-
PDF
- Split View
-
Views
-
Cite
Cite
Hans-Christian Sigle, Sascha Thewes, Markus Niewerth, Hans Christian Korting, Monika Schäfer-Korting, Bernhard Hube, Oxygen accessibility and iron levels are critical factors for the antifungal action of ciclopirox against Candida albicans, Journal of Antimicrobial Chemotherapy, Volume 55, Issue 5, May 2005, Pages 663–673, https://doi.org/10.1093/jac/dki089
Close - Share Icon Share
Abstract
Objectives: Ciclopirox is a topical antifungal agent of the hydroxypyridone class whose mode of action is poorly understood. In order to elucidate the mechanism of action of ciclopirox, we analysed the growth, cellular integrity, biochemical properties, viability and transcriptional profile of the polymorphic yeast Candida albicans following exposure to this antifungal agent.
Methods: Multiple biochemical assays served to identify factors that were critical for antifungal activity and to identify proteins whose activities changed in drug-exposed cells. Genome-wide transcriptional profiling was used to identify genes that were up-regulated in response to the cellular effects of the drug.
Results: Ciclopirox inhibited growth of C. albicans yeast and hyphal cells in a dose-dependent manner. This effect was reduced (i) by the addition of iron ions or the metabolic inhibitor 2-deoxy-D-glucose to growth media, (ii) in media that lacked glucose, and (iii) for cells that were pre-incubated with hydrogen peroxide or menadione [which caused induction of proteins involved in detoxification of reactive oxygen species (ROS)]. In contrast, cells pre-cultured under poor oxygen conditions (which had decreased activity of proteins involved in ROS detoxification) were more susceptible to ciclopirox. Treatment with ciclopirox did not directly cause cell membrane damage and did not change intracellular levels of ATP. Finally, the transcriptional profiling pattern of drug-treated cells strongly resembled iron-limited conditions.
Conclusions: These data indicate that metabolic activity, oxygen accessibility and iron levels are critical parameters in the mode of action of ciclopirox olamine.
Introduction
Hydroxypyridones are effective topically used antifungal agents with a very broad spectrum against dermatophytes, yeasts, filamentous fungi and bacteria. Despite the fact that hydroxypyridones have been in clinical use for more than 20 years, their mode of action is poorly understood. This is in contrast to other antimycotics such as polyenes, azoles, flucytosine or echinocandins.1,2
The minimal inhibitory concentration (MIC) of the hydroxypyridone ciclopirox olamine [ethanolamine salt of 6-cyclohexyl-1-hydroxy-4-methyl-2(1H)-pyridone, in the following text referred to as ‘ciclopirox’] was found to be between 0.49 and 3.9 mg/L for many human pathogenic fungi.3 Iwata & Yamaguchi4 showed that even low concentrations of ciclopirox inhibit the cellular uptake of amino acids, potassium and phosphate ions. It was concluded that the reduced biosynthesis and respiration of ciclopirox-treated cells resulted from the reduced ability to store these essential components. In contrast to polyenes and azoles, however, cell membranes were damaged only when very high concentrations (50–200 mg/L) of hydroxypyridones were added to the cells.5–7 Furthermore, studies using Candida albicans protoplasts and membrane models with ergosterol-containing liposomes showed that ciclopirox does not directly damage or interact with cell membranes.4 Since azole-resistant strains are still susceptible to ciclopirox, the inhibition of ergosterol biosynthesis can be excluded as the principal mode of action of ciclopirox.8 Rilopirox, another hydroxypyridone, reduces the respiratory activity of Saccharomyces cerevisiae possibly by influencing the uptake or metabolism of glucose. Hydroxypyridones have also been shown to inhibit the activity of the metal ion containing enzymes NADH-ubiquinone-oxidoreductase (complex I) and catalase.9 Since numerous cellular proteins depend on metal ions as co-factors, binding of metal ions may be of major importance in the mode of action of hydroxypyridone antimycotics.
In this study, we analysed the effect of ciclopirox on the growth, cellular integrity and viability of C. albicans cells and used a series of biochemical assays to identify parameters that are critical for antifungal activity and to identify proteins whose activities are modulated in drug-exposed cells. Furthermore, we used genome-wide transcriptional profiling to identify genes that may be up-regulated in response to the drug.
Materials and methods
Strains and growth conditions
Growth and biochemical assays were carried out with the type strain C. albicans CBS 562 (kindly provided by H. J. Tietz, Berlin, Germany) and the clinical isolate 5342 ZN (Dermatologische Klinik, Munich, Germany). Strain CBS 562, a relatively poor germ tube producer, was preferred to strain 5342 ZN in experiments when yeast growth was favoured, for example to monitor viability. For transcriptional profiling experiments, we used the strain SC5314,10 which was used for the Stanford sequencing project (http://www-sequence.stanford.edu/group/candida). Susceptibility of SC5314 to ciclopirox was investigated as described recently.11
Semi-synchronized cells were used to standardize growth experiments. To semi-synchronize the cells,12 25 mL of modified Sabouraud (0.2%)13 glucose medium was inoculated with 2 × 105 cells/mL and incubated at 150 rpm for 16 h at room temperature. Cells were harvested and inoculated (4 × 106 cells/mL) into fresh medium and shaken for 24 h at 37°C. Cells were again harvested, washed three times with phosphate-buffered saline (PBS) pH 7.4 and used in further experiments.
To analyse the susceptibility of C. albicans to antifungal drugs (IC30), cells were grown in Yeast Nitrogen Base (YNB) (Difco Laboratories) and Lee's medium14 and tested in a microdilution test. Drug solutions (100 μL), 90 μL of medium and 10 μL of semi-synchronized cells (107 cells/mL) were added to the wells of a 96-well plate. Final drug concentrations were between 0.0037 and 128 mg/L. Controls included medium without any antifungal agent and medium containing drug solution without fungi. Sealed plates were incubated for 24 h at 37°C and the OD540 was measured using an ELISA reader. The calculated IC30 value was the lowest drug concentration that caused a 70% reduction in the OD540 of the untreated control. The IC30 value was preferred to the minimal inhibitory concentration (MIC) since the IC30 value is considered to be more reliable and independent of the inoculum size.15,16
For hyphal induction, semi-synchronized cells (107 cells/mL) were incubated in Lee's medium pH 6.8, 5 mM ornithine/10 mM glucose in PBS, 5 mM N-acetylglucosamine (GlcNAc)/10 mM glucose in PBS or 10% human serum at 37°C. To analyse the inhibition of germ tube formation by drugs (G-IC30 value), 100 μL of drug solution, 90 μL of medium and 10 μL of semi-synchronized cells (2 × 108 cells/mL) were added to the wells of a 96-well plate. Final drug concentrations were between 0.125 and 64 mg/L. Plates were incubated for 4 h at 37°C. Fixation solution (40 μL, 10% SDS/10% formalin) was added to the wells and the percentage of germ tubes was counted microscopically (three times 100 randomly chosen cells). The G-IC30 value was the lowest drug concentration that caused a 70% reduction in the germ tube formation of the untreated control.
To analyse the influence of metal ions on the inhibitory effect of ciclopirox (acid, Aventis) to germ tube formation, we mixed 11.1 mM proline, 22.2 mM glucose and 222 μM of metal ions (MgSO4, CaCl2, MnCl2, FeCl2 and FeCl3) in PBS pH 7.4. The ciclopirox stock solution was diluted with PBS pH 7.4 to give concentrations from 0.5 to 64 mg/L. One hundred microlitres of drug solution, 90 μL of test medium, and 10 μL of semi-synchronized cells (2 × 108/mL) were mixed and incubated as described above and the G-IC30 value was calculated.
To measure the viability of cells exposed to drugs, treated or untreated cells were diluted and plated on modified Sabouraud glucose agar or mixed into sterile 40°C cooled agar and colony forming units (cfu) were counted after 48–72 h incubation at room temperature. The cfu was calculated relative to the viability of untreated cells at the start of the experiments.
