | 24 880 | C/T | Intron7 | rs1612859 | 0.458 | |
| 331 | SLC21A9S11 | 25 005 | T/A | Intron7 | rs1789693 | 0.401 | |
| 332 | SLC21A9S13 | 26 261 | T/G | Intron7 | New | 0.376 | |
| 333 | SLC21A9S14 | 39 441 | G/A | Intron8 | rs1789692 | 0.479 | |
| 334 | SLC21A9S15 | 43 575 | C/T | Intron9 | New | 0.430 | |
| 335 | SLC21A9S16 | 45 422 | C/T | Exon10 | New | 0.432 | |
| 336 | STAT2 | STAT2S1 | 10 525 | A/G | Intron14 | rs2020854 | 0.021 |
| 337 | STAT4 | STAT4S1 | 5564 | C/A | Intron3 | rs1031509 | 0.396 |
| 338 | STAT4S2 | 19 235 | G/A | Intron3 | rs1551443 | 0.214 | |
| 339 | STAT4S3 | 91 147 | A/T | Intron10 | New | 0.516 | |
| 340 | STAT4S4 | 100 038 | G/C | Intron14 | rs1400655 | 0.095 | |
| 341 | STAT4S5 | 118 213 | A/G | Intron21 | rs925847 | 0.553 | |
| 342 | STAT4S6 | 119 708 | A/C | Intron22 | rs1517351 | 0.495 | |
| 343 | STAT4S7 | 119 711 | G/A | Intron22 | New | 0.165 | |
| 344 | TBX21 | TBX21S1 | 8941 | T/C | Intron1 | rs2158079 | 0.201 |
| 345 | TBXA2R | TBXA2RS1 | 10937 | T/C | Exon3-synonymous | rs4523 | 0.184 |
| 346 | TXAs | TXAsS2 | 17778 | A/C | Intron1 | rs41708 | 0.302 |
| 347 | TXAsS3 | 24573 | T/C | Intron1 | rs41706 | 0.355 | |
| 348 | TXAsS4 | 29218 | A/T | Intron1 | rs194150 | 0.443 | |
| 349 | TXAsS5 | 57546 | T/G | Intron3 | rs1015571 | 0.447 | |
| 350 | TXAsS7 | 63115 | C/A | Intron3 | rs2013219 | 0.426 | |
| 351 | TXAsS9 | 69322 | G/A | Intron3 | rs757762 | 0.447 | |
| 352 | TXAsS11 | 80249 | C/T | Intron3 | rs1978180 | 0.028 | |
| 353 | TXAsS13 | 1 06 385 | T/C | Intron4 | rs41733 | 0.117 | |
| 354 | TXAsS14 | 1 06 401 | G/A | Intron4 | rs41732 | 0.117 | |
| 355 | TXAsS15 | 1 12 521 | T/C | Intron5 | New | 0.247 | |
| 356 | TXAsS16 | 1 12 694 | C/T | Intron5 | rs42335 | 0.134 | |
| 357 | TXAsS17 | 1 15 453 | G/A | Intron5 | New | 0.011 | |
| 358 | TXAsS21 | 1 28 260 | G/T | Intron7 | New | 0.165 | |
| 359 | TXAsS22 | 1 39 370 | T/C | Intron9 | rs41718 | 0.146 | |
| 360 | TXAsS23 | 1 46 442 | T/C | Intron9 | rs740150 | 0.389 | |
| 361 | TXAsS24 | 1 52 303 | A/G | Intron9 | rs193949 | 0.177 | |
| 362 | TXAsS25 | 1 52 455 | C/T | Intron9 | New | 0.308 | |
| 363 | TXAsS29 | 1 78 072 | G/A | Intron10 | rs740204 | 0.234 | |
| 364 | TXAsS30 | 1 86 274 | G/A | Intron10 | New | 0.479 | |
| 365 | UPAR | UPARS1 | 11 475 | A/C | Intron3 | New | 0.019 |
| 366 | UPARS2 | 11 639 | G/A | Intron3 | rs2283628 | 0.335 | |
| 367 | UPARS3 | 11 667 | A/C | Intron3 | rs2239373 | 0.385 | |
| 368 | UPARS4 | 11 746 | C/T | Intron3 | rs2239372 | 0.375 | |
| 369 | UPARS5 | 18 100 | C/T | Intron5 | rs2302525 | 0.074 | |
| 370 | UPARS6 | 18 228 | A/G | Exon6-non-synonymous (K220R) | rs2302524 | 0.114 |
| No. . | Gene . | SNP . | Sequence position . | Variation . | Localization . | rs ID . | Minor allele frequncies (96 CTR) . |
|---|---|---|---|---|---|---|---|
| 1 | ADAM33 | ADAM33S1 | 11 513 | C/A | Intron19 | rs44707 | 0.467 |
| 2 | ADAM33S2 | 12 505 | T/C | Exon20-non-synonymous (M764T) | rs2280091 | 0.098 | |
| 3 | ADAM33S3 | 12 534 | T/C | Exon20-non-synonymous (M774S) | rs2280090 | 0.102 | |
| 4 | ADAM33S4 | 12 612 | T/C | Intron20 | rs2280089 | 0.092 | |
| 5 | ADAM33S5 | 13 018 | G/A | Intron21 | rs628977 | 0.362 | |
| 6 | ADAM33S6 | 13 026 | T/C | Intron21 | rs628965 | 0.362 | |
| 7 | ADAM33S7 | 13 060 | A/C | Intron21 | rs543749 | 0.181 | |
| 8 | ALOX12B | ALOX12BS1 | 1963 | G/A | Intron2 | rs3027303 | 0.189 |
| 9 | ALOX12BS4 | 3276 | G/T | Intron2 | rs3027294 | 0.391 | |
| 10 | ALOX12BS5 | 7053 | G/C | Intron4 | rs2304908 | 0.392 | |
| 11 | ALOX12BS6 | 7223 | C/T | Intron4 | rs2304907 | 0.396 | |
| 12 | ALOX12BS7 | 7341 | C/T | Intron4 | rs2304906 | 0.130 | |
| 13 | ALOX5 | ALOX5S1 | 9223 | A/G | Intron2 | rs4769060 | 0.151 |
| 14 | ALOX5S2 | 26 904 | G/A | Intron3 | New | 0.401 | |
| 15 | ALOX5S3 | 50 760 | G/A | Intron4 | New | 0.198 | |
| 16 | ALOX5S4 | 59 443 | A/G | Intron6 | New | 0.193 | |
| 17 | ALOX5S5 | 69 751 | G/C | Intron7 | New | 0.354 | |
| 18 | CLP | CLPS1 | 1152 | G/A | Intron2 | rs2966305 | 0.401 |
| 19 | CLPS2 | 3607 | C/A | Intron2 | New | 0.032 | |
| 20 | CLPS3 | 3688 | A/G | Intron2 | rs2967871 | 0.315 | |
| 21 | CLPS4 | 9097 | C/T | Intron2 | rs2925050 | 0.495 | |
| 22 | CLPS5 | 9197 | T/G | Intron2 | New | 0.457 | |
| 23 | CLPS6 | 11 744 | T/C | Intron2 | rs1835156 | 0.333 | |
| 24 | CLPS7 | 17 612 | T/C | Intron2 | New | 0.229 | |
| 25 | CLPS8 | 17 865 | G/C | Intron2 | New | 0.356 | |
| 26 | CLPS9 | 17 923 | G/C | Intron2 | rs2967876 | 0.284 | |
| 27 | CLPS10 | 20 308 | C/T | Intron2 | rs2925064 | 0.263 | |
| 28 | CLPS11 | 20 380 | T/C | Intron2 | rs934166 | 0.468 | |
| 29 | CLPS12 | 20 412 | G/A | Intron2 | rs201854 | 0.447 | |
| 30 | CLPS13 | 23 054 | T/C | Intron2 | New | 0.104 | |
| 31 | CLPS17 | 27 698 | A/G | Intron2 | rs2288584 | 0.250 | |
| 32 | CLPS18 | 31 174 | T/C | Intron3 | rs2967855 | 0.283 | |
| 33 | CLPS19 | 31 262 | C/G | Intron3 | New | 0.214 | |
| 34 | CLPS20 | 31 615 | C/T | Intron3 | New | 0.208 | |
| 35 | CLPS21 | 43 186 | A/T | Intron3 | rs2914823 | 0.185 | |
