-
PDF
- Split View
-
Views
-
Cite
Cite
M. A. Zeitlinger, R. Sauermann, F. Traunmüller, A. Georgopoulos, M. Müller, C. Joukhadar, Impact of plasma protein binding on antimicrobial activity using time–killing curves, Journal of Antimicrobial Chemotherapy, Volume 54, Issue 5, November 2004, Pages 876–880, https://doi.org/10.1093/jac/dkh443
Close - Share Icon Share
Abstract
Objectives: Plasma protein binding (PPB) is known to impair the antimicrobial activity of β-lactams, but its impact on the activity of other classes of antimicrobials such as fluoroquinolones is controversial. This study was undertaken to investigate the effect of PPB on bacterial killing by selected antibiotics and moxifloxacin, which served as a model compound for the class of fluoroquinolones.
Methods: Bacterial time–killing curves were employed in the absence and presence of physiological albumin concentrations (40 g/L). Moxifloxacin, ampicillin and oxacillin were investigated. Fosfomycin, a non-protein bound antibiotic was used for comparison. Simulations were carried out by employing concentrations of antibiotics of one-fourth of the minimal inhibitory concentration (MIC), equal to the MIC and four-fold the MIC of one select bacterial strain (Staphylococcus aureus ATCC 29213). To correlate bacterial killing to the extent of PPB, bacterial time–killing curves were plotted using the calculated free and the total drug concentration.
Results: Bacterial killing by fosfomycin was not affected by the addition of albumin. The antimicrobial activity of oxacillin and ampicillin was reduced in the presence of albumin as expected by the calculation of the free fraction of these antibiotics. Adding albumin to moxifloxacin resulted in a significant decrease in bacterial killing of more than 1 log10 cfu/mL after a period of 8 h when the moxifloxacin concentration was equal to the respective MIC.
Conclusions: Our data confirm the view that albumin substantially impairs the antimicrobial activity of antibiotics including moxifloxacin, a member of the class of fluoroquinolones.
Introduction
Plasma protein binding (PPB) has been shown to substantially affect tissue penetration, elimination half-life and the volume of distribution of antimicrobial agents.1–3 Additionally, the impact of PPB on the microbial effect of antibiotics has been investigated by several in vitro studies.4,5 In general, PPB reduces the free fraction of drug available for bacterial killing, since only the non-protein bound antibiotic is pharmacologically active.6 Although, the effect of PPB on bacterial killing is well documented for β-lactams, data on other classes of antimicrobial agents are still rare.7–10 Therefore, it is doubtful whether findings obtained from β-lactams can be extrapolated to other classes of antibiotics such as the fluoroquinolones.11
Currently, most available data related to this topic are exclusively based on in vitro studies employing the minimal inhibitory concentration (MIC) method. This established approach, however, describes bacterial growth inhibition after 18–24 h only, and does not represent bactericidal activity nor can it provide information on the time versus killing course of antimicrobials. In contrast, time–killing curves demonstrate better correlations with the in vivo efficacy than other microbiological methods and therefore have been currently suggested for the exploration of the impact of PPB on antimicrobial activity.12,13
In this study, we set out to test the hypothesis that the antimicrobial activity of moxifloxacin is substantially affected by PPB. For that reason, we used bacterial time–killing curves in the absence and presence of albumin in the growth media. Ampicillin and oxacillin, two well-investigated β-lactam antibiotics with low and high PPB, respectively, were used for comparison. The non-plasma protein bound antibiotic fosfomycin was used to preclude other interactions of antibiotic and albumin that might influence bacterial killing.
Materials and methods
Organism
Staphylococcus aureus was obtained from the American Type Culture Collection (ATCC 29213). Between experiments, S. aureus was stored frozen in liquid nitrogen until it was used.