Treatment with inhibitors and inducers
For incubation in 2-deoxy-D-glucose-supplemented and glucose-free buffer, semi-synchronized cells (107 cells/mL) were inoculated into 2 mL Eppendorf tubes containing either PBS, 10 mM glucose in PBS or 10 mM 2-deoxy-D-glucose in PBS supplemented with or without ciclopirox (32 or 64 mg/L). Incubation was stopped by 10 000 × dilution with glucose-free buffer after 4 h at 37°C and the number of cfu was counted.
For pre-treatment with either hydrogen peroxide (H2O2) or menadione, cells were cultivated in 800 mL of Sabouraud 2% glucose to exponential phase (5 × 106−1 × 107 cells/mL) and divided into 50 mL portions. To each portion, 50 μL of 100 mM menadione in DMSO or 50 μL of 1 M H2O2 was added. Control cultures contained the corresponding concentration of DMSO. Cultures were incubated for 60 min (H2O2) or 90 min (menadione) at 37°C, harvested and washed in PBS pH 7.5. Washed cells (final density 107 cells/mL) were then inoculated into media supplemented with ciclopirox as described above.
For depletion of cellular catalase activity 4 × 106 cells/mL were pre-incubated in modified Sabouraud glucose for 16 h at 37°C supplemented with 20 or 40 mM of the catalase inhibitor 3-amino-1,2,4-triazole. Cells were then washed in PBS and exposed to ciclopirox as described above.
Cell membrane damage assay
To analyse the effect of various antifungal agents on cell membrane integrity, potassium concentrations were measured in the supernatant of cultures. Semi-synchronized cells (109 cells/mL) were washed three times with 20 mM sodium phosphate buffer pH 7.2 and added to 1 mL of buffer containing 16 and 32 mg/L ciclopirox (final cell number 108 cells/mL). Controls contained no drug. The maximum possible potassium concentration was measured from the supernatant of 108 untreated cells boiled for 5 min. The potassium concentration was obtained by flame photometry using CsCl as an internal standard and defined concentrations of potassium (0–2 mM) in 20 mM sodium phosphate buffer pH 7.2.
Intracellular ATP concentrations
In order to measure the intracellular ATP concentration, semi-synchronized cells (107) were incubated in 5 mM proline/10 mM glucose/PBS supplemented with ciclopirox at 37°C for 30 min. The cell suspension (1 mL) was diluted with 25 mM Tris–EDTA pH 7.8 and heated at 100°C for 4 min. The suspension was cooled on ice, centrifuged and the ATP content of the supernatant was measured. Controls contained no drug. ATP was measured using the luciferin–luciferase bioluminescence-based ‘ATP Determination Kit’ (Molecular Probes) according to the manufacturer's instructions.
Activity of catalase, glucose-6-phosphate-dehydrogenase, cytochrome c peroxidase, and superoxide dismutase
To measure catalase, glucose-6-phosphate-dehydrogenase (G6P-DH), cytochrome c peroxidase, and superoxide dismutase (Sod) activity, cells were grown in 800 mL of Sabouraud 2% glucose to exponential phase (5 × 106−1 × 107 cells/mL) and divided into 50 mL portions supplemented with ciclopirox and incubated (150 rpm) for 90 min at 37°C. Cells were harvested and washed twice with PBS. For cytochrome c peroxidase activity, cells (2 × 105) were incubated in Sabouraud glucose for 24 h, harvested, washed in PBS resuspended in fresh medium (5 × 107 cells/mL), and exposed to ciclopirox for 90 min as described above. Cell pellets were lysed with a bead mill in 500 μL of PBS twice for 3 min on ice and the extracts were centrifuged for 1 min at maximum speed in a benchtop centrifuge.
The protein content of the supernatants was measured17 to give specific enzyme activities for catalase,18 G6P-DH,19 cytochrome c peroxidase,20 and Sod.21 In order to discriminate between total Sod and Mn-Sod activity, we used KCN (6.1 mM) to inhibit any CuZn-Sod activity.
RNA extraction and microarray analysis
RNA was extracted as described previously.11 mRNA was translated into cDNA and labelled as follows. Oligo dT (2 μg) was added to 30 μg of total RNA and brought up to 25 μL with RNase-free water. After denaturation at 70°C for 10 min, the RNA was incubated for 1 min on ice and 30 μL of labelling master mix (1 × RT buffer (Invitrogen), 1 mM DTT, 500 μM dATP, dCTP, dGTP, and 100 μM dTTP) was added. Cy3 dUTP (3 μL, Amersham) was then added to one sample and Cy5 dUTP (3 μL, Amersham) to the other sample and cDNA synthesis was initiated with 2 μL of Superscript II reverse transcriptase (400 U) (Invitrogen). After 2 h at 42°C, 3 μL of 20 μM EDTA was added to stop the reaction and 3 μL of 500 mM NaOH was added to degrade the RNA for 10 min at 70°C. Hydrochloric acid (3 μL, 500 mM) was added to neutralize the reaction. The labelled cDNA was purified using spin columns (Macherey–Nagel), dried and resuspended in 20 μL of water.
For transcriptional profiling, we used C. albicans microarrays (Eurogentec) containing 6039 open reading frames (ORFs) and 27 control genes spotted in duplicate on a glass slide. The C. albicans MicroArray has been developed in collaboration with the European Galar Fungail Consortium (www.pasteur.fr/recherche/unites/Galar_Fungail). The Stanford Genome Technology Center generated the nucleotide sequence data for C. albicans with funding from NIDCR, NIH and the Burroughs Wellcome Fund. Information about coding sequences and proteins was obtained from the CandidaDB database (www.pasteur.fr/recherche/unites/Galar_Fungail/CandidaDB), which has been developed by the Galar Fungail European Consortium. Arrays were designed as described under http://www.pasteur.fr/recherche/unites/Galar_Fungail/arrays.html.
For array hybridization, C. albicans MicroArrays were pre-hybridized for 45 min at 42°C in pre-hybridization mix [5 × sodium citrate (SSC), 1% sodium dodecyl sulphate (SDS), 1% bovine serum albumin (BSA)] and washed with water and isopropanol. Hybridization solution (25 μL, 50% formamide/10× SSC/0.2% SDS) was mixed with 25 μL of the purified Cy3- and Cy5-labelled cDNA mix, boiled for 3 min and spotted on the MicroArray slide. The array was covered with a 25 × 44 mm LifterSlip (Erie Scientific) and incubated overnight at 42°C in a hybridization chamber (Corning). Arrays were washed in 2× SSC, 1% SDS for 15 min, in 1× SSC, 0.2% SDS for 8 min, and in 0.1× SSC, 0.2% SDS for 5 min at room temperature with agitation. Slides were dried in a 50 mL Falcon tube by centrifugation at low speed for 4 min.
Hybridized slides were scanned using an Axon 4000B scanner. Data were extracted using GenePix 4.1 (Axon). Before the extracted data were normalized, data were adjusted by background correction (data transformation). Signals with the same intensity as the local background were neglected.22,23
This transformation was followed by an intensity-dependent per spot and per chip normalization (LOWESS). Data analysis was carried out using GeneSpring 5.0 (Silicon Genetics) from triplicate (two biological independent experiments and one dye swap) experiments.
Statistical analysis
All data presented are given as the average of n ≥ 3 samples. Standard deviations (SD) are indicated in each case. All experiments were repeated at least once. Shapiro–Wilk-, F- and Student's t-test were carried out to test statistical significance. A value of P ≤ 0.05 was taken as significant.