| 36 | CLPS22 | 43 360 | T/G | Intron3 | New | 0.404 | |
| 37 | CLPS23 | 43 394 | A/G | Intron3 | New | 0.250 | |
| 38 | CLPS24 | 47 961 | G/A | Intron3 | New | 0.116 | |
| 39 | CLPS25 | 48 577 | C/T | Intron3 | New | 0.347 | |
| 40 | CLPS26 | 51794 | C/T | Exon4-UTR | rs247862 | 0.297 | |
| 41 | CNOT3 | CNOT3S1 | 328 | C/T | Intron1 | rs42318 | 0.115 |
| 42 | CTSG | CTSGS1 | 308 | G/A | Intron1 | rs2236742 | 0.212 |
| 43 | CTSGS2 | 527 | A/G | Intron1 | New | 0.034 | |
| 44 | CTSGS3 | 726 | A/G | Intron1 | rs1957523 | 0.477 | |
| 45 | CTSGS4 | 1226 | C/T | Intron2 | New | 0.034 | |
| 46 | CTSGS5 | 1349 | G/A | Intron2 | rs2070697 | 0.314 | |
| 47 | CTSGS6 | 1767 | A/G | Exon4-synonymous | New | 0.182 | |
| 48 | CYP4F2 | CYP4F2S1 | 4937 | A/T | Intron1 | New | 0.349 |
| 49 | CYP4F2S4 | 18 733 | T/C | Intron1 | rs2072269 | 0.335 | |
| 50 | CYP4F2S5 | 18 810 | G/C | Intron1 | rs1064796 | 0.495 | |
| 51 | CYP4F2S6 | 30 715 | G/A | Intron1 | rs2018460 | 0.170 | |
| 52 | CYP4F3 | CYP4F3S0 | 3187 | G/A | Intron2 | rs2203998 | 0.328 |
| 53 | CYP4F3S0.5 | 6221 | C/T | Intron4 | rs1290626 | 0.141 | |
| 54 | CYP4F3S1 | 8507 | C/T | Intron6 | rs2283612 | 0.189 | |
| 55 | CYP4F3S2 | 11 871 | A/C | Intron8 | rs2733750 | 0.183 | |
| 56 | CYP4F3S3 | 12 015 | A/G | Exon9-synonymous | New | 0.363 | |
| 57 | CYP4F3S4 | 12 808 | G/A | Intron9 | rs2733752 | 0.136 | |
| 58 | CYP4F3S5 | 12 877 | T/A | Intron9 | New | 0.140 | |
| 59 | CYP4F3S6 | 17 772 | T/C | Intron11 | New | 0.453 | |
| 60 | CYP4F8 | CYP4F8S1 | 3610 | C/T | Intron2 | rs2072599 | 0.168 |
| 61 | CYP4F8S2 | 6771 | G/T | Intron5 | rs2072601 | 0.276 | |
| 62 | CYP4F8S3 | 13 568 | C/T | Intron11 | rs2239366 | 0.276 | |
| 63 | CYSLT1R | CYSLT1RS1 | 927 | C/T | Exon1-synonymous | rs320995 | 0.474 |
| 64 | CYSLT2R | CYSLT2RS1 | 2797 | A/G | Exon1-UTR | rs912277 | 0.417 |
| 65 | CYSLT2RS2 | 3078 | A/C | Exon1-UTR | rs1323552 | 0.489 | |
| 66 | CYSLT2RS3 | 3105 | A/G | Exon1-UTR | New | 0.116 | |
| 67 | FLAP | FLAPS1 | 162 | C/A | Intron1 | rs4769055 | 0.522 |
| 68 | FLAPS2 | 838 | T/G | Intron1 | rs9579645 | 0.096 | |
| 69 | FLAPS3 | 7272 | A/G | Intron1 | rs9551960 | 0.339 | |
| 70 | FLAPS4 | 8640 | A/C | Intron2 | rs3803277 | 0.406 | |
| 71 | FLAPS5 | 8733 | T/C | Intron2 | rs3803278 | 0.370 | |
| 72 | FLAPS6 | 13 674 | G/A | Intron2 | rs4075692 | 0.286 | |
| 73 | FLAPS7 | 20 616 | G/C | Intron3 | New | 0.286 | |
| 74 | FLAPS8 | 20 648 | C/T | Intron3 | rs4468448 | 0.240 | |
| 75 | FLAPS9 | 23 849 | T/A | Intron3 | rs9551964 | 0.277 | |
| 76 | FLAPS9.5 | 24 348 | G/A | Intron3 | New | 0.006 | |
| 77 | FLAPS10 | 28 209 | A/G | Intron3 | rs4769060 | 0.401 | |
| 78 | HPGD | HPGDS1 | 464 | G/A | Exon2-synonymous | rs1050145 | 0.220 |
| 79 | HPGDS2 | 798 | A/G | Intron2 | rs1365613 | 0.447 | |
| 80 | HPGDS3 | 13 332 | A/C | Intron3 | rs2555629 | 0.405 | |
| 81 | HPGDS4 | 13 430 | A/T | Intron3 | New | 0.253 | |
| 82 | HPGDS5 | 20 442 | A/G | Intron4 | New | 0.446 | |
| 83 | HPGDS6 | 20 516 | G/A | Intron4 | New | 0.110 | |
| 84 | IGF1 | IGF1S1 | 3083 | A/G | Intron2 | rs2162679 | 0.368 |
| 85 | IGF1S2 | 9917 | G/T | Intron3 | rs1019731 | 0.021 | |
| 86 | IGF1S3 | 17 640 | G/C | Intron3 | rs2195239 | 0.426 | |
| 87 | IGF1S4 | 17 695 | T/C | Intron3 | rs2195240 | 0.443 | |
| 88 | IGF1S5 | 49 421 | G/A | Intron3 | rs972936 | 0.484 | |
| 89 | IGF1S6 | 60 710 | G/A | Intron3 | rs2072592 | 0.245 | |
| 90 | IGF1S7 | 72 103 | G/A | Intron5 | rs978458 | 0.135 | |
| 91 | IGF1S8 | 84 150 | G/A | Exon6-UTR | rs6219 | 0.271 | |
| 92 | IL13 | IL13S1 | −978 | C/T | Promoter | rs11575055 | 0.179 |
| 93 | IL13S2 | 571 | C/A | Intron1 | rs2066960 | 0.281 | |
| 94 | IL13S3 | 598 | G/C | Intron1 | rs1295987 | 0.120 | |
| 95 | IL13S4 | 805 | C/T | Intron1 | rs2069744 | 0.120 | |
| 96 | IL13S5 | 2100 | G/A | Exon4-non-synonymous (R144Q) | rs20541 | 0.333 | |
| 97 | IL4 | IL4S1 | −219 | T/C | Promoter | rs2243250 | 0.328 |
| 98 | IL4S2 | 3353 | A/C | Intron2 | rs2227284 | 0.255 | |
| 99 | IL4S3 | 3927 | C/G | Intron2 | rs2243263 | 0.073 | |
| 100 | LTA4H | LTA4HS1 | 9153 | A/G | Intron3 | rs763842 | 0.226 |
| 101 | LTA4HS4 | 20 165 | T/C | Intron11 | rs1978331 | 0.335 | |
| 102 | LTB4DH | LTB4DHS1 | 4435 | A/T | Intron2 | New | 0.506 |
| 103 | LTB4DHS2 | 8896 | A/G | Intron4 | New | 0.283 | |
| 104 | LTB4DHS2.5 | 9182 | A/G | Intron4 | New | 0.092 | |
| 105 | LTB4DHS5 | 9502 | C/T | Intron4 | rs1053968 | 0.005 | |
| 106 | LTB4DHS6 | 13 234 | C/G | Intron4 | New | 0.319 | |
| 107 | LTB4DHS6.5 | 13 249 | A/G | Intron4 | New | 0.231 | |
| 108 | LTB4DHS7 | 20 718 | G/A | Intron6 | New | 0.253 | |
| 109 | LTB4DHS8 | 25 971 | A/G | Intron8 | rs1322258 | 0.347 | |
| 110 | LTB4DHS9 | 26 221 | A/G | Intron8 | New | 0.406 | |
| 111 | LTB4DHS10 | 35 589 | A/C | Intron9 | rs2146078 | 0.229 | |
| 112 | LTB4DHS11 | 35 819 | G/A | Intron9 | New | 0.354 | |
| 113 | LTB4R | LTB4RS1 | 1165 | G/C | Exon1-UTR | New | 0.140 |
| 114 | LTC4S | LTC4SS1.5 | −348 | A/C | Promoter | New | 0.208 |