Antibiotics
Oxacillin, ampicillin and fosfomycin were obtained from Sigma Aldrich (Germany); moxifloxacin was obtained from Bayer (Germany). All antimicrobial agents were prepared and stored throughout the investigations following the manufacturers' recommendations. In all experiments with fosfomycin, glucose-6-phosphate (Boehringer-Mannheim, Germany) was added at 25 mg/L according to the NCCLS guidelines.14
In vitro susceptibility tests
MIC values were determined by a two-fold serial Mueller–Hinton microdilution method, according to the NCCLS criteria.14 Therefore, S. aureus was precultured overnight on a Columbia agar plate (Columbia + 5% sheep blood, BioMérieux, France) and was then introduced at an initial inoculum of approximately 5 × 105 cfu/mL into Mueller–Hinton broth (MHB, Mikrobiologie Mueller–Hinton Bouillon, Merck, Germany) or MHB containing 40 g/L human albumin (Baxter, Vienna, albumin level of normal serum 35–52 g/L), respectively. Growth media contained defined concentrations of oxacillin, ampicillin, fosfomycin and moxifloxacin in decreasing two-fold steps. The lowest concentration of antibiotic that inhibited visible bacterial growth after 20 h of incubation at 37°C was defined as the respective MIC value. For each antibiotic and growth medium, the MIC was determined five times.
Time–killing curves
Bacterial killing curves were carried out by inoculating S. aureus with antibiotic concentrations of one fourth of the respective MIC, equal to the MIC and four-fold the MIC. Each concentration was simulated in MHB, MHB containing 40 g/L human albumin and in MHB containing the calculated free concentration of the antibiotic. The free fraction of an antibiotic was calculated as described below according to PPB data reported in the literature. Simulations were carried out six and four times (fosfomycin), respectively.
In brief, antibiotic concentrations in the flask were adjusted in MHB or MHB containing 40 g/L human albumin according to the desired concentration. Culture tubes containing 4 mL aliquots were kept in a water bath at 37°C to allow protein binding to take place. After 30 min, tubes were inoculated with S. aureus 29213 at an approximate inoculum of 5 × 105 cfu/mL. Samples were drawn and bacteria were counted at 0, 4 and 8 h of incubation at 37°C. Therefore, after vortexing the culture tubes, two 50 μL samples were removed and serially diluted with 0.9% sodium chloride. After each dilution step, 20 μL was plated onto Columbia agar plates, which were incubated for 24 h at 37°C. Afterwards the colonies were counted and back-extrapolated to the original volume to determine cfu/mL.
Controls present bacterial growth in MHB or MHB containing 40 g/L human albumin when no antibiotic was added. Time–killing curves were plotted as log10 differences of cfu/mL versus time.
Calculating the free fraction of an antibiotic
Statistical calculations
For statistical analysis, Wilcoxon matched pair test was carried out using a commercially available computer program (Statistica, StatSoft, Inc., Tulsa, USA). A two-sided P value < 0.05 was considered the level of significance.
Results
This study tested the effect of PPB of different classes of antibiotics on their activity against the model strain S. aureus (ATCC 29213) using time–killing curves. Table 1 compares MIC values of oxacillin, ampicillin, fosfomycin and moxifloxacin against the model strain in MHB in the absence and presence of physiological albumin concentrations. As indicated by the ratios of MIC values for MHB with albumin and without albumin (4 and 2 mg/L for oxacillin and ampicillin, respectively), the MIC method detected a significant (P < 0.05) impact of PPB for both β-lactams, but was unable to detect an impairment of antimicrobial activity for moxifloxacin (ratio = 1). The MIC of fosfomycin was not affected by the presence of physiological albumin concentrations (P > 0.05).