Results
Antifungal activity of ciclopirox is media dependent
In order to evaluate the antifungal effects of ciclopirox on C. albicans, we used IC30 values (lowest drug concentration which caused 70% reduction in the OD540 of the untreated control), G-IC30 values (lowest drug concentration which caused 70% reduction in the germ tube formation of the untreated control) and a viability test (cfu). Two C. albicans strains (5342 ZN and CBS 562) were tested in various media (Lee's medium, YNB and YNB-4% human serum albumin) to quantify the IC30 values for ciclopirox. The susceptibility of both strains was similar; however, the IC30 values were strongly dependent on the media used. The IC30 values for both strains were 2 mg/L for Lee's medium and between 8 and 16 mg/L for YNB. Addition of human serum albumin further enhanced this effect (128 mg/L for 5342 ZN and 32 mg/L for CBS 562) suggesting that human serum albumin may bind to the drug and reduce its activity. Addition of DMSO, which was used to prepare the ciclopirox stock solution, did not influence the IC30 values at concentrations up to 2.5%. Final concentrations of DMSO did not exceed 1% in any experiment indicating that the solvent did not cause any inhibitory effects.
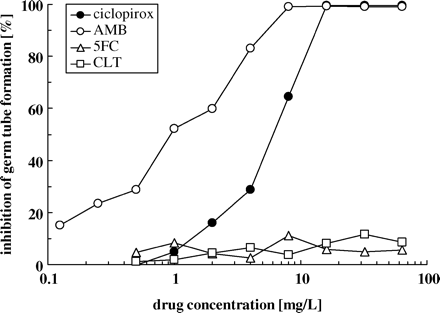
Ciclopirox inhibits germ tube formation of C. albicans
C. albicans is able to grow in two main morphological forms, as a spherical yeast or as a long filamentous hyphal cell. In order to investigate the influence of ciclopirox on growth during the yeast to hyphal transition, semi-synchronized yeast cells of strain 5342 ZN were incubated at 37°C in Lee's medium pH 6.8 containing ciclopirox at concentrations ranging from 0.125 to 64 mg/L and the percent inhibition of hyphae formation was monitored after 4 h of incubation. In parallel, cells were incubated under the same conditions in media containing amphotericin B, flucytosine and clotrimazole. As with amphotericin B, hyphal formation was inhibited by ciclopirox in a concentration-dependent manner (Figure 1). In contrast, flucytosine and clotrimazole did not inhibit germ tube formation even at the highest concentrations tested. These results were confirmed for cells grown in hyphal inducing media containing 5 mM proline/10 mM glucose, 5 mM ornithine/10 mM glucose and 5 mM GlcNAc/10 mM glucose (data not shown).
Addition of iron ions to culture media reverses the inhibitory effect of ciclopirox
Since the effect of hydroxypyridones may be caused by the binding of cellular metal ions,8 we investigated whether the addition of metal ions to culture media may influence the inhibitory effect of ciclopirox. First, semi-synchronized cells of strain 5342 ZN (107 cell/mL) were incubated in 5 mM proline/10 mM glucose/PBS supplemented with 100 μM Ca2+, Mg2+, Mn2+, Fe2+ or Fe3 + and the percentage of germ tubes was determined. None of the metal ions significantly reduced the ability to produce germ tubes (data not shown). Furthermore, the addition of Ca2+, Mg2+ and Mn2+ did not influence the G-IC30 of ciclopirox in the same medium; however, the addition of both Fe2+ and Fe3 + (G-IC30 64 mg/L) clearly reduced the effect of ciclopirox compared with cultures without the addition of metal ions (G-IC30 8 mg/L) indicating that these ions possibly interacted with the drug and thus prevented the inhibition of hyphal formation.
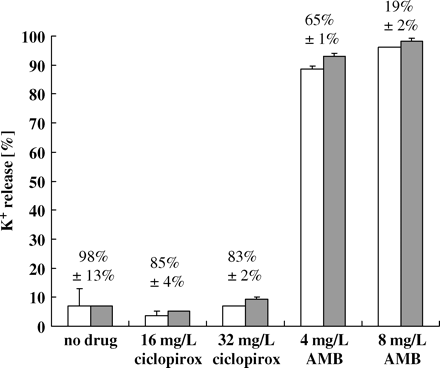
Treatment with ciclopirox does not cause cell membrane damage
Antifungal drugs such as polyenes and azoles act primarily on the cell membrane of fungi. In order to investigate whether ciclopirox has an effect on the cell membrane of C. albicans, we tested the cell membrane integrity of strain CBS 562 by measuring the extracellular potassium concentration of drug-treated cells. Damage of the cell membrane should increase the extracellular concentration of potassium in the medium. Semi-synchronized cells of CBS 562 (108 cells/mL) were left untreated or exposed to 16 and 32 mg/L ciclopirox or 4 and 8 mg/L amphotericin B in 20 mM sodium phosphate buffer pH 7.2 for 30 and 60 min (Figure 2). The maximum possible potassium concentration (100% in Figure 2) was measured as 0.60 ± 0.01 mM from boiled untreated cells. Amphotericin B at both concentrations caused more than 90% release of potassium, but the viability still exceeded 50% after 60 min. In contrast, exposure to ciclopirox reduced the cell viability to 20% without causing any release of potassium into the culture medium compared with untreated cells, indicating that cell membrane damage was unlikely.
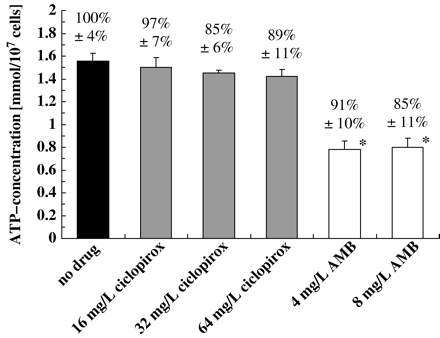
Treatment with ciclopirox does not change intracellular levels of ATP
It has been postulated that hydroxypyridones inhibit essential cellular processes possibly by blocking the uptake of macromolecule precursors and essential ions into the fungal cell.4 Such a mode of action would probably cause a direct or indirect depletion of cellular energy. In order to investigate this hypothesis, we measured the intracellular level of ATP after cells were exposed to ciclopirox. Semi-synchronized cells (107 cells/mL) of strain 5342 ZN were incubated in 5 mM proline/10 mM glucose/PBS in the presence and absence of 16, 32, or 64 mg/L ciclopirox and 4 or 8 mg/L amphotericin B for 30 min. Cells were extracted with hot buffer and the ATP content was measured (Figure 3). Treatment with amphotericin B significantly reduced the cellular content of ATP at both concentrations whereas ciclopirox had no effect. Similar results were obtained after prolonged incubations and in media lacking glucose (data not shown). The ATP concentration of the culture supernatant was not more than 5% of the total content. Therefore, the antifungal activity of ciclopirox was not caused by a lack of intracellular ATP.
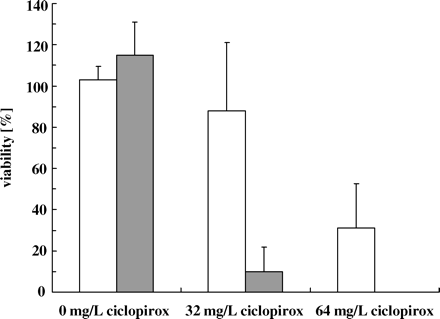
Oxygen influences the fungicidal effect of ciclopirox
Previous data from our group indicate that ciclopirox causes increased sensitivity to oxidative stress.11 To investigate the influence of oxygen on the fungicidal activity of ciclopirox, we pre-cultivated cells of C. albicans strain CBS 562 under reduced oxygen conditions and then exposed these cells to ciclopirox. Cells were pre-incubated in modified Sabouraud glucose in batch cultures (4 × 106 cells/mL) with and without shaking at 37°C for 24 h, then harvested and incubated in 5 mM proline/10 mM glucose/PBS supplemented with 32 or 64 mg/L ciclopirox under aerobic conditions (Figure 4). The viability of cells in control cultures without drug was similar for cells pre-cultured under oxygen-rich (with shaking) or reduced oxygen (without shaking) conditions; however, cells pre-incubated under reduced oxygen conditions were much more susceptible to ciclopirox compared with cells grown under oxygen-rich conditions.
