-
PDF
- Split View
-
Views
-
Cite
Cite
Hans Christian Erichsen, Stephanie A. Mulherin Engel, Peter K. Eck, Robert Welch, Meredith Yeager, Mark Levine, Anna Maria Siega-Riz, Andrew F. Olshan, Stephen J. Chanock, Genetic Variation in the Sodium-dependent Vitamin C Transporters, SLC23A1, and SLC23A2 and Risk for Preterm Delivery, American Journal of Epidemiology, Volume 163, Issue 3, 1 February 2006, Pages 245–254, https://doi.org/10.1093/aje/kwj035
Close - Share Icon Share
Abstract
Vitamin C has been the focus of epidemiologic investigation in preterm delivery (<37 weeks' gestation), which is a leading cause of neonatal mortality and birth-related morbidity. There are two sodium-dependent membrane transporters encoded by SLC23A1 and SLC23A2, which have key roles in human vitamin C metabolism and which control dietary uptake, reabsorption, and tissue distribution of vitamin C. Using maternal DNA, the authors evaluated common single-nucleotide polymorphisms (SNPs) in SLC23A1 and SLC23A2 in a nested case-control analysis of the Pregnancy, Infection, and Nutrition Study (1995–2000) cohort. Of the associations observed for both haplotypes in SLC23A1 and individual SNPs in SLC23A2, the most robust finding is with an intron 2 variant in SLC23A2. Heterozygotes and homozygotes for this variant had a 1.7-fold (95% confidence interval: 0.9, 3.3) and a 2.7-fold (95% confidence interval: 1.2, 6.3) elevation in the risk of spontaneous preterm birth, respectively. Semi-Bayesian hierarchical regression analysis, which simultaneously adjusted for multiple SNPs within the same gene, gave comparable results. The authors' findings link genetic variants in the vitamin C transporters to spontaneous preterm birth, which may explain previous dietary associations. If the findings from this study are confirmed, they may serve as the foundation for genetic risk assessment of nutritional pathways in preterm birth.
Preterm delivery (<37 completed weeks' gestation) is a leading cause of neonatal mortality and birth-related morbidity (1). Over 40 years ago, dietary intake was hypothesized to play a role in premature rupture of membranes, one of the possible clinical presentations of preterm delivery (2). Because vitamin C is necessary for maintenance of collagen (3) and was speculated to be related to membrane tensile strength (2), a number of studies have investigated the role of vitamin C in preterm delivery (2, 4–10). The emphasis of most studies has been on the measurement of ascorbic acid concentrations in serum, leukocytes, or cord blood (2, 7–9). Other studies have examined dietary intake. In addition, we recently reported that dietary vitamin C intake is specifically associated with preterm birth via premature rupture of membranes (10). Since vitamin C is absorbed from dietary sources, such as fruits and vegetables together with additional nutrients, assessment of the effect of vitamin C is indirect. Consequently, it is possible that additional components of fruits and vegetables could be active.
On the basis of the observation that vitamin C ingestion and concentrations in body fluids could be associated with premature rupture of membranes, it is possible that common genetic variants in vitamin C transport could contribute to the risk for preterm birth-related outcomes. Vitamin C is accumulated in human tissues by two general mechanisms: 1) transport of vitamin C itself as the substrate and 2) transport of oxidized vitamin C (dehydroascorbic acid) followed by intracellular reduction. The first mechanism is that vitamin C itself is transported by one of two sodium-dependent vitamin C transporters (SVCT1, encoded by SLC23A1 and mapped to 5q31.2; and SVCT2, encoded by SLC23A2 and mapped to 20p13) (11, 12). The tissue distribution of the two membrane transporters differs: SLC23A1 is expressed in tissues critical for absorption and reabsorption of vitamin C (kidney, intestinal, and hepatic tissues), whereas SLC23A2 is expressed in nearly all cell types and probably contributes to vitamin C accumulation in most tissue compartments (13). The second mechanism of vitamin C accumulation includes genes that transport dehydroascorbic acid, the oxidized form of ascorbate. Once inside cells, dehydroascorbic acid is reduced back to ascorbic acid (14, 15). This mechanism, termed ascorbate recycling, is sodium independent, is mediated by a subset of the family of glucose transporters, and is utilized in circumstances of high oxidant stress, such as uptake in polymorphonuclear leukocytes, particularly when activated (16–20). However, the dominant mechanism of vitamin C accumulation in most tissues is sodium-dependent transport by SVCT1 and SVCT2, based on evidence that ascorbic acid is found in blood, but dehydroascorbic acid is not (21), and that vitamin C accumulation is virtually eliminated in most tissues in the slc23a2 (e.g., svct2) knock-out mouse model (22).