| 115 | LTC4SS2 | 289 | G/C | Intron1 | New | 0.026 | |
| 116 | LTC4SS3 | 1296 | C/T | Intron1 | New | 0.208 | |
| 117 | LTC4SS4 | 2659 | A/G | Intron5 | New | 0.201 | |
| 118 | MGST2 | MGST2S1 | 815 | A/G | Intron1 | rs1000222 | 0.292 |
| 119 | MGST2S2 | 12 971 | C/T | Intron2 | rs795589 | 0.198 | |
| 120 | MGST2S3 | 12 977 | A/C | Intron2 | rs795588 | 0.077 | |
| 121 | MMP1 | MMP1S1 | 193 | G/A | Intron1 | rs470358 | 0.292 |
| 122 | MMP1S2 | 2579 | G/A | Exon5-synonymous | rs470558 | 0.130 | |
| 123 | MMP1S3 | 7300 | T/C | Intron8 | rs470747 | 0.137 | |
| 124 | MMP1S4 | 7815 | C/T | Exon10-UTR | rs2239008 | 0.319 | |
| 125 | MMP1S5 | 7936 | T/C | Exon10-UTR | rs2071230 | 0.298 | |
| 126 | MMP2 | MMP2S1 | 3606 | A/T | Intron1 | rs857403 | 0.214 |
| 127 | MMP2S2 | 3665 | G/A | Intron1 | rs1030868 | 0.229 | |
| 128 | MMP2S3 | 6505 | C/T | Exon5-synonymous | rs1053605 | 0.208 | |
| 129 | MMP2S4 | 6730 | T/G | Intron5 | New | 0.064 | |
| 130 | MMP2S5 | 6958 | A/G | Intron5 | rs866770 | 0.151 | |
| 131 | MMP2S6 | 10 603 | C/T | Exon7-synonymous | rs243849 | 0.151 | |
| 132 | MMP2S7 | 10 896 | T/C | Intron7 | rs243847 | 0.443 | |
| 133 | MMP2S8 | 14 011 | G/A | Exon9-synonymous | rs2287074 | 0.326 | |
| 134 | MMP2S9 | 14 196 | G/A | Intron9 | rs243843 | 0.306 | |
| 135 | MMP2S10 | 14 281 | G/A | Intron9 | New | 0.065 | |
| 136 | MMP2S11 | 14 320 | T/C | Intron9 | rs243842 | 0.364 | |
| 137 | MMP2S12 | 17 660 | G/T | Intron9 | rs171498 | 0.363 | |
| 138 | MMP2S13 | 17 670 | G/T | Intron9 | rs243838 | 0.153 | |
| 139 | MMP2S14 | 17 928 | C/T | Intron10 | New | 0.057 | |
| 140 | MMP2S15 | 20 976 | G/A | Intron11 | New | 0.115 | |
| 141 | MMP2S16 | 21 134 | G/A | Intron11 | rs243836 | 0.386 | |
| 142 | MMP2S17 | 26 345 | C/T | Exon13-UTR | New | 0.021 | |
| 143 | MMP2S18 | 26 512 | A/C | Exon13-UTR | rs7201 | 0.234 | |
| 144 | MMP8 | MMP8S1 | 524 | G/A | Intron1 | rs1939012 | 0.359 |
| 145 | MMP8S2 | 5458 | A/G | Intron4 | rs1940051 | 0.266 | |
| 146 | MMP8S3 | 11 357 | T/A | Intron9 | New | 0.287 | |
| 147 | MMP8S4 | 11 473 | T/C | Exon10-synonymous | New | 0.016 | |
| 148 | MMP9 | MMP9S1 | 2679 | G/A | Exon6-non-synonymous (R279Q) | rs2664538 | 0.382 |
| 149 | MMP9S2 | 3029 | A/C | Intron6 | rs2236416 | 0.179 | |
| 150 | MMP9S3 | 5565 | G/A | Exon12-non-synonymous (R668Q) | rs2274756 | 0.214 | |
| 151 | MMP9S4 | 7419 | G/A | Exon13-synonymous | rs13925 | 0.292 | |
| 152 | P2Y10 | P2Y10S1 | 1192 | A/G | Intron1 | New | 0.016 |
| 153 | P2Y10S2 | 5030 | C/T | Intron1 | rs2858570 | 0.110 | |
| 154 | P2Y10S3 | 5318 | A/T | Intron1 | rs2251477 | 0.184 | |
| 155 | P2Y10S4 | 9417 | T/C | Intron1 | rs2742205 | 0.115 | |
| 156 | PAFAH1B1 | PAFAH1B1S2 | 12 145 | G/T | Intron1 | New | 0.099 |
| 157 | PAFAH1B1S3 | 21 146 | C/G | Intron1 | New | 0.080 | |
| 158 | PAFAH1B1S4 | 37 780 | A/G | Intron1 | rs1266474 | 0.084 | |
| 159 | PAFAH1B1S5 | 38 033 | C/T | Intron1 | New | 0.047 | |
| 160 | PAFAH1B1S6 | 40 522 | G/A | Intron1 | New | 0.130 | |
| 161 | PAFAH1B1S7 | 60 034 | C/T | Intron2 | New | 0.266 | |
| 162 | PAFAH1B1S8 | 68 545 | G/C | Intron2 | New | 0.078 | |
| 163 | PAFAH1B1S9 | 76 403 | T/G | Intron5 | New | 0.032 | |
| 164 | PAFAH1B2 | PAFAH1B2S1 | 3971 | T/C | Intron1 | rs2008908 | 0.446 |
| 165 | PAFAH1B3 | PAFAH1B3S1 | 2538 | C/T | Exon4-synonymous | New | 0.016 |
| 166 | PAFAH2 | PAFAH2S1 | 1279 | G/T | Intron1 | rs3008423 | 0.172 |
| 167 | PAFAH2S2 | 4171 | C/T | Intron1 | New | 0.036 | |
| 168 | PAFAH2S3 | 15456 | G/T | Intron5 | rs1469512 | 0.214 | |
| 169 | PAI1m2 | PAI1m2S1 | 4484 | G/A | Intron1 | rs840088 | 0.401 |
| 170 | PAI1m2S2 | 5094 | A/G | Intron1 | New | 0.089 | |
| 171 | PAI1m2S3 | 19 022 | C/A | Intron1 | rs2099601 | 0.201 | |
| 172 | PAI1m2S4 | 19 047 | A/C | Intron1 | New | 0.201 | |
| 173 | PAI1m2S5 | 19 120 | T/C | Intron1 | rs2083120 | 0.201 | |
| 174 | PAI1m2S6 | 19 348 | A/C | Intron1 | New | 0.393 | |
| 175 | PAI1m2S7 | 62 178 | A/G | Intron8 | New | 0.271 | |
| 176 | PDGFB | PDGFBS1 | 581 | G/A | Intron1 | rs758588 | 0.184 |
| 177 | PDGFBS3 | 20 237 | C/T | Intron3 | rs740750 | 0.310 | |
| 178 | PDGFBS4 | 26 041 | C/T | Intron10 | rs1864972 | 0.198 | |
| 179 | PDGFBS5 | 35 170 | G/A | Intron18 | rs1432878 | 0.108 | |
| 180 | PDGFRL | PDGFRLS1 | 978 | G/C | Intron1 | rs2720576 | 0.365 |
| 181 | PDGFRLS2 | 1187 | C/T | Intron1 | rs2588164 | 0.245 | |
| 182 | PDGFRLS3 | 1416 | T/C | Intron1 | rs2588163 | 0.391 | |
| 183 | PDGFRLS4 | 2963 | G/C | Intron1 | rs2517267 | 0.458 | |
| 184 | PDGFRLS5 | 3794 | C/G | Intron1 | rs2517268 | 0.394 | |
| 185 | PDGFRLS6 | 3882 | G/A | Intron1 | New | 0.216 | |
| 186 | PDGFRLS7 | 3999 | A/G | Intron1 | rs2517269 | 0.220 | |
| 187 | PDGFRLS8 | 8893 | G/A | Intron1 | New | 0.226 | |
| 188 | PDGFRLS9 | 9922 | G/A | Intron1 | New | 0.232 | |
| 189 | PDGFRLS10 | 14 053 | G/C | Intron2 | rs2720583 | 0.167 | |
| 190 | PDGFRLS11 | 14 194 | A/C | Intron2 | New | 0.396 | |
| 191 | PDGFRLS12 | 18 220 | T/C | Intron2 | rs2427709 | 0.140 | |