In order to clarify to what degree the oxygen content of the pre-cultures influenced intracellular enzymes involved in oxidative stress, we measured the specific activity of catalase, G6P-DH and cytochrome c peroxidase in pre-cultured cells. The specific activity of catalase was almost twice as high in the oxygen-rich pre-cultured cells compared with the low oxygen pre-cultured cells (97.8 ± 16.9 versus 46.0 ± 13.8 U/mL of protein; n=8; P ≤ 0.05), but the specific activity of G6P-DH (145.8 ± 7.8% versus 100 ± 8.1%; n=2; P ≤ 0.05) and cytochrome c peroxidase (0.239 ± 0.015 versus 0.076 ± 0.002 ΔA460 nm × min−1 × mg−1; n=3; P ≤ 0.05) was also higher. Therefore, poor oxygen supply reduced the specific activity of certain antioxidant enzymes, which in turn, may influence the susceptibility of cells to ciclopirox.
Addition of 2-deoxy-D-glucose reduces whereas addition of glucose enhances the effect of ciclopirox
Since oxygen seems to play an essential role in the mode of action of ciclopirox, we questioned whether metabolic activity is also necessary for the antifungal activity of this drug.
The metabolic inhibitor 2-deoxy-D-glucose prevents the import of glucose into fungal cells,24 but neither the lack of glucose in the incubation buffer nor the addition of 2-deoxy-d-glucose influenced the viability of C. albicans CBS 562 cells (Figure 5). However, cells that were incubated in the absence of glucose or in the presence of 10 mM 2-deoxy-D-glucose had a higher viability when exposed to ciclopirox compared with cells incubated in glucose-containing medium supplemented with ciclopirox (Figure 5) suggesting that 2-deoxy-d-glucose reduced and glucose enhanced the effect of the drug.
Pre-incubation of C. albicans cells with hydrogen peroxide or menadione caused higher tolerance to ciclopirox
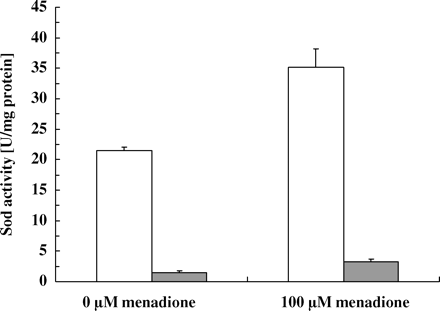
The production of reactive oxygen species (ROS) may play an important role in the mode of action of ciclopirox, since oxygen and metabolic activity are both required for the drug to be fungicidal. In this case, one would expect that the susceptibility of fungal cells to this drug was related to the accessibility and activity of enzymes involved in detoxification of ROS. In order to modulate the intracellular activity of ROS-detoxifying enzymes, we exposed exponential growing cells of C. albicans strain CBS 562 to hydrogen peroxide (H2O2) and menadione for 60 and 90 min. Sublethal concentrations of H2O2 (0.5 and 1 mM) reduced cell growth and enhanced intracellular catalase activity 11 and 17 times compared with untreated cells (Figure 6a). Menadione (100 μM) doubled the catalase activity within 90 min (Figure 6b). Menadione also increased the activity of total Sod activity by one-third compared with untreated cells (Figure 7). Since higher concentrations of the inducers were toxic to the cells, we used 1 mM H2O2 and 100 μM menadione for further experiments. Since G6P-DH provides essential co-factors for the detoxification of ROS, we also measured the activity of this enzyme in cells exposed to H2O2 and menadione. In fact, treatment with H2O2 almost doubled the activity of G6P-DH after 1 h treatment with 1 mM H2O2 compared with untreated cells (188 ± 2% versus 100 ± 6%; n=2; P ≤ 0.05). Similarly, treatment with 100 μM menadione enhanced the activity of this enzyme (146 ± 18% versus 100 ± 7%; n=3; P ≤ 0.05).
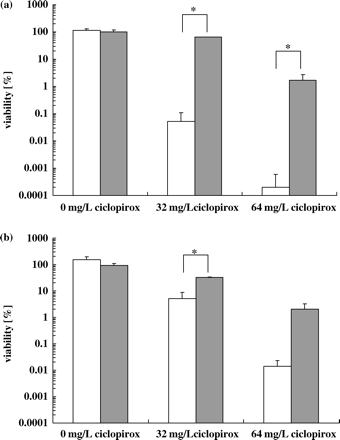
When C. albicans cells pre-treated with H2O2 or menadione were washed and incubated in 5 mM proline/10 mM glucose/PBS supplemented with ciclopirox, we observed a clear protective effect in these pre-treated cells compared with control cultures (Figure 8). Pre-treatment with H2O2 or menadione reduced the growth, but did not kill the cells. The viability of cells pre-incubated for 60 min with hydrogen peroxide (1 mM) was 114 ± 15% whereas the viability of control culture without inducer was 186 ± 26%. When cells were pre-incubated for 90 min with menadione (100 μM), the viability was 131 ± 26% and 176 ± 36% for an untreated pre-culture. Therefore, the induction of enzymes involved in the detoxification of ROS caused a higher tolerance to ciclopirox.
Ciclopirox induces intracellular glucose-6-phosphate-dehydrogenase, but reduces catalase and does not influence superoxide dismutase activity
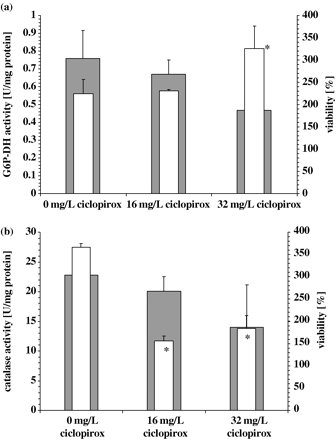
Since higher levels of ROS-detoxifying enzymes had a protective effect for cells treated with ciclopirox, it may be possible that this drug may itself cause the production of ROS. This may in turn cause the production or the induction of the activity of ROS-detoxifying enzymes. To prove this hypothesis, we investigated the influence of ciclopirox on the activity of G6P-DH, catalase and Sod in exponential growing cells of C. albicans strain CBS 562. Treatment with ciclopirox induced the activity of G6P-DH only at higher concentrations (32 mg/L) (Figure 9a), reduced the activity of catalase at 16 and 32 mg/L by ∼50% (Figure 9b) and had only a minor effect on the Sod activity at both concentrations (not shown).
Inhibition of catalase does not influence cellular viability and does not increase the fungicidal effect of ciclopirox
As shown above, ciclopirox did not induce, but rather reduced catalase activity. Therefore, catalase itself may be a target of ciclopirox and the inhibition of this enzyme may explain the mode of action of this drug. In order to prove whether reduced catalase activity may influence cell growth and viability of C. albicans and to investigate the influence of ciclopirox on cells with depleted catalase activity, we pre-incubated cells (CBS 562) with the catalase inhibitor 3-amino-1,2,4-triazole and exposed these cells to ciclopirox. As expected, 3-amino-1,2,4-triazole (20 and 40 mM) reduced the catalase activity of pre-incubated cells. The catalase activity was 65 ± 13 U/mL of protein in the untreated control culture, but only 1.7 ± 0.7 (20 mM) and 1.4 ± 0.7 (40 mM) U/mL of protein in the cultures treated with 3-amino-1,2,4-triazole; however, pre-incubation with the catalase inhibitor did not influence the viability of the cells. Furthermore, lower levels of catalase did not increase the antifungal activity of ciclopirox. Therefore, the fungicidal activity of ciclopirox appears to be independent of the intracellular activity of catalase in C. albicans.