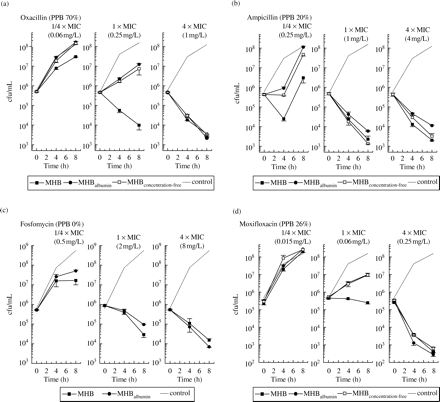
Using time–killing curves, the antimicrobial activity of oxacillin and ampicillin in the presence of albumin resembled the activity of the calculated free fraction of the antibiotic (Figure 1a and b). For moxifloxacin, a significant decrease in bacterial killing of 1.5 log10 cfu/mL (P < 0.05) after a period of 8 h was observed in the presence of albumin (Figure 1d) if the concentration of moxifloxacin was close to the pathogen's MIC. Bacterial killing of fosfomycin was not affected by the presence of physiological albumin concentrations (P > 0.05; Figure 1c). The median log10-differences of bacterial cfu/mL between the initial inoculum and 8 h after exposure to oxacillin, ampicillin, fosfomycin and moxifloxacin at concentrations of one-fourth of the MIC, equal to the MIC and four-fold the MIC are listed in Table 2. Data are shown for simulations in MHB, MHBalbumin and MHBconcentration-free.
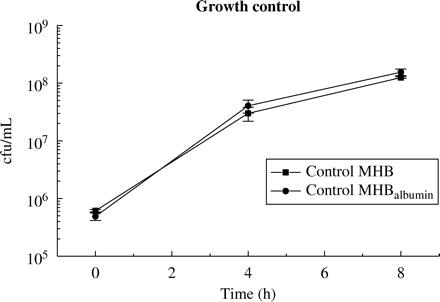
As depicted in Figure 2, the addition of 40 g/L human albumin to MHB did not result in any difference in bacterial growth compared to MHB without albumin during the observation period of 8 h.
Discussion
It is well documented by in vitro experiments that PPB significantly impairs the antimicrobial activity of β-lactams.9,10,19 In contrast, the impact of PPB on the pharmacodynamics of fluoroquinolones is still questioned.11 Those doubts are mainly based on theoretical considerations or on data derived from experiments determining the effect of PPB on changes of the pathogen's MIC.17 However, the MIC method, which only describes visible bacterial growth or growth inhibition within two-fold concentrations steps seems inadequate to detect small changes of antimicrobial effect. This becomes particularly relevant if considering that most fluoroquinolones have only low to moderate PPB and the addition of serum will most probably not result in an increase in the respective MIC.11,20 Small changes in the MIC value might therefore be easily overlooked within two-fold dilutions steps.
In this study, we tested the influence of PPB on antimicrobial activity of moxifloxacin, a model compound for the class of fluoroquinolones, by employing dynamic bacterial time–killing curves. Ampicillin, oxacillin and fosfomycin were used as comparators. Bacterial killing curves were carried out at concentrations of one-fourth the respective MIC, equal to the MIC and four-fold the MIC to allow for better characterization of the concentration range at which PPB affects bacterial killing.
In previous studies, differences in bacterial growth inhibition between broth and serum have been shown to be significant for select antibiotics.5 Albumin is considered to be the principal binding site of β-lactam antibiotics and fluoroquinolones, although binding to other serum proteins might also exist.11 Therefore, the addition of albumin instead of serum was chosen in our experiments to avoid differences in bacterial growth between media used. A separate experiment has shown that no difference in bacterial growth is detectable between the used media MHB and MHB with albumin (Figure 2).