| 192 | PDGFRLS13 | 18 473 | G/A | Intron2 | rs2588144 | 0.068 | |
| 193 | PDGFRLS15 | 19 592 | T/G | Intron2 | rs2246488 | 0.339 | |
| 194 | PDGFRLS16 | 28 052 | G/A | Intron2 | rs2517187 | 0.104 | |
| 195 | PDGFRLS17 | 28 449 | C/T | Intron2 | rs2237823 | 0.104 | |
| 196 | PDGFRLS18 | 31 111 | T/C | Intron2 | rs2517198 | 0.094 | |
| 197 | PDGFRLS19 | 31 230 | A/T | Intron2 | New | 0.260 | |
| 198 | PDGFRLS20 | 31 258 | A/G | Intron2 | New | 0.031 | |
| 199 | PDGFRLS21 | 31 499 | T/C | Intron2 | rs2427715 | 0.094 | |
| 200 | PDGFRLS22 | 31 600 | T/C | Intron2 | New | 0.167 | |
| 201 | PDGFRLS23 | 33 975 | G/C | Intron2 | rs2517208 | 0.037 | |
| 202 | PDGFRLS24 | 41 991 | A/G | Intron2 | rs2237831 | 0.189 | |
| 203 | PDGFRLS25 | 42 131 | G/A | Intron2 | rs2237831 | 0.205 | |
| 204 | PDGFRLS26 | 43 810 | G/T | Intron2 | New | 0.073 | |
| 205 | PDGFRLS27 | 46 759 | T/C | Intron3 | New | 0.120 | |
| 206 | PDGFRLS28 | 46 888 | G/A | Intron3 | rs2237835 | 0.226 | |
| 207 | PDGFRLS29 | 46 936 | C/G | Intron3 | New | 0.074 | |
| 208 | PDGFRLS30 | 47 097 | C/G | Intron3 | New | 0.197 | |
| 209 | PDGFRLS31 | 49 411 | A/C | Intron3 | rs2237836 | 0.548 | |
| 210 | PDGFRLS32 | 49 937 | G/T | Intron3 | rs2237837 | 0.103 | |
| 211 | PDGFRLS33 | 53 887 | T/C | Intron4 | rs2237842 | 0.188 | |
| 212 | PDGFRLS34 | 56 021 | C/T | Intron4 | rs2427719 | 0.063 | |
| 213 | PDGFRLS35 | 56 410 | T/C | Intron4 | rs2237845 | 0.240 | |
| 214 | PDGFRLS36 | 65 408 | C/T | Intron5 | New | 0.083 | |
| 215 | PDGFRLS37 | 65 512 | T/C | Exon6-synonymous | rs4705 | 0.521 | |
| 216 | PGDS | PGDSS1 | 5446 | T/C | Intron1 | rs2129595 | 0.191 |
| 217 | PGIS | PGISS1 | 4650 | T/C | Intron1 | rs477627 | 0.063 |
| 218 | PGISS2 | 6734 | A/C | Intron1 | rs927068 | 0.226 | |
| 219 | PGISS3 | 6840 | T/C | Intron1 | New | 0.226 | |
| 220 | PGISS4 | 6870 | G/C | Intron1 | rs498646 | 0.276 | |
| 221 | PGISS4.5 | 6924 | T/C | Intron1 | rs476496 | 0.280 | |
| 222 | PGISS6 | 28 941 | A/G | Intron5 | rs501908 | 0.033 | |
| 223 | PGISS7 | 41 990 | A/G | Intron5 | New | 0.093 | |
| 224 | PGISS8 | 44 026 | G/A | Exon6-synonymous | rs5628 | 0.036 | |
| 225 | PGISS9 | 50 926 | C/T | Intron6 | New | 0.104 | |
| 226 | PGISS10 | 55 002 | C/A | Intron8 | rs5629 | 0.281 | |
| 227 | PGISS11 | 57 532 | A/G | Intron9 | rs729824 | 0.226 | |
| 228 | PGISS12 | 62 730 | T/C | Exon10-UTR | rs5602 | 0.438 | |
| 229 | PLA2G7 | PLA2G7S1 | 2007 | G/T | Intron1 | New | 0.214 |
| 230 | PLA2G7S2 | 2338 | G/A | Intron1 | rs1421369 | 0.468 | |
| 231 | PLA2G7S3 | 6203 | G/A | Intron1 | New | 0.141 | |
| 232 | PLA2G7S4 | 20 965 | G/T | Intron5 | rs1362931 | 0.042 | |
| 233 | PLA2G7S5 | 23 741 | T/C | Exon7-non-synonymous (I198T) | rs1805018 | 0.281 | |
| 234 | PLA2G7S6 | 27 025 | C/G | Intron9 | rs2216465 | 0.542 | |
| 235 | PLA2G7S7 | 30 101 | C/T | Exon11-non-synonymous (A379V) | rs1051931 | 0.042 | |
| 236 | PTAFR | PTAFRS3 | 36 031 | C/A | Exon2-non-synonymous (A224D) | rs5938 | 0.068 |
| 237 | PTGDR | PTGDRS1 | 408 | G/A | Intron1 | New | 0.021 |
| 238 | PTGDRS2 | 3290 | A/G | Intron1 | rs1254609 | 0.194 | |
| 239 | PTGDRS3 | 5754 | T/C | Intron1 | New | 0.075 | |
| 240 | PTGDRS4 | 5793 | A/G | Intron1 | rs708486 | 0.081 | |
| 241 | PTGDS | PTGDSS1 | 4186 | C/A | Exon7-UTR | rs6926 | 0.198 |
| 242 | EP1 | PTGER1S1 | −267 | G/C | Promoter | New | 0.109 |
| 243 | EP2 | PTGER2uS1 | −26 643 | T/C | 5′-Upstream | rs988209 | 0.232 |
| 244 | PTGER2uS2 | −18 461 | T/G | 5′-Upstream | rs1390375 | 0.077 | |
| 245 | PTGER2uS3 | −18 360 | G/A | 5′-Upstream | rs1390374 | 0.022 | |
| 246 | PTGER2uS4 | −15 332 | A/T | 5′-Upstream | rs708490 | 0.444 | |
| 247 | PTGER2uS5 | −12 813 | G/A | 5′-Upstream | New | 0.223 | |
| 248 | PTGER2uS6 | −10 918 | A/G | 5′-Upstream | New | 0.449 | |
| 249 | PTGER2uS7 | −10 814 | T/A | 5′-upstream | New | 0.427 | |
| 250 | PTGER2uS8 | −10 250 | A/G | 5′-upstream | rs714366 | 0.433 | |
| 251 | PTGER2uS9 | −7075 | A/G | 5′-upstream | New | 0.452 | |
| 252 | PTGER2uS10 | −6179 | A/G | 5′-upstream | New | 0.479 | |
| 253 | PTGER2S1 | −609 | G/A | Promoter | rs1254601 | 0.288 | |
| 254 | PTGER2S2 | 300 | G/A | Exon1-UTR | rs1254600 | 0.462 | |
| 255 | PTGER2S3 | 498 | C/G | Exon1-UTR | rs2075797 | 0.441 | |
| 256 | PTGER2S4 | 948 | A/G | Exon1-UTR | rs1353411 | 0.417 | |
| 257 | PTGER2S5 | 1042 | G/A | Exon1-UTR | rs1254598 | 0.350 | |
| 258 | PTGER2S6 | 2803 | G/A | Intron1 | New | 0.164 | |
| 259 | PTGER2S7 | 2988 | C/T | Intron1 | New | 0.339 | |
| 260 | PTGER2S8 | 6063 | C/T | Intron1 | New | 0.116 | |
| 261 | PTGER2S9 | 10 927 | T/G | Intron1 | rs1254585 | 0.446 | |
| 262 | PTGER2S10 | 14 081 | C/T | Exon2-UTR | rs708502 | 0.398 | |
| 263 | PTGER2dS1 | 29 784 | C/T | 3′-Downstream | rs708511 | 0.219 | |
| 264 | PTGER2dS2 | 47 461 | A/G | 3′-Downstream | New | 0.226 | |
| 265 | PTGER2dS3 | 57 931 | C/T | 3′-Downstream | rs708531 | 0.750 | |
| 266 | PTGER2dS4 | 58 051 | C/T | 3′-Downstream | rs708532 | 0.329 | |