Genome-wide transcriptional profiling of ciclopirox-exposed cells
In order to investigate the transcriptional profiling of drug-treated cells, we grew cells under subinhibitory conditions (0.6 mg/L) in modified Sabouraud glucose medium11 and isolated RNA as described previously.11 Control cultures were left untreated and RNA was isolated at the same time point. mRNA was translated into cDNA and labelled with Cy3 and Cy5. Labelled cDNA from drug-treated and control cultures were used to hybridize microarrays representing PCR fragments of the complete genome of C. albicans (6039 genes).
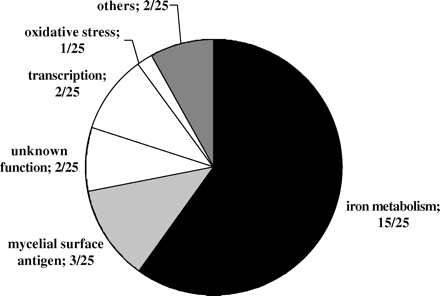
Out of the 6039 genes of the C. albicans genome, only 25 were found to be significantly up-regulated ( > 2-fold) and only 21 genes were down-regulated ( > 2-fold) in drug-exposed cells under subinhibitory conditions. The vast majority of the up-regulated genes (15 genes) were involved in iron metabolism (Figure 10 and Table 1). These included known genes encoding iron reductases (CFL1), iron permeases and transporters (FTR1, FTR2 and FTH1) which were previously found up- (FTR1, FTH1) or down- (FTR2) regulated by semi-quantitative RT–PCR and northern analysis in ciclopirox-exposed cells,11 confirming the quality of the array data. In addition, we also found a number of strongly up-regulated genes possibly involved in iron metabolism (by homology) that have not yet been described in C. albicans (CFL2, CFL12, FET5, FET32, FET33, FET34, FRE5, FRE31, FRE32). Furthermore, a number of genes encoding proteins similar to the GPI-protein Rbt5 (RBT5, RBT2, IPF12101, CSA1), two genes encoding proteins with unknown functions, two genes encoding transcriptional factors, one gene encoding a RNA binding protein, one gene encoding NADP-glutamate-dehydrogenase and one gene (IPF1218/SOD4) encoding the superoxide dismutase Sod4 were up-regulated (Figure 10).
Down-regulated genes included genes with unknown functions (9/21), general stress response genes (3/21), cell elongation/invasive growth genes (2/21), phosphate uptake genes (2/21) and genes with other functions (4/21). Interestingly, one of the strongly down-regulated genes was CTA1/CAT1, known to encode catalase.
Discussion
Ciclopirox acts on yeast and hyphal cells of C. albicans
Hydroxypyridones are widely used topical antifungal drugs with a broad antimicrobial spectrum including the yeast C. albicans. In this study, we showed that not only the yeast growth form of C. albicans, but also germ tube formation is strongly inhibited by ciclopirox at concentrations higher than 1 mg/L (Figure 1). This was possibly due to a general block of growth rather than a specific inhibition of the morphological transition as subinhibitory concentrations of 0.6 mg/L did not influence hyphal formation.11 Both hyphal and yeast forms are found in mucosal infections of this fungus and it is assumed that the hyphal form is essential for invasive growth. Therefore, inhibition of hyphae formation by ciclopirox may not only stop proliferation on the epithelial surface, but most importantly may also prevent penetration of this fungus into deeper tissues. Being exclusively used as a topical drug, the cutaneous absorption is known to be very low and ciclopirox is quickly eliminated from the bloodstream. Not only poor uptake and rapid elimination but also serum proteins possibly interfering with antifungal activity may limit the use of this drug to topical treatment.
Ciclopirox does not damage fungal cell membranes
Despite the fact that hydroxypyridones have been in clinical use for more than two decades, their mode of action is still not clear. Early studies suggested that the hydroxypyridone ciclopirox inhibits the energy production of fungal cells consequently blocking the uptake of components essential for growth.4S. cerevisiae cells exposed to another hydroxypyridone, rilopirox, showed a reduced respiratory activity, which may have been caused by the reduced uptake or decreased metabolism of glucose or by a direct attack on the mitochondrial electron transport chain. This may be explained by the high affinity of hydroxypyridones for metal ions; rilopirox has been shown to inhibit the activity of NADH-ubiquinone-oxidoreductase.9 If energy depletion was the principal mode of action, one would expect a slow onset of antifungal activity; however, our study showed that ciclopirox acts instantly on proliferating and non-proliferating cells which is in disagreement with the view that energy depletion is the main antifungal principle. Furthermore, the ATP concentration of cells treated with ciclopirox did not change, suggesting that a lack of energy resources as the principal mode of action is unlikely. In contrast to ciclopirox, treatment with amphotericin B caused reduced intracellular ATP concentrations possibly by damaging the cellular or mitochondrial membranes. This together with the fact that cells incubated with ciclopirox do not result in the release of potassium ions into the culture medium strongly suggests that this drug does not directly damage fungal membranes although modification of the cell membrane has been observed in cells treated with ciclopirox for longer periods.11 However, this damage is likely to be due to secondary effects.
It is unlikely that the mode of action of ciclopirox is based on the cellular uptake of amino acids, potassium- and phosphate ions as suggested by Iwata & Yamaguchi.4 Starvation of amino acids and phosphate would probably be reflected in the transcriptional up-regulation of amino acid and phosphate transporters; however, none of the 28 identified genes encoding phosphate or amino acid transporters or permeases were found to be up-regulated in cells exposed to subinhibitory concentrations of ciclopirox (data not shown).
Role of oxygen and metabolism
C. albicans cells pre-cultivated in static cultures were found to be more susceptible to ciclopirox compared with cells pre-cultivated under aerobic conditions suggesting that oxygen plays a major role in the mode of action of this drug. This may be linked to the production of reactive oxygen species (ROS) and/or a reduced cellular detoxification of these molecules. Static cultures of S. cerevisiae have been found to produce more peroxides when transferred to an oxygen-rich environment.25 This was thought to be due to redox active components, which were reduced under oxygen-poor conditions. Since the redox status was not changed immediately when cultures were exposed to oxygen again, molecular oxygen was only partially reduced and the production of reactive oxygen species was fostered.26,27 Furthermore, proteins that are involved in the detoxification of ROS, were less induced. For example, cytochrome c peroxidase activity was low under reduced oxygen conditions, but induced in oxygen-rich conditions.28 When exposed to pure oxygen, S. cerevisiae produced significantly more Sod and catalase compared with anaerobic cultures.29 This is in accordance with our data which show that the activity of enzymes directly or indirectly involved in the metabolism of ROS such as G6P-DH, catalase, cytochrome c peroxidase and Sod were reduced in C. albicans cells grown under oxygen-poor conditions and this may in turn have caused the higher susceptibility to ciclopirox. In contrast, cells pre-incubated with hydrogen peroxide or menadione had significantly higher levels of antioxidant enzymes (G6P-DH, catalase and Sod) and were more tolerant to ciclopirox.
In addition to oxygen, it seems that an active metabolism is also required, because both the depletion of glucose in incubation media and the addition of the glucose antagonist 2-deoxy-D-glucose strongly reduced the antifungal activity of ciclopirox; however, it is unlikely that an active metabolism is essential for the uptake of ciclopirox since it has been shown that ciclopirox enters the cell and accumulates intracellularly in an ATP-independent manner.6,7
Evidence that ciclopirox acts via the binding of iron ions
Two independent experiments provide strong evidence that ciclopirox acts principally via the binding of iron. First, the addition of iron ions (Fe2+ and Fe3 + ) strongly reduced the inhibitory effect of ciclopirox on germ tube formation. Such an effect was not seen for other metal ions such as Ca2+, Mg2+ and Mn2+. Ciclopirox–Fe complexes possibly caused the yellow coloration regularly seen in cultures supplemented with ciclopirox and iron ions. Secondly, genome-wide transcriptional profiling of cells exposed to ciclopirox clearly reflected that the cells lacked iron: 60% (15/25) of all strongly (more than 2-fold) up-regulated genes are involved in iron uptake and metabolism. These include genes such as CFL1 (iron reductase), FTR1 and FTH1 (iron permeases and transporters) that were previously found to be up-regulated by semi-quantitative RT–PCR and northern analysis in ciclopirox-exposed cells.11 It should be noted that the high-affinity iron permease gene FTR1 and the low-affinity iron permease gene FTR2 are highly similar. Therefore, spots on the microarray representing these two genes are likely to hybridize with either FTR1 or FTR2 probes. Since we previously showed that FTR1 was up-regulated in cells treated with ciclopirox, whereas FTR2 was down-regulated,11 we concluded that the signals observed for FTR2 (Table 1) were due to cross-hybridizations with probes representing FTR1 mRNAs.