Whereas antimicrobial killing of fosfomycin was not impaired by the presence of physiological albumin concentrations (40 g/L), a significant impact of albumin was demonstrated for antibiotics displaying PPB (Figure 1 and Table 2). This finding confirms the assumption that the influence of PPB on bacterial killing of antibiotics is predominantly based on the reduction in the free drug, rather than on other interactions. The antimicrobial activity of oxacillin and ampicillin in the presence of albumin was in accordance with corresponding simulations using the calculated free fraction of these antibiotics in time–killing curves (Figure 1a and b) and the MIC method (Table 1). Therefore, previous speculations of a close relation between the microbial activity of antibiotics in the presence of albumin and the free-drug fraction are clearly confirmed by our data.5,19
A similar pattern was observed for quinolones regarding time–killing curves of moxifloxacin (Figure 1d). After 8 h of incubation, significant (P < 0.05) differences in bacterial growth inhibition of 1.5 log10 cfu/mL were detected for the simulations using MHB with and without albumin (Table 2, Figure 1d). In contrast, no increase in the MIC value was seen by adding albumin (Table 1). For time-killing curves an effect was only observed in the simulation with the moxifloxacin concentration equal to the pathogen's MIC. These data underline the view of a higher specificity and sensitivity of the time–killing curve method compared with the MIC approach.13
Until now, only one study has investigated the relevance of albumin on bacterial killing of moxifloxacin using time–killing curves.21 The authors published bacterial-killing curves in the absence and presence of 50% albumin and found that no difference of bacterial killing of Streptococcus pneumoniae is detectable after 24 h when the moxifloxacin concentration was equal to one-half of the pathogen's MIC, concluding that albumin had no impact on the antimicrobial activity of moxifloxacin. This is not contradictory to our results, showing the maximum inhibitory effect of albumin at antibiotic concentrations equal to the pathogen's MIC. Nevertheless, the question needs to be addressed why concentrations of fluoroquinolones below the MIC have reduced the final inoculum by about 2 log10 compared to baseline after 24 h of incubation.21 Indeed, it is generally expected that concentrations below the MIC result in an increase in bacterial counts up to a visible turbidity of broth (usually approximately 5 × 107 cfu/mL) according to the definition of the MIC. Therefore, these data are difficult to explain and we decided to repeat the experiment by Rubinstein et al. according to NCCLS guidelines.21 In contrast to the previous work, we found an increase in about 2 log10 in cfu/mL of bacteria compared to baseline after exposure to moxifloxacin in MHB with and without albumin.
In the present study, the model strain S. aureus 29213 was chosen because it is easily accessible in all microbiological laboratories and is recommended by the NCCLS as a control strain for the determination of MICs for all antibiotics investigated in the present work.14 The use of one bacterial strain only limits the uncritical extrapolation of our data to clinical practice. However, the objective of this study was to develop an in vitro model for testing the impact of PPB on microbial effects of antibiotics including the class of fluoroquinolones rather than determining clinical outcome. All the antimicrobials used in this study exert good activity against this strain, though not all of them would be chosen as first line therapy against infections caused by S. aureus.
In conclusion, this study confirmed that plasma protein binding hampers bacterial killing by antimicrobial agents including the class of fluoroquinolones. The microbial activity of non-plasma protein bound antibiotics, like fosfomycin, appeared to be unaffected by the presence of albumin. However, the significance of PPB for antimicrobial and clinical outcome remains subject to further investigations in patient populations.
Bacterial time–killing profiles of S. aureus (ATCC 29213) after exposure to oxacillin (a), ampicillin (b), fosfomycin (c) and moxifloxacin (d). The left column shows time–killing profiles at concentrations of one-fourth of the MIC, equal to the MIC (middle column) and four-fold the MIC (right column). Each simulation was carried out using MHB only (filled squares), MHB containing 40 g/L of human albumin (filled circles) and MHB using the calculated free concentration of the respective antibiotic (open squares). Controls show bacterial growth in the absence of antibiotic. Data are shown as mean ± S.E.M.
Bacterial growth of S. aureus (ATCC 29213) in different media. Bacteria were incubated in MHB (squares) and MHB containing 40 g/L human albumin (circles). Data are shown as mean ± S.E.M.
MIC values of oxacillin, ampicillin, fosfomycin and moxifloxacin against S. aureus 29213 in MHB and MHB containing 40 g/L human albumin (MHBalbumin) (data are shown as median and range, n=5) and ratios of mean MIC values in MHB containing 40 g/L human albumin to those in MHB without albumin
| . | MIC (mg/L) [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | oxacillin . | ampicillin . | fosfomycin . | moxifloxacin . | |||
| MHB | 0.25 (0.25–0.25) | 1 (1–1) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin | 1* (1–1) | 2* (2–2) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin/MHB | 4 | 2 | 1 | 1 | |||
| . | MIC (mg/L) [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | oxacillin . | ampicillin . | fosfomycin . | moxifloxacin . | |||
| MHB | 0.25 (0.25–0.25) | 1 (1–1) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin | 1* (1–1) | 2* (2–2) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin/MHB | 4 | 2 | 1 | 1 | |||
MHB, Mueller–Hinton broth; MIC, minimal inhibitory concentration.