| 267 | EP3 | PTGER3S1 | 27 837 | G/A | Intron1 | rs1008484 | 0.058 |
| 268 | PTGER3S2 | 28 078 | G/A | Intron1 | rs1569593 | 0.258 | |
| 269 | PTGER3S5 | 36 177 | A/T | Intron2 | rs5680 | 0.313 | |
| 270 | PTGER3S7 | 44 999 | T/G | Intron2 | rs1983588 | 0.268 | |
| 271 | PTGER3S8 | 45 040 | A/C | Intron2 | rs1983587 | 0.059 | |
| 272 | PTGER3S9 | 54 444 | G/C | Intron2 | rs1883461 | 0.054 | |
| 273 | PTGER3S10 | 54 634 | T/A | Intron2 | rs1883460 | 0.094 | |
| 274 | PTGER3S11 | 58 525 | C/T | Intron2 | New | 0.271 | |
| 275 | PTGER3S12 | 65 699 | G/C | Intron2 | rs647921 | 0.048 | |
| 276 | PTGER3S13 | 65 973 | A/T | Intron2 | rs646621 | 0.443 | |
| 277 | PTGER3S14 | 70 342 | G/A | Intron2 | rs909842 | 0.442 | |
| 278 | PTGER3S15 | 70 357 | A/C | Intron2 | New | 0.300 | |
| 279 | PTGER3S17 | 70 409 | G/A | Intron2 | New | 0.442 | |
| 280 | PTGER3S20 | 70 755 | A/T | Intron2 | rs484675 | 0.441 | |
| 281 | PTGER3S24 | 86 923 | A/G | Intron2 | rs573688 | 0.082 | |
| 282 | PTGER3S25 | 95 146 | A/C | Intron3 | New | 0.037 | |
| 283 | PTGER3S27 | 1 00 052 | T/C | Intron3 | rs1409984 | 0.255 | |
| 284 | PTGER3S29 | 1 14 078 | G/A | Intron3 | rs625617 | 0.371 | |
| 285 | PTGER3S30 | 1 21 798 | C/A | Intron3 | New | 0.328 | |
| 286 | PTGER3S31 | 1 21 846 | G/A | Intron3 | rs602383 | 0.297 | |
| 287 | PTGER3S32 | 1 49 402 | C/T | Intron3 | rs1409165 | 0.370 | |
| 288 | PTGER3S33 | 1 49 626 | G/A | Intron3 | rs1409166 | 0.398 | |
| 289 | PTGER3S35 | 1 60 372 | A/G | Intron3 | New | 0.214 | |
| 290 | PTGER3S36 | 1 60 403 | A/G | Intron3 | rs1409978 | 0.319 | |
| 291 | PTGER3S37 | 1 60 432 | C/G | Intron3 | New | 0.214 | |
| 292 | EP4 | PTGER4S1 | 5748 | C/T | Intron2 | New | 0.389 |
| 293 | PTGER4S2 | 7984 | A/G | Intron2 | New | 0.198 | |
| 294 | PTGER4S3 | 8012 | G/A | Intron2 | New | 0.385 | |
| 295 | PTGES | PTGESS1 | 214 | A/G | Intron1 | rs2241271 | 0.135 |
| 296 | PTGESS2 | 406 | G/A | Intron1 | rs2241270 | 0.120 | |
| 297 | PTGESS3 | 9636 | T/C | Intron2 | New | 0.326 | |
| 298 | PTGESS4 | 9669 | G/A | Intron2 | New | 0.152 | |
| 299 | PTGFR | PTGFRS1 | 714 | A/C | Intron1 | rs3766355 | 0.425 |
| 300 | PTGFRS2 | 823 | G/A | Intron1 | rs3766354 | 0.204 | |
| 301 | PTGFRS3 | 1132 | G/T | Intron1 | rs3766353 | 0.242 | |
| 302 | PTGFRS4 | 3617 | G/A | Intron2 | rs1830763 | 0.489 | |
| 303 | PTGFRS5 | 6578 | A/C | Intron2 | rs1322935 | 0.031 | |
| 304 | PTGFRS6 | 8734 | A/G | Intron2 | rs2057423 | 0.452 | |
| 305 | PTGFRS10 | 24 552 | G/A | Intron2 | rs3766346 | 0.036 | |
| 306 | PTGFRS12 | 33 123 | A/C | Intron2 | rs520171 | 0.276 | |
| 307 | PTGFRS13 | 33 548 | G/C | Intron2 | rs3766345 | 0.516 | |
| 308 | PTGFRS14 | 38 195 | T/C | Intron2 | rs3766338 | 0.266 | |
| 309 | PTGFRS15 | 38 202 | C/T | Intron2 | rs590309 | 0.511 | |
| 310 | PTGFRS16 | 40 734 | G/C | Intron2 | rs622346 | 0.202 | |
| 311 | PTGFRS19 | 46 572 | A/G | Exon3-UTR | rs3766331 | 0.115 | |
| 312 | PTGFRS20 | 46 628 | A/G | Exon3-UTR | rs899 | 0.005 | |
| 313 | PTGS1 | PTGS1S1 | 2850 | C/T | Intron2 | rs1213264 | 0.063 |
| 314 | PTGS1S2 | 3219 | T/C | Intron2 | rs1213265 | 0.067 | |
| 315 | PTGS1S2.5 | 7468 | C/G | Intron3 | rs2282169 | 0.079 | |
| 316 | PTGS1S4 | 23 970 | C/A | Exon11-UTR | rs10306194 | 0.028 | |
| 317 | SCYA5 | SCYA5S1 | 328 | T/C | Intron1 | rs2280789 | 0.349 |
| 318 | SCYB14 | SCYB14S1 | 313 | T/C | Intron1 | rs2072347 | 0.311 |
| 319 | SCYB14S2 | 2099 | G/C | Intron2 | rs2237062 | 0.307 | |
| 320 | SCYB14S3 | 6261 | C/T | Intron3 | rs1016666 | 0.370 | |
| 321 | SLC21A9 | SLC21A9S1 | 196 | T/C | Exon1-UTR | New | 0.158 |
| 322 | SLC21A9S2 | 231 | G/A | Exon1-UTR | rs1944612 | 0.055 | |
| 323 | SLC21A9S3 | 4589 | C/T | Intron1 | New | 0.443 | |
| 324 | SLC21A9S5 | 10 724 | A/G | Intron1 | New | 0.374 | |
| 325 | SLC21A9S6 | 13 387 | G/C | Intron3 | New | 0.453 | |
| 326 | SLC21A9S7 | 15 413 | G/A | Intron4 | rs949069 | 0.391 | |
| 327 | SLC21A9S8 | 15 916 | A/G | Intron4 | New | 0.161 | |
| 328 | SLC21A9S9 | 17 493 | G/A | Intron4 | rs1676878 | 0.136 | |
| 329 | SLC21A9S10 | 24 698 | A/G | Intron7 | rs1676881 | 0.226 | |
| 330 | SLC21A9S10.5 | 24 880 | C/T | Intron7 | rs1612859 | 0.458 | |
| 331 | SLC21A9S11 | 25 005 | T/A | Intron7 | rs1789693 | 0.401 | |
| 332 | SLC21A9S13 | 26 261 | T/G | Intron7 | New | 0.376 | |
| 333 | SLC21A9S14 | 39 441 | G/A | Intron8 | rs1789692 | 0.479 | |
| 334 | SLC21A9S15 | 43 575 | C/T | Intron9 | New | 0.430 | |
| 335 | SLC21A9S16 | 45 422 | C/T | Exon10 | New | 0.432 | |
| 336 | STAT2 | STAT2S1 | 10 525 | A/G | Intron14 | rs2020854 | 0.021 |
| 337 | STAT4 | STAT4S1 | 5564 | C/A | Intron3 | rs1031509 | 0.396 |
| 338 | STAT4S2 | 19 235 | G/A | Intron3 | rs1551443 | 0.214 | |
| 339 | STAT4S3 | 91 147 | A/T | Intron10 | New | 0.516 | |
| 340 | STAT4S4 | 100 038 | G/C | Intron14 | rs1400655 | 0.095 | |
| 341 | STAT4S5 | 118 213 | A/G | Intron21 | rs925847 | 0.553 | |
| 342 | STAT4S6 | 119 708 | A/C | Intron22 | rs1517351 | 0.495 | |
| 343 | STAT4S7 | 119 711 | G/A | Intron22 | New | 0.165 | |