In addition, we also found a number of strongly up-regulated genes possibly involved in iron metabolism (by homology) that have not yet been described in C. albicans (CFL2, CFL12, FET5, FET32, FET33, FET34, FRE5, FRE31, FRE32, CCC2). An orthologue of the C. albicansFRE family has recently been identified in a screen for genes involved in response to ciclopirox treatment of S. cerevisiae.30 Therefore, transcriptional profiling clearly reflects iron-limiting conditions providing valuable information regarding the mode of action of ciclopirox. The observed transcriptional profile of ciclopirox-exposed cells is unlikely to be a general phenomenon of drug treatment as cells exposed to azoles exhibit a gene expression pattern which clearly differs from the data presented here.11,31
Iron limitation may have multiple effects on the fungal cell. One likely effect is the reduced activity of proteins that require iron ions as an essential cofactor. This would include iron–sulphur (Fe/S) proteins such as succinate dehydrogenase and aconitase, haem proteins such as peroxidases, oxidases, cytochrome P450, and non-haem and non-Fe/S proteins such as dioxygenase, oxygenase, lipoxygenases and ribonucleotide reductase. The loss of sufficient activities of all these iron-dependent enzymes may cause a defined or cumulative or synergic effect resulting in cell death.
One further possible consequence of iron limitation (caused by ciclopirox) and oxygen exposure may be an enhanced level of ROS. This may be due to a higher production of ROS and/or a reduced detoxification via a blocked activity of antioxidant enzymes. Since iron is an essential co-factor for the antioxidant enzyme catalase, it is possible that the activity of this enzyme is reduced under iron-limited conditions; however, it should also be noted that high iron levels may itself stimulate or cause the production of ROS. Therefore, cells need to regulate the concentration of iron since there is a delicate balance between levels that are essential and those that are toxic.
Our data show that the enzyme activity of two key antioxidant proteins, catalase and Sod, and/or the gene expression levels of their corresponding genes are altered in ciclopirox-exposed cells. Out of the 6039 genes of the C. albicans genome, only 25 including IPF1218, which encodes a putative Sod, were found to be up-regulated ( > 2-fold). However, the total Sod activity in ciclopirox-treated cells was not altered compared with control cells suggesting that either other Sods cause the major Sod activity and/or that post-transcriptional modifications or co-factors are responsible for the reduced activity.
Of the 21 genes which were found to be down-regulated ( > 2-fold) in drug-exposed cells, the catalase gene CAT1 (CTA1)32 was 2.5-fold down-regulated. In addition, the catalase activity was significantly reduced in drug-exposed cells. Since catalases belong to the group of iron-dependent enzymes, we concluded that the reduced activity is due to both a reduction at the transcriptional level and reduced accessibility to the co-factor iron. These data confirm our previous observation that the sensitivity of ciclopirox-exposed cells to oxidative stress increased dramatically.11 However, the reduction in catalase activity is unlikely to be the principal mode of action which causes cell death since8 catalase inhibitors did not reduce fungal growth and did not add to the antifungal activity of ciclopirox (see above) and18 mutants lacking CAT1 were still viable.32
In summary, this is the first study combining microbiological, biochemical and genome-wide transcriptional profiling analyses to elucidate the mode of action of an antifungal drug. The data obtained in this study indicate that oxygen accessibility and iron levels are critical parameters in the antifungal activity of ciclopirox possibly by influencing the level of reactive oxygen species.
Influence of ciclopirox and other antifungals on germ tube formation of C. albicans. Semi-synchronized yeast cells of strain 5342 ZN (107 cells/mL) were incubated at 37°C in Lee's medium containing ciclopirox, amphotericin B (AMB), flucytosine (5FC) or clotrimazole (CLT) at concentrations ranging from 0.125 to 64 mg/L. The percentage inhibition of hyphal formation was monitored after 4 h of incubation. Values represent the averages of at least four different samples (n=4–8).
Release of potassium ions from C. albicans strain CBS 562 following 30 min (open bars) and 60 min (grey bars) incubation with ciclopirox (16 and 32 mg/L) and amphotericin B (4 and 8 mg/L). The maximal release of potassium (=100%) was defined as the amount of potassium released by boiling untreated cells for 5 min. The viability was 100% at time 0. The viability after 60 min is indicated at the top of each bar as the average ± SD from three independent experiments. Control, no drug.
Influence of ciclopirox (grey bars) (16, 32 or 64 mg/L) and amphotericin B (open bars) (4 and 8 mg/L) on the intracellular ATP concentration of semi-synchronized cells (107 cells/mL) of C. albicans strain 5342 ZN after 30 min of incubation in 5 mM proline/10 mM glucose/PBS. The viability was 100% at time 0. The viability after 30 min is indicated at the top of each bar as the average ± SD from three independent experiments. Control, no drug. *Significantly (P ≤ 0.05) different from the ATP content of the control.
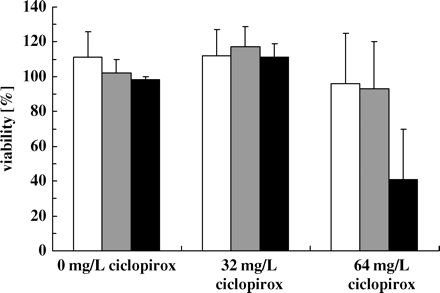
Influence of oxygen on the susceptibility of cells to ciclopirox. C. albicans CBS 562 was pre-cultured in modified Sabouraud glucose medium with and without shaking. Pre-cultured cells were incubated in 5 mM proline/10 mM glucose/PBS supplemented with 32 or 64 mg/L ciclopirox or without drug (control) for 4 h (oxygen-rich conditions, open bars; reduced oxygen conditions, grey bars). The viability for each culture is indicated as the average ± SD from five independent experiments. The viability for cells pre-cultured under reduced oxygen conditions and incubated with 64 mg/L ciclopirox was 0 ± 0.084%.
Influence of ciclopirox (32 or 64 mg/L) on the viability of semi-synchronized C. albicans CBS 562 (107 cells/mL) in PBS (open bars), 10 mM 2-deoxy-D-glucose in PBS (grey bars) and 10 mM glucose in PBS (filled bars). The viability was 100% at time 0. The viability after 4 h incubated is shown as the average ± SD for five independent experiments. Control, no drug.
Induction of intracellular catalase activity with hydrogen peroxide (H2O2) and menadione. Exponential growing cells of C. albicans strain CBS 562 (5 × 106 cells/mL) were exposed to H2O2 for 60 min (a) and menadione for 90 min (b). Catalase activity (U/mL of protein) is shown as open bars and viability [cfu after incubation compared with cfu at time point 0 (100%)] is shown as grey bars. Values represent the averages ± SD for three independent experiments. *Significantly (P ≤ 0.05) different from the activity of the control. Control, no drug.
Induction of intracellular superoxide dismutase (Sod) activity with menadione. Exponential grown cells of C. albicans strain CBS 562 (5 × 106 cells/mL) were exposed to 100 μM menadione for 90 min. Total Sod (open bars) and Mn-Sod (grey bars) activity of 50 μL (250 mL of protein/L) cell extracts were measured as U/mL of protein. Values represent the averages ± SD of three samples. Control, no drug.