P < 0.05 compared with MHB.
MIC values of oxacillin, ampicillin, fosfomycin and moxifloxacin against S. aureus 29213 in MHB and MHB containing 40 g/L human albumin (MHBalbumin) (data are shown as median and range, n=5) and ratios of mean MIC values in MHB containing 40 g/L human albumin to those in MHB without albumin
| . | MIC (mg/L) [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | oxacillin . | ampicillin . | fosfomycin . | moxifloxacin . | |||
| MHB | 0.25 (0.25–0.25) | 1 (1–1) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin | 1* (1–1) | 2* (2–2) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin/MHB | 4 | 2 | 1 | 1 | |||
| . | MIC (mg/L) [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | oxacillin . | ampicillin . | fosfomycin . | moxifloxacin . | |||
| MHB | 0.25 (0.25–0.25) | 1 (1–1) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin | 1* (1–1) | 2* (2–2) | 2 (2–4) | 0.125 (0.125–0.125) | |||
| MHBalbumin/MHB | 4 | 2 | 1 | 1 | |||
MHB, Mueller–Hinton broth; MIC, minimal inhibitory concentration.
P < 0.05 compared with MHB.
Log10 differences of bacterial cfu/mL between initial inoculum and 8 h after exposure to oxacillin, ampicillin, fosfomycin and moxifloxacin at concentrations one-fourth of the MIC, equal to the MIC and four-fold the MIC
| . | Change in bacterial counts, log10 cfu/mL [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | . | ¼× MIC . | 1× MIC . | 4× MIC . | |||
| Oxacillin (n=6) | MHB | 1.78 (1.75–1.82) | −1.79 (−2.06 to −1.41) | −2.14 (−2.14 to −2.08) | |||
| MHBalbumin | 2.47* (2.47–2.60) | 1.42* (1.33–1.51) | −2.38 (−2.62 to −1.82) | ||||
| MHBconcentration-free | 2.44 (2.38–2.53) | 0.92 (0.86–1.51) | −2.22 (−2.22 to −2.03) | ||||
| Ampicillin (n=6) | MHB | 0.31 (–0.52–1.14) | −2.50 (−2.68 to −2.04) | −2.34 (−2.38 to −2.28) | |||
| MHBalbumin | 2.44* (2.29–2.48) | −1.88* (−1.95 to −1.88) | −1.60* (−1.64 to −1.53) | ||||
| MHBconcentration-free | 2.04 (1.88–2.14) | −2.56 (−2.56 to −2.50) | −2.16 (−2.53 to −1.90) | ||||
| Fosfomycin (n=4) | MHB | 1.36 (1.28–1.76) | −1.51 (−1.77 to −1.29) | −1.52 (−1.74 to −1.48) | |||
| MHBalbumin | 2.00 (1.88–2.04) | −0.96 (−1.02 to −0.94) | −1.87 (−1.95 to −1.86) | ||||
| Moxifloxacin (n=6) | MHB | 2.88 (2.88–3.11) | −0.27 (−0.30 to –0.22) | −2.88 (−2.89 to −2.70) | |||
| MHBalbumin | 3.04 (2.94–3.14) | 1.37* (1.13–1.43) | −3.17 (−3.31 to −2.51) | ||||
| MHBconcentration-free | 2.96 (2.87–3.05) | 1.27 (1.13–1.33) | −2.79 (−2.86 to −2.54) | ||||
| . | Change in bacterial counts, log10 cfu/mL [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | . | ¼× MIC . | 1× MIC . | 4× MIC . | |||