| 344 | TBX21 | TBX21S1 | 8941 | T/C | Intron1 | rs2158079 | 0.201 |
| 345 | TBXA2R | TBXA2RS1 | 10937 | T/C | Exon3-synonymous | rs4523 | 0.184 |
| 346 | TXAs | TXAsS2 | 17778 | A/C | Intron1 | rs41708 | 0.302 |
| 347 | TXAsS3 | 24573 | T/C | Intron1 | rs41706 | 0.355 | |
| 348 | TXAsS4 | 29218 | A/T | Intron1 | rs194150 | 0.443 | |
| 349 | TXAsS5 | 57546 | T/G | Intron3 | rs1015571 | 0.447 | |
| 350 | TXAsS7 | 63115 | C/A | Intron3 | rs2013219 | 0.426 | |
| 351 | TXAsS9 | 69322 | G/A | Intron3 | rs757762 | 0.447 | |
| 352 | TXAsS11 | 80249 | C/T | Intron3 | rs1978180 | 0.028 | |
| 353 | TXAsS13 | 1 06 385 | T/C | Intron4 | rs41733 | 0.117 | |
| 354 | TXAsS14 | 1 06 401 | G/A | Intron4 | rs41732 | 0.117 | |
| 355 | TXAsS15 | 1 12 521 | T/C | Intron5 | New | 0.247 | |
| 356 | TXAsS16 | 1 12 694 | C/T | Intron5 | rs42335 | 0.134 | |
| 357 | TXAsS17 | 1 15 453 | G/A | Intron5 | New | 0.011 | |
| 358 | TXAsS21 | 1 28 260 | G/T | Intron7 | New | 0.165 | |
| 359 | TXAsS22 | 1 39 370 | T/C | Intron9 | rs41718 | 0.146 | |
| 360 | TXAsS23 | 1 46 442 | T/C | Intron9 | rs740150 | 0.389 | |
| 361 | TXAsS24 | 1 52 303 | A/G | Intron9 | rs193949 | 0.177 | |
| 362 | TXAsS25 | 1 52 455 | C/T | Intron9 | New | 0.308 | |
| 363 | TXAsS29 | 1 78 072 | G/A | Intron10 | rs740204 | 0.234 | |
| 364 | TXAsS30 | 1 86 274 | G/A | Intron10 | New | 0.479 | |
| 365 | UPAR | UPARS1 | 11 475 | A/C | Intron3 | New | 0.019 |
| 366 | UPARS2 | 11 639 | G/A | Intron3 | rs2283628 | 0.335 | |
| 367 | UPARS3 | 11 667 | A/C | Intron3 | rs2239373 | 0.385 | |
| 368 | UPARS4 | 11 746 | C/T | Intron3 | rs2239372 | 0.375 | |
| 369 | UPARS5 | 18 100 | C/T | Intron5 | rs2302525 | 0.074 | |
| 370 | UPARS6 | 18 228 | A/G | Exon6-non-synonymous (K220R) | rs2302524 | 0.114 |
The SNPs applied to the first screening are listed. The sequence position indicates the location of the SNP relative to the transcription initiation site of exon1of each gene. The variation of allele and the localization in gene structure are shown. Each given SNP is referenced an accession number in dbSNP created by NCBI, rs ID (http://www.ncbi.nlm.nih.gov/SNP/). The New findings are mentioned as ‘new’. The minor allele frequency of each SNP typed in the control sample of 96 individuals is shown.
Allele frequencies of SNPs of EP2 and comparisons between AIA and ATA and AIA and CTR
| . | Position . | Variation . | Localization . | Allele count . | Minor allele frequency . | Permutation P value . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | AIA . | CTR . | ATA . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| uS5 | −12 813 | G/A | 5′-Upstream | 396 | 548 | 564 | 0.311 | 0.221 | 0.222 | 0.0016** | 0.0017** |
| uS6 | −10 918 | A/G | 5′-Upstream | 392 | 528 | 526 | 0.395 | 0.449 | 0.451 | 0.0844 | 0.0912 |
| uS7 | −10 814 | T/A | 5′-Upstream | 396 | 540 | 562 | 0.492 | 0.428 | 0.397 | 0.0393* | 0.0025** |
| uS8 | −10 250 | A/G | 5′-Upstream | 384 | 522 | 516 | 0.393 | 0.433 | 0.438 | 0.1767 | 0.1716 |
| uS9 | −7075 | A/G | 5′-Upstream | 384 | 504 | 522 | 0.432 | 0.452 | 0.469 | 0.5329 | 0.2594 |
| uS10 | −6179 | A/G | 5′-Upstream | 374 | 526 | 564 | 0.374 | 0.401 | 0.452 | 0.4213 | 0.0199* |
| S1 | −609 | G/A | Promoter | 396 | 538 | 564 | 0.348 | 0.288 | 0.303 | 0.0554 | 0.1175 |
| S2 | 300 | G/A | Exon1(UTR) | 384 | 530 | 558 | 0.445 | 0.462 | 0.504 | 0.6037 | 0.0757 |
| S3 | 498 | C/G | Exon1(UTR) | 392 | 534 | 558 | 0.401 | 0.427 | 0.468 | 0.4121 | 0.0393* |
| S4 | 948 | A/G | Exon1(UTR) | 396 | 532 | 564 | 0.477 | 0.417 | 0.415 | 0.0665 | 0.0480* |
| S5 | 1042 | G/A | Exon1(UTR) | 396 | 514 | 564 | 0.422 | 0.350 | 0.367 | 0.0276* | 0.0844 |
| S6 | 2803 | G/A | Intron1 | 390 | 538 | 554 | 0.141 | 0.164 | 0.132 | 0.3396 | 0.6785 |
| S7 | 2988 | C/T | Intron1 | 388 | 528 | 554 | 0.327 | 0.339 | 0.377 | 0.7125 | 0.1169 |
| S8 | 6063 | C/T | Intron1 | 396 | 536 | 562 | 0.109 | 0.119 | 0.117 | 0.5876 | 0.6338 |
| S9 | 10 927 | T/G | Intron1 | 390 | 534 | 548 | 0.474 | 0.446 | 0.451 | 0.3808 | 0.4641 |
| S10 | 14 081 | C/T | Exon2(UTR) | 378 | 516 | 554 | 0.373 | 0.399 | 0.419 | 0.4484 | 0.1632 |
| dS1 | 29 784 | C/T | 3′-Downstream | 372 | 522 | 542 | 0.194 | 0.218 | 0.173 | 0.3565 | 0.4316 |
| . | Position . | Variation . | Localization . | Allele count . | Minor allele frequency . | Permutation P value . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | AIA . | CTR . | ATA . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| uS5 | −12 813 | G/A | 5′-Upstream | 396 | 548 | 564 | 0.311 | 0.221 | 0.222 | 0.0016** | 0.0017** |
| uS6 | −10 918 | A/G | 5′-Upstream | 392 | 528 | 526 | 0.395 | 0.449 | 0.451 | 0.0844 | 0.0912 |
| uS7 | −10 814 | T/A | 5′-Upstream | 396 | 540 | 562 | 0.492 | 0.428 | 0.397 | 0.0393* | 0.0025** |
| uS8 | −10 250 | A/G | 5′-Upstream | 384 | 522 | 516 | 0.393 | 0.433 | 0.438 | 0.1767 | 0.1716 |
| uS9 | −7075 | A/G | 5′-Upstream | 384 | 504 | 522 | 0.432 | 0.452 | 0.469 | 0.5329 | 0.2594 |
| uS10 | −6179 | A/G | 5′-Upstream | 374 | 526 | 564 | 0.374 | 0.401 | 0.452 | 0.4213 | 0.0199* |