Fungicidal effect of ciclopirox (32 and 64 mg/L) on the viability of exponential grown C. albicans CBS 562 (107 cells/mL) pre-incubated with 1 mM H2O2 (a) or 100 μM menadione (b) (grey bars) compared with untreated cultures (open bars). Values represent averages ± SD (n=3). *Significant (P ≤ 0.05) difference between samples. Control, no drug.
Effect of ciclopirox (16 or 32 mg/L) after 90 min incubation on the viability and activity of (a) glucose-6-phosphate-dehydrogenase and (b) catalase of exponential growing cells of C. albicans strains CBS 562. Enzyme activity is shown as open bars, viability compared with time point 0 as grey bars. Values represent averages ± SD (n=3). Control, no drug. *Significantly (P ≤ 0.05) different from the activity of the control culture.
Known and putative functions of genes up-regulated in cells exposed to subinhibitory concentrations of ciclopirox (0.6 mg/L).
List of up-regulated genes in cells exposed to subinhibitory concentrations of ciclopirox (0.6 mg/L) as identified by genome-wide transcriptional profiling using microarrays
| Systematic . | Normalized fold up-regulateda . | P valueb . | Known or putative function (reference) . |
|---|---|---|---|
| RBT5 | 19.78 | 0.002 | repressed by TUP1 protein 5 (homology to CSA1) (4) |
| CFL2 | 8.98 | 1.69×10−6 | ferric reductase (by homology) |
| IPF13493 | 8.81 | 3.96×10−7 | unknown function |
| IPF12101 | 7.75 | 0.006 | mycelial surface antigen precursor (by homology) |
| RBT2 | 7.17 | 2.39×10−7 | ferric reductase (by homology) (4) |
| FRE5 | 7.05 | 0.029 | ferric reductase transmembrane component (by homology) |
| FTR1 | 6.61 | 1.96×10−7 | high affinity iron permease (26) |
| FRE31 | 6.61 | 9.47×10−7 | ferric reductase (by homology) |
| FET34.3eoc | 6.39 | 1.94×10−5 | iron transport multicopper oxidase, 3-prime end (by homology) |
| CSA1 | 5.94 | 1.01×10−5 | mycelial surface antigen (by homology) (19) |
| FRE32 | 5.75 | 1.26×10−5 | ferric reductase (by homology) |
| FET32 | 5.48 | 0.004 | cell surface ferroxidase (by homology) |
| FET5 | 5.19 | 1.93×10−6 | multicopy oxidase (by homology) |
| GDH3 | 4.09 | 0.047 | NADP-glutamate dehydrogenase (by homology) |
| FTH1 | 3.81 | 0.011 | iron transporter (34) |
| FET33 | 3.76 | 1.55×10−4 | cell surface ferroxidase (by homology) |
| FTR2 | 3.13 | 8.17×10−5 | low affinity iron permease (26) |
| IPF7711 | 3.05 | 0.002 | related to Neurospora crassa AP-1-like transcription factor (by homology) |
| CCC2 | 2.91 | 0.021 | putative copper-transporting ATPase (by homology) (36) |
| IPF9315 | 2.70 | 0.057 | putative CCAAT-binding factor subunit (by homology) |
| IPF1218/SOD4 | 2.64 | 0.048 | similar to superoxide dismutase (by homology) |
| SNM1 | 2.47 | 2.42×10−4 | RNA binding protein of RNase MRP (by homology) |
| IPF7023.3 | 2.47 | 0.052 | unknown function, 3-prime end |
| CFL1 | 2.31 | 9.30×10−4 | ferric reductase (12) |
| CFL12 | 2.06 | 0.024 | similar to ferric reductase Fre2p (by homology) |
| Systematic . | Normalized fold up-regulateda . | P valueb . | Known or putative function (reference) . |
|---|---|---|---|
| RBT5 | 19.78 | 0.002 | repressed by TUP1 protein 5 (homology to CSA1) (4) |
| CFL2 | 8.98 | 1.69×10−6 | ferric reductase (by homology) |
| IPF13493 | 8.81 | 3.96×10−7 | unknown function |
| IPF12101 | 7.75 | 0.006 | mycelial surface antigen precursor (by homology) |
| RBT2 | 7.17 | 2.39×10−7 | ferric reductase (by homology) (4) |
| FRE5 | 7.05 | 0.029 | ferric reductase transmembrane component (by homology) |
| FTR1 | 6.61 | 1.96×10−7 | high affinity iron permease (26) |
| FRE31 | 6.61 | 9.47×10−7 | ferric reductase (by homology) |
| FET34.3eoc | 6.39 | 1.94×10−5 | iron transport multicopper oxidase, 3-prime end (by homology) |
| CSA1 | 5.94 | 1.01×10−5 | mycelial surface antigen (by homology) (19) |
| FRE32 | 5.75 | 1.26×10−5 | ferric reductase (by homology) |
| FET32 | 5.48 | 0.004 | cell surface ferroxidase (by homology) |
| FET5 | 5.19 | 1.93×10−6 | multicopy oxidase (by homology) |
| GDH3 | 4.09 | 0.047 | NADP-glutamate dehydrogenase (by homology) |
| FTH1 | 3.81 | 0.011 | iron transporter (34) |
| FET33 | 3.76 | 1.55×10−4 | cell surface ferroxidase (by homology) |
| FTR2 | 3.13 | 8.17×10−5 | low affinity iron permease (26) |
| IPF7711 | 3.05 | 0.002 | related to Neurospora crassa AP-1-like transcription factor (by homology) |
| CCC2 | 2.91 | 0.021 | putative copper-transporting ATPase (by homology) (36) |
| IPF9315 | 2.70 | 0.057 | putative CCAAT-binding factor subunit (by homology) |
| IPF1218/SOD4 | 2.64 | 0.048 | similar to superoxide dismutase (by homology) |
| SNM1 | 2.47 | 2.42×10−4 | RNA binding protein of RNase MRP (by homology) |
| IPF7023.3 | 2.47 | 0.052 | unknown function, 3-prime end |
| CFL1 | 2.31 | 9.30×10−4 | ferric reductase (12) |
| CFL12 | 2.06 | 0.024 | similar to ferric reductase Fre2p (by homology) |
As compared with untreated control.
Student's t-test.