| Oxacillin (n=6) | MHB | 1.78 (1.75–1.82) | −1.79 (−2.06 to −1.41) | −2.14 (−2.14 to −2.08) | |||
| MHBalbumin | 2.47* (2.47–2.60) | 1.42* (1.33–1.51) | −2.38 (−2.62 to −1.82) | ||||
| MHBconcentration-free | 2.44 (2.38–2.53) | 0.92 (0.86–1.51) | −2.22 (−2.22 to −2.03) | ||||
| Ampicillin (n=6) | MHB | 0.31 (–0.52–1.14) | −2.50 (−2.68 to −2.04) | −2.34 (−2.38 to −2.28) | |||
| MHBalbumin | 2.44* (2.29–2.48) | −1.88* (−1.95 to −1.88) | −1.60* (−1.64 to −1.53) | ||||
| MHBconcentration-free | 2.04 (1.88–2.14) | −2.56 (−2.56 to −2.50) | −2.16 (−2.53 to −1.90) | ||||
| Fosfomycin (n=4) | MHB | 1.36 (1.28–1.76) | −1.51 (−1.77 to −1.29) | −1.52 (−1.74 to −1.48) | |||
| MHBalbumin | 2.00 (1.88–2.04) | −0.96 (−1.02 to −0.94) | −1.87 (−1.95 to −1.86) | ||||
| Moxifloxacin (n=6) | MHB | 2.88 (2.88–3.11) | −0.27 (−0.30 to –0.22) | −2.88 (−2.89 to −2.70) | |||
| MHBalbumin | 3.04 (2.94–3.14) | 1.37* (1.13–1.43) | −3.17 (−3.31 to −2.51) | ||||
| MHBconcentration-free | 2.96 (2.87–3.05) | 1.27 (1.13–1.33) | −2.79 (−2.86 to −2.54) | ||||
Data are shown for simulations in MHB, MHB containing 40 g/L human albumin (MHBalbumin) and in MHB using the calculated free concentration (MHBconcentration-free). MHB, Mueller–Hinton broth; MIC, minimal inhibitory concentration.
P < 0.05 compared with MHB.
Log10 differences of bacterial cfu/mL between initial inoculum and 8 h after exposure to oxacillin, ampicillin, fosfomycin and moxifloxacin at concentrations one-fourth of the MIC, equal to the MIC and four-fold the MIC
| . | Change in bacterial counts, log10 cfu/mL [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | . | ¼× MIC . | 1× MIC . | 4× MIC . | |||
| Oxacillin (n=6) | MHB | 1.78 (1.75–1.82) | −1.79 (−2.06 to −1.41) | −2.14 (−2.14 to −2.08) | |||
| MHBalbumin | 2.47* (2.47–2.60) | 1.42* (1.33–1.51) | −2.38 (−2.62 to −1.82) | ||||
| MHBconcentration-free | 2.44 (2.38–2.53) | 0.92 (0.86–1.51) | −2.22 (−2.22 to −2.03) | ||||
| Ampicillin (n=6) | MHB | 0.31 (–0.52–1.14) | −2.50 (−2.68 to −2.04) | −2.34 (−2.38 to −2.28) | |||
| MHBalbumin | 2.44* (2.29–2.48) | −1.88* (−1.95 to −1.88) | −1.60* (−1.64 to −1.53) | ||||
| MHBconcentration-free | 2.04 (1.88–2.14) | −2.56 (−2.56 to −2.50) | −2.16 (−2.53 to −1.90) | ||||
| Fosfomycin (n=4) | MHB | 1.36 (1.28–1.76) | −1.51 (−1.77 to −1.29) | −1.52 (−1.74 to −1.48) | |||
| MHBalbumin | 2.00 (1.88–2.04) | −0.96 (−1.02 to −0.94) | −1.87 (−1.95 to −1.86) | ||||
| Moxifloxacin (n=6) | MHB | 2.88 (2.88–3.11) | −0.27 (−0.30 to –0.22) | −2.88 (−2.89 to −2.70) | |||
| MHBalbumin | 3.04 (2.94–3.14) | 1.37* (1.13–1.43) | −3.17 (−3.31 to −2.51) | ||||