| S1 | −609 | G/A | Promoter | 396 | 538 | 564 | 0.348 | 0.288 | 0.303 | 0.0554 | 0.1175 |
| S2 | 300 | G/A | Exon1(UTR) | 384 | 530 | 558 | 0.445 | 0.462 | 0.504 | 0.6037 | 0.0757 |
| S3 | 498 | C/G | Exon1(UTR) | 392 | 534 | 558 | 0.401 | 0.427 | 0.468 | 0.4121 | 0.0393* |
| S4 | 948 | A/G | Exon1(UTR) | 396 | 532 | 564 | 0.477 | 0.417 | 0.415 | 0.0665 | 0.0480* |
| S5 | 1042 | G/A | Exon1(UTR) | 396 | 514 | 564 | 0.422 | 0.350 | 0.367 | 0.0276* | 0.0844 |
| S6 | 2803 | G/A | Intron1 | 390 | 538 | 554 | 0.141 | 0.164 | 0.132 | 0.3396 | 0.6785 |
| S7 | 2988 | C/T | Intron1 | 388 | 528 | 554 | 0.327 | 0.339 | 0.377 | 0.7125 | 0.1169 |
| S8 | 6063 | C/T | Intron1 | 396 | 536 | 562 | 0.109 | 0.119 | 0.117 | 0.5876 | 0.6338 |
| S9 | 10 927 | T/G | Intron1 | 390 | 534 | 548 | 0.474 | 0.446 | 0.451 | 0.3808 | 0.4641 |
| S10 | 14 081 | C/T | Exon2(UTR) | 378 | 516 | 554 | 0.373 | 0.399 | 0.419 | 0.4484 | 0.1632 |
| dS1 | 29 784 | C/T | 3′-Downstream | 372 | 522 | 542 | 0.194 | 0.218 | 0.173 | 0.3565 | 0.4316 |
*Significant difference at the 5% level, **At the 1% level.
Allele frequencies of SNPs of EP2 and comparisons between AIA and ATA and AIA and CTR
| . | Position . | Variation . | Localization . | Allele count . | Minor allele frequency . | Permutation P value . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | AIA . | CTR . | ATA . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| uS5 | −12 813 | G/A | 5′-Upstream | 396 | 548 | 564 | 0.311 | 0.221 | 0.222 | 0.0016** | 0.0017** |
| uS6 | −10 918 | A/G | 5′-Upstream | 392 | 528 | 526 | 0.395 | 0.449 | 0.451 | 0.0844 | 0.0912 |
| uS7 | −10 814 | T/A | 5′-Upstream | 396 | 540 | 562 | 0.492 | 0.428 | 0.397 | 0.0393* | 0.0025** |
| uS8 | −10 250 | A/G | 5′-Upstream | 384 | 522 | 516 | 0.393 | 0.433 | 0.438 | 0.1767 | 0.1716 |
| uS9 | −7075 | A/G | 5′-Upstream | 384 | 504 | 522 | 0.432 | 0.452 | 0.469 | 0.5329 | 0.2594 |
| uS10 | −6179 | A/G | 5′-Upstream | 374 | 526 | 564 | 0.374 | 0.401 | 0.452 | 0.4213 | 0.0199* |
| S1 | −609 | G/A | Promoter | 396 | 538 | 564 | 0.348 | 0.288 | 0.303 | 0.0554 | 0.1175 |
| S2 | 300 | G/A | Exon1(UTR) | 384 | 530 | 558 | 0.445 | 0.462 | 0.504 | 0.6037 | 0.0757 |
| S3 | 498 | C/G | Exon1(UTR) | 392 | 534 | 558 | 0.401 | 0.427 | 0.468 | 0.4121 | 0.0393* |
| S4 | 948 | A/G | Exon1(UTR) | 396 | 532 | 564 | 0.477 | 0.417 | 0.415 | 0.0665 | 0.0480* |
| S5 | 1042 | G/A | Exon1(UTR) | 396 | 514 | 564 | 0.422 | 0.350 | 0.367 | 0.0276* | 0.0844 |
| S6 | 2803 | G/A | Intron1 | 390 | 538 | 554 | 0.141 | 0.164 | 0.132 | 0.3396 | 0.6785 |
| S7 | 2988 | C/T | Intron1 | 388 | 528 | 554 | 0.327 | 0.339 | 0.377 | 0.7125 | 0.1169 |
| S8 | 6063 | C/T | Intron1 | 396 | 536 | 562 | 0.109 | 0.119 | 0.117 | 0.5876 | 0.6338 |
| S9 | 10 927 | T/G | Intron1 | 390 | 534 | 548 | 0.474 | 0.446 | 0.451 | 0.3808 | 0.4641 |
| S10 | 14 081 | C/T | Exon2(UTR) | 378 | 516 | 554 | 0.373 | 0.399 | 0.419 | 0.4484 | 0.1632 |
| dS1 | 29 784 | C/T | 3′-Downstream | 372 | 522 | 542 | 0.194 | 0.218 | 0.173 | 0.3565 | 0.4316 |
| . | Position . | Variation . | Localization . | Allele count . | Minor allele frequency . | Permutation P value . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | AIA . | CTR . | ATA . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| uS5 | −12 813 | G/A | 5′-Upstream | 396 | 548 | 564 | 0.311 | 0.221 | 0.222 | 0.0016** | 0.0017** |
| uS6 | −10 918 | A/G | 5′-Upstream | 392 | 528 | 526 | 0.395 | 0.449 | 0.451 | 0.0844 | 0.0912 |
| uS7 | −10 814 | T/A | 5′-Upstream | 396 | 540 | 562 | 0.492 | 0.428 | 0.397 | 0.0393* | 0.0025** |
| uS8 | −10 250 | A/G | 5′-Upstream | 384 | 522 | 516 | 0.393 | 0.433 | 0.438 | 0.1767 | 0.1716 |
| uS9 | −7075 | A/G | 5′-Upstream | 384 | 504 | 522 | 0.432 | 0.452 | 0.469 | 0.5329 | 0.2594 |
| uS10 | −6179 | A/G | 5′-Upstream | 374 | 526 | 564 | 0.374 | 0.401 | 0.452 | 0.4213 | 0.0199* |
| S1 | −609 | G/A | Promoter | 396 | 538 | 564 | 0.348 | 0.288 | 0.303 | 0.0554 | 0.1175 |
| S2 | 300 | G/A | Exon1(UTR) | 384 | 530 | 558 | 0.445 | 0.462 | 0.504 | 0.6037 | 0.0757 |
| S3 | 498 | C/G | Exon1(UTR) | 392 | 534 | 558 | 0.401 | 0.427 | 0.468 | 0.4121 | 0.0393* |
| S4 | 948 | A/G | Exon1(UTR) | 396 | 532 | 564 | 0.477 | 0.417 | 0.415 | 0.0665 | 0.0480* |
| S5 | 1042 | G/A | Exon1(UTR) | 396 | 514 | 564 | 0.422 | 0.350 | 0.367 | 0.0276* | 0.0844 |
| S6 | 2803 | G/A | Intron1 | 390 | 538 | 554 | 0.141 | 0.164 | 0.132 | 0.3396 | 0.6785 |
| S7 | 2988 | C/T | Intron1 | 388 | 528 | 554 | 0.327 | 0.339 | 0.377 | 0.7125 | 0.1169 |
| S8 | 6063 | C/T | Intron1 | 396 | 536 | 562 | 0.109 | 0.119 | 0.117 | 0.5876 | 0.6338 |
| S9 | 10 927 | T/G | Intron1 | 390 | 534 | 548 | 0.474 | 0.446 | 0.451 | 0.3808 | 0.4641 |
| S10 | 14 081 | C/T | Exon2(UTR) | 378 | 516 | 554 | 0.373 | 0.399 | 0.419 | 0.4484 | 0.1632 |
| dS1 | 29 784 | C/T | 3′-Downstream | 372 | 522 | 542 | 0.194 | 0.218 | 0.173 | 0.3565 | 0.4316 |
*Significant difference at the 5% level, **At the 1% level.