List of up-regulated genes in cells exposed to subinhibitory concentrations of ciclopirox (0.6 mg/L) as identified by genome-wide transcriptional profiling using microarrays
| Systematic . | Normalized fold up-regulateda . | P valueb . | Known or putative function (reference) . |
|---|---|---|---|
| RBT5 | 19.78 | 0.002 | repressed by TUP1 protein 5 (homology to CSA1) (4) |
| CFL2 | 8.98 | 1.69×10−6 | ferric reductase (by homology) |
| IPF13493 | 8.81 | 3.96×10−7 | unknown function |
| IPF12101 | 7.75 | 0.006 | mycelial surface antigen precursor (by homology) |
| RBT2 | 7.17 | 2.39×10−7 | ferric reductase (by homology) (4) |
| FRE5 | 7.05 | 0.029 | ferric reductase transmembrane component (by homology) |
| FTR1 | 6.61 | 1.96×10−7 | high affinity iron permease (26) |
| FRE31 | 6.61 | 9.47×10−7 | ferric reductase (by homology) |
| FET34.3eoc | 6.39 | 1.94×10−5 | iron transport multicopper oxidase, 3-prime end (by homology) |
| CSA1 | 5.94 | 1.01×10−5 | mycelial surface antigen (by homology) (19) |
| FRE32 | 5.75 | 1.26×10−5 | ferric reductase (by homology) |
| FET32 | 5.48 | 0.004 | cell surface ferroxidase (by homology) |
| FET5 | 5.19 | 1.93×10−6 | multicopy oxidase (by homology) |
| GDH3 | 4.09 | 0.047 | NADP-glutamate dehydrogenase (by homology) |
| FTH1 | 3.81 | 0.011 | iron transporter (34) |
| FET33 | 3.76 | 1.55×10−4 | cell surface ferroxidase (by homology) |
| FTR2 | 3.13 | 8.17×10−5 | low affinity iron permease (26) |
| IPF7711 | 3.05 | 0.002 | related to Neurospora crassa AP-1-like transcription factor (by homology) |
| CCC2 | 2.91 | 0.021 | putative copper-transporting ATPase (by homology) (36) |
| IPF9315 | 2.70 | 0.057 | putative CCAAT-binding factor subunit (by homology) |
| IPF1218/SOD4 | 2.64 | 0.048 | similar to superoxide dismutase (by homology) |
| SNM1 | 2.47 | 2.42×10−4 | RNA binding protein of RNase MRP (by homology) |
| IPF7023.3 | 2.47 | 0.052 | unknown function, 3-prime end |
| CFL1 | 2.31 | 9.30×10−4 | ferric reductase (12) |
| CFL12 | 2.06 | 0.024 | similar to ferric reductase Fre2p (by homology) |
| Systematic . | Normalized fold up-regulateda . | P valueb . | Known or putative function (reference) . |
|---|---|---|---|
| RBT5 | 19.78 | 0.002 | repressed by TUP1 protein 5 (homology to CSA1) (4) |
| CFL2 | 8.98 | 1.69×10−6 | ferric reductase (by homology) |
| IPF13493 | 8.81 | 3.96×10−7 | unknown function |
| IPF12101 | 7.75 | 0.006 | mycelial surface antigen precursor (by homology) |
| RBT2 | 7.17 | 2.39×10−7 | ferric reductase (by homology) (4) |
| FRE5 | 7.05 | 0.029 | ferric reductase transmembrane component (by homology) |
| FTR1 | 6.61 | 1.96×10−7 | high affinity iron permease (26) |
| FRE31 | 6.61 | 9.47×10−7 | ferric reductase (by homology) |
| FET34.3eoc | 6.39 | 1.94×10−5 | iron transport multicopper oxidase, 3-prime end (by homology) |
| CSA1 | 5.94 | 1.01×10−5 | mycelial surface antigen (by homology) (19) |
| FRE32 | 5.75 | 1.26×10−5 | ferric reductase (by homology) |
| FET32 | 5.48 | 0.004 | cell surface ferroxidase (by homology) |
| FET5 | 5.19 | 1.93×10−6 | multicopy oxidase (by homology) |
| GDH3 | 4.09 | 0.047 | NADP-glutamate dehydrogenase (by homology) |
| FTH1 | 3.81 | 0.011 | iron transporter (34) |
| FET33 | 3.76 | 1.55×10−4 | cell surface ferroxidase (by homology) |
| FTR2 | 3.13 | 8.17×10−5 | low affinity iron permease (26) |
| IPF7711 | 3.05 | 0.002 | related to Neurospora crassa AP-1-like transcription factor (by homology) |
| CCC2 | 2.91 | 0.021 | putative copper-transporting ATPase (by homology) (36) |
| IPF9315 | 2.70 | 0.057 | putative CCAAT-binding factor subunit (by homology) |
| IPF1218/SOD4 | 2.64 | 0.048 | similar to superoxide dismutase (by homology) |
| SNM1 | 2.47 | 2.42×10−4 | RNA binding protein of RNase MRP (by homology) |
| IPF7023.3 | 2.47 | 0.052 | unknown function, 3-prime end |
| CFL1 | 2.31 | 9.30×10−4 | ferric reductase (12) |
| CFL12 | 2.06 | 0.024 | similar to ferric reductase Fre2p (by homology) |
As compared with untreated control.
Student's t-test.
We wish to thank Donika Kunze, Robert Koch-Institut, Berlin, Germany, for preparing RNA samples, Florian Wagner, German Resource Center for Genome Research, Berlin, Germany, for technical help with scanning microarrays and Caroline Westwater, Medical University of South Carolina, Charleston, SC, USA, for critical reading of the manuscript. This work has been supported by the Free University of Berlin and the Robert Koch-Institute.
References
Dittmar, W. & Lohaus, G. (
Dittmar, W., Grau, W., Raether, W. et al. (
Iwata, K. & Yamaguchi, H. (
Iwata, K. & Yamaguchi, H. (
Sakurai, K., Sakaguchi, T., Yamaguchi, H. et al. (
Sakurai, K., Sakaguchi, T., Yamaguchi, H. et al. (
Abrams, B. B., Hanel, H. & Hoehler, T. (
Kruse, R., Hengstenberg, W., Hanel, H. et al. (
Gillum, A. M., Tsay, E. Y. & Kirsch, D. R. (
Niewerth, M., Kunze, D., Seibold, M. et al. (
Johnson, E. M., Richardson, M. D. & Warnock, D. W. (
Evans, E. G., Odds, F. C., Richardson, M. D. et al. (
Lee, K. L., Buckley, H. R. & Campbell, C. C. (
Hughes, C. E., Bennett, R. L. & Beggs, W. H. (
Johnson, E. M., Richardson, M. D. & Warnock, D. W. (
Smith, P. K., Krohn, R. I., Hermanson, G. T. et al. (
Löhr, G. W. & Waller, H. D. (
Valdivia, E., Martinez, J., Ortega, J. M. et al. (
Oberley, L. W. & Spitz, D. R. (
Lashkari, D. A., DeRisi, J. L., McCusker, J. H. et al. (
Tao, H., Bausch, C., Richmond, C. et al. (
Shepherd, M. G. & Sullivan, P. A. (
Yurkow, E. J. & McKenzie, M. A. (
Bast, A., Haenen, G. R. & Doelman, C. J. (
Dawson, T. L., Gores, G. J., Nieminen, A. L. et al. (
Yonetani, T. & Ohnishi, T. (
Gregory, E. M., Goscin, S. A. & Fridovich, I. (
Leem, S. H., Park, J. E., Kim, I. S. et al. (
Rogers, P. D. & Barker, K. S. (
Author notes
1Institut für Pharmazie, Abteilung für Pharmakologie und Toxikologie, Freie Universität Berlin, Königin-Luise-Str. 2 + 4, D-14195 Berlin, Germany;
2Robert Koch-Institut, Nordufer 20, D-13353 Berlin, Germany;
3Dermatologische Klinik und Poliklinik, Universität München, Frauenlobstraβe 9–11, D-80337 Munich, Germany



![Induction of intracellular catalase activity with hydrogen peroxide (H2O2) and menadione. Exponential growing cells of C. albicans strain CBS 562 (5 × 106 cells/mL) were exposed to H2O2 for 60 min (a) and menadione for 90 min (b). Catalase activity (U/mL of protein) is shown as open bars and viability [cfu after incubation compared with cfu at time point 0 (100%)] is shown as grey bars. Values represent the averages ± SD for three independent experiments. *Significantly (P ≤ 0.05) different from the activity of the control. Control, no drug.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/jac/55/5/10.1093/jac/dki089/2/m_138029f6.gif?Expires=1716845502&Signature=kHwf2J7uccd6s0KywCl2-Vhq15~Zx~ujcbMloRsIg0Yaqkyvtw8xJrDHHhI5l~MZBc-YLVzonsxRi-c5UhMxfoLeSeNJwycj8fBvkfJ3Zl9g0fbdLdsQ6BR91IBb-QB-WjffRflmqvgv~32bzoLN2x~bH4mYOjK~d4L8MfJ~RJzcNb5Us3IBfH7JOYNOKDzQOwepddGDC7vSfB-H-3qeaGpLGOwSdGDykeytgPWlhwq8WdD8ieiU-Kk8YLcf7qhxmgl48ChcyYNDG1MT6IM3vONsTatCt36MKTNZmxs6jTZqPwshUxeawB1lfKLMeh0ElDr~-Nz6mfjnYBWDojoyUg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)