| MHBconcentration-free | 2.96 (2.87–3.05) | 1.27 (1.13–1.33) | −2.79 (−2.86 to −2.54) | ||||
| . | Change in bacterial counts, log10 cfu/mL [median (range)] . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| . | . | ¼× MIC . | 1× MIC . | 4× MIC . | |||
| Oxacillin (n=6) | MHB | 1.78 (1.75–1.82) | −1.79 (−2.06 to −1.41) | −2.14 (−2.14 to −2.08) | |||
| MHBalbumin | 2.47* (2.47–2.60) | 1.42* (1.33–1.51) | −2.38 (−2.62 to −1.82) | ||||
| MHBconcentration-free | 2.44 (2.38–2.53) | 0.92 (0.86–1.51) | −2.22 (−2.22 to −2.03) | ||||
| Ampicillin (n=6) | MHB | 0.31 (–0.52–1.14) | −2.50 (−2.68 to −2.04) | −2.34 (−2.38 to −2.28) | |||
| MHBalbumin | 2.44* (2.29–2.48) | −1.88* (−1.95 to −1.88) | −1.60* (−1.64 to −1.53) | ||||
| MHBconcentration-free | 2.04 (1.88–2.14) | −2.56 (−2.56 to −2.50) | −2.16 (−2.53 to −1.90) | ||||
| Fosfomycin (n=4) | MHB | 1.36 (1.28–1.76) | −1.51 (−1.77 to −1.29) | −1.52 (−1.74 to −1.48) | |||
| MHBalbumin | 2.00 (1.88–2.04) | −0.96 (−1.02 to −0.94) | −1.87 (−1.95 to −1.86) | ||||
| Moxifloxacin (n=6) | MHB | 2.88 (2.88–3.11) | −0.27 (−0.30 to –0.22) | −2.88 (−2.89 to −2.70) | |||
| MHBalbumin | 3.04 (2.94–3.14) | 1.37* (1.13–1.43) | −3.17 (−3.31 to −2.51) | ||||
| MHBconcentration-free | 2.96 (2.87–3.05) | 1.27 (1.13–1.33) | −2.79 (−2.86 to −2.54) | ||||
Data are shown for simulations in MHB, MHB containing 40 g/L human albumin (MHBalbumin) and in MHB using the calculated free concentration (MHBconcentration-free). MHB, Mueller–Hinton broth; MIC, minimal inhibitory concentration.
P < 0.05 compared with MHB.
References
Peterson, L. R. & Gerding, D. N. (
Wong, B. K., Bruhin, P. J. & Lin, J. H. (
Stoeckel, K. (
Boswell, F. J., Ashby, J. P., Andrews, J. M. et al. (
Kunin, C. M. (
Craig, W. A. & Ebert, S. C. (
Van der Auwera, P. & Klastersky, J. (
Leggett, J. E. & Craig, W. A. (
Merrikin, D. J., Briant, J. & Rolinson, G. N. (
Jones, R. N. & Barry, A. L. (
Bergogne-Berezin, E. (
National Committee for Clinical Laboratory Standards. (
Mueller, M., de la Pena, A. & Derendorf, H. (
National Committee for Clinical Laboratory Standards. (
Bennett, J. V. & Kirby, W. M. (
Ullmann, U. (
Woodcock, J. M., Andrews, J. M., Boswell, F. J. et al. (
Kunin, C. M., Craig, W. A., Kornguth, M. et al. (
Turnidge, J. (
Rubinstein, E., Diamantstein, L., Yoseph, G. et al. (
Author notes
1Department of Clinical Pharmacology, Division of Clinical Pharmacokinetics; 2Department of Internal Medicine I, Division of Infectious Diseases and Chemotherapy; 3Institute of Pharmacology, Medical University of Vienna, Waehringer Guertel 18–20, 1090 Vienna, Austria