Haplotype-based association study with AIA
| Haplotype: uS5/uS7/S1/S3/S6/S8 . | Haplotype frequency . | Permutation P value . | ||||
|---|---|---|---|---|---|---|
| . | Total . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| M/M/M/m/M/M | 0.381 | 0.341 | 0.375 | 0.415 | 0.3074 | 0.0295* |
| m/m/m/M/M/M | 0.160 | 0.212 | 0.130 | 0.155 | 0.0012** | 0.0259* |
| M/m/M/M/m/M | 0.104 | 0.101 | 0.120 | 0.092 | 0.3515 | 0.6218 |
| M/m/M/M/M/M | 0.064 | 0.062 | 0.072 | 0.057 | 0.5298 | 0.7388 |
| M/M/M/M/M/M | 0.059 | 0.056 | 0.060 | 0.058 | 0.8965 | 0.9032 |
| M/M/m/M/M/m | 0.055 | 0.034 | 0.061 | 0.066 | 0.0636 | 0.0488* |
| Haplotype: uS5/uS7/S1/S3/S6/S8 . | Haplotype frequency . | Permutation P value . | ||||
|---|---|---|---|---|---|---|
| . | Total . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| M/M/M/m/M/M | 0.381 | 0.341 | 0.375 | 0.415 | 0.3074 | 0.0295* |
| m/m/m/M/M/M | 0.160 | 0.212 | 0.130 | 0.155 | 0.0012** | 0.0259* |
| M/m/M/M/m/M | 0.104 | 0.101 | 0.120 | 0.092 | 0.3515 | 0.6218 |
| M/m/M/M/M/M | 0.064 | 0.062 | 0.072 | 0.057 | 0.5298 | 0.7388 |
| M/M/M/M/M/M | 0.059 | 0.056 | 0.060 | 0.058 | 0.8965 | 0.9032 |
| M/M/m/M/M/m | 0.055 | 0.034 | 0.061 | 0.066 | 0.0636 | 0.0488* |
M and m denotes major and minor alleles, respectively.
*Significant difference at the 5% level, **At the 1% level.
Haplotype-based association study with AIA
| Haplotype: uS5/uS7/S1/S3/S6/S8 . | Haplotype frequency . | Permutation P value . | ||||
|---|---|---|---|---|---|---|
| . | Total . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| M/M/M/m/M/M | 0.381 | 0.341 | 0.375 | 0.415 | 0.3074 | 0.0295* |
| m/m/m/M/M/M | 0.160 | 0.212 | 0.130 | 0.155 | 0.0012** | 0.0259* |
| M/m/M/M/m/M | 0.104 | 0.101 | 0.120 | 0.092 | 0.3515 | 0.6218 |
| M/m/M/M/M/M | 0.064 | 0.062 | 0.072 | 0.057 | 0.5298 | 0.7388 |
| M/M/M/M/M/M | 0.059 | 0.056 | 0.060 | 0.058 | 0.8965 | 0.9032 |
| M/M/m/M/M/m | 0.055 | 0.034 | 0.061 | 0.066 | 0.0636 | 0.0488* |
| Haplotype: uS5/uS7/S1/S3/S6/S8 . | Haplotype frequency . | Permutation P value . | ||||
|---|---|---|---|---|---|---|
| . | Total . | AIA . | CTR . | ATA . | AIA versus CTR . | AIA versus ATA . |
| M/M/M/m/M/M | 0.381 | 0.341 | 0.375 | 0.415 | 0.3074 | 0.0295* |
| m/m/m/M/M/M | 0.160 | 0.212 | 0.130 | 0.155 | 0.0012** | 0.0259* |
| M/m/M/M/m/M | 0.104 | 0.101 | 0.120 | 0.092 | 0.3515 | 0.6218 |
| M/m/M/M/M/M | 0.064 | 0.062 | 0.072 | 0.057 | 0.5298 | 0.7388 |
| M/M/M/M/M/M | 0.059 | 0.056 | 0.060 | 0.058 | 0.8965 | 0.9032 |
| M/M/m/M/M/m | 0.055 | 0.034 | 0.061 | 0.066 | 0.0636 | 0.0488* |
M and m denotes major and minor alleles, respectively.
*Significant difference at the 5% level, **At the 1% level.
References
Szczeklik, A. and Stevenson, D.D. (
Sampson, A., Holgate, S., Austen, K.F. and Szczeklik, A. (
Szczeklik, A., Sanak, M., Nizankowska-Mogilnicka, E. and Kielbasa, B. (
Pierzchalska, M., Szabo, Z., Sanak, M., Soja, J. and Szczeklik, A. (
Cowburn, A.S., Sladek, K., Soja, J., Adamek, L., Nizankowska, E., Szczeklik, A., Lam, B.K., Penrose, J.F., Austen, F.K., Holgate, S.T. et al. (
Lam, B.K. and Frank Austen, K. (
Szczeklik, A., Nizankowska, E. and Duplaga, M. (
Sanak, M., Simon, H.U. and Szczeklik, A. (
Sanak, M., Pierzchalska, M., Bazan-Socha, S. and Szczeklik, A. (
Van Sambeek, R., Stevenson, D.D., Baldasaro, M., Lam, B.K., Zhao, J., Yoshida, S., Yandora, C., Drazen, J.M. and Penrose, J.F. (
Kawagishi, Y., Mita, H., Taniguchi, M., Maruyama, M., Oosaki, R., Higashi, N., Kashii, T., Kobayashi, M. and Akiyama, K. (
Choi, J.H., Park, H.S., Oh, H.B., Lee, J.H., Suh, Y.J., Park, C.S. and Shin, H.D. (
Nei, M. and Kumar, S. (
Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A.E., Kel, O.V., Ignatieva, E.V., Ananko, E.A., Podkolodnaya, O.A., Kolpakov, F.A. et al. (
Kotanides, H. and Reich, N.C. (
Sampson, A.P., Cowburn, A.S., Sladek, K., Adamek, L., Nizankowska, E., Szczeklik, A., Lam, B.K., Penrose, J.F., Austen, K.F. and Holgate, S.T. (
Pierzchalska, M., Mastalerz, L., Sanak, M., Zazula, M. and Szczeklik, A. (
Antczak, A., Montuschi, P., Kharitonov, S., Gorski, P. and Barnes, P.J. (
Sousa, A.R., Parikh, A., Scadding, G., Corrigan, C.J. and Lee, T.H. (
Steinke, J.W., Bradley, D., Arango, P., Crouse, C.D., Frierson, H., Kountakis, S.E., Kraft, M. and Borish, L. (
Celik, G., Bavbek, S., Misirligil, Z. and Melli, M. (
Sestini, P., Armetti, L., Gambaro, G., Pieroni, M.G., Refini, R.M., Sala, A., Vaghi, A., Folco, G.C., Bianco, S. and Robuschi, M. (
Tenor, H., Hatzelmann, A., Church, M.K., Schudt, C. and Shute, J.K. (
Coleman, R.A., Smith, W.L. and Narumiya, S. (
Kennedy, C.R., Zhang, Y., Brandon, S., Guan, Y., Coffee, K., Funk, C.D., Magnuson, M.A., Oates, J.A., Breyer, M.D. and Breyer, R.M. (
Tilley, S.L., Audoly, L.P., Hicks, E.H., Kim, H.S., Flannery, P.J., Coffman, T.M. and Koller, B.H. (
Jenkins, C., Costello, J. and Hodge, L. (
National Heart, Lung and Blood Institute (
Zhao, J.H., Curtis, D. and Sham, P.C. (
Hill, W.G. and Robertson, A. (