Recently, the pattern of common genetic variants has been characterized for both SLC23A1 and SLC23A2, which share common intron/exon borders, have related coding sequence, but differ greatly in size and linkage disequilibrium patterns (23). These observations serve as the basis for investigating the possible contribution of genetic variation in vitamin C transport to select pregnancy outcomes such as preterm birth. If there is an association between common genetic variants in vitamin C transporters crucial for dietary uptake and tissue accumulation, then it would be possible to establish an indirect link to the role of vitamin C. To this purpose, a genetic association study examining common genetic variants in SLC23A1 and SLC23A2 was conducted in a nested case-control sample from the Pregnancy, Infection, and Nutrition Study cohort (24).
MATERIALS AND METHODS
Study population and design
We utilized a nested case-control sample of the Pregnancy, Infection, and Nutrition Study cohort, which prospectively enrolled women between 24 and 29 weeks' gestation from the Wake County Human Services Department and the Wake Medical Center/Wake Area Health Education Center from February 1996 through June 1998 and the University of North Carolina, Chapel Hill, obstetric private and resident clinics from August 1995 through June 2000 (24). During the intake visit, maternal peripheral blood samples were obtained. Exclusion criteria are detailed elsewhere (24). The control group was randomly selected at enrollment to represent the exposure distribution of the cohort, at an approximate 1:1.5 preterm case:control ratio. Our analysis population consisted of 129 African-American cases of spontaneous preterm birth and 237 African-American term controls (a total of 366 individuals), as well as 142 Caucasian spontaneous preterm birth cases and 335 Caucasian term controls (a total of 477 individuals).
Gestational age was assigned by ultrasound completed before 22 weeks' gestation; in the absence of an ultrasound examination, the last menstrual period date was used. Subjects were classified as having a spontaneous preterm birth if birth occurred before completion of 37 weeks' gestation and had a clinical presentation of either premature rupture of the membranes or idiopathic preterm labor. “Premature rupture of the membranes” was defined as spontaneous membrane rupture occurring at least 4 hours before the onset of labor, and “idiopathic preterm labor” was defined as spontaneous labor occurring within 4 hours of membrane rupture. Study obstetricians reviewed records of cases to determine preterm clinical presentation based on a systematic algorithm previously described (25). A second study obstetrician, blinded to the first assessment, reviewed ambiguous charts plus a 10 percent random sample of all preterm births.
Genes and single-nucleotide polymorphism selection
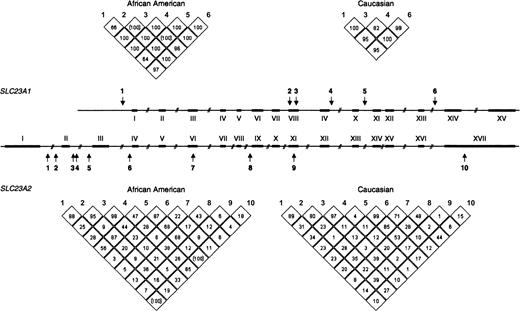
As mentioned above, SLC23A1 and SLC23A2 are crucial for vitamin C metabolism in humans. The two genes arose from a duplication event estimated to have taken place at least 450 million years ago, and they may have evolved under different selective pressures (23). Because the two genes differ 10-fold in size and have marked differences in their patterns of linkage disequilibrium, our analysis approach differed for the two genes. For the smaller gene, SLC23A1, we estimated risk of spontaneous preterm birth by use of haplotypes as exposure variables. Based on the pattern of linkage disequilibrium in this gene, these haplotypes were estimated to capture more than 90 percent of ancestral, common haplotypes within the gene. Haplotypes were preferable to genotypes as the unit of analysis for this gene because of the strong association between individual variants. For SLC23A2, single-nucleotide polymorphisms (SNPs) spaced throughout the gene were used as genetic markers and were analyzed as individual exposure variables because of the complex block structure (23). The SNPs assayed in both genes are depicted in figure 1.
Single-nucleotide polymorphism positions and linkage disequilibrium, Pregnancy, Infection, and Nutrition Study, 1995–2000. The two genes SLC23A1 and SLC23A2 differ 10-fold in size (16 kb vs. 160 kb, respectively), but exon/intron architecture is very similar. Arrows indicate the positions of the single-nucleotide polymorphisms used for the association studies. The degree of linkage disequilibrium in African-American and Caucasian populations in the two genes is represented in the triangular figures. D′ values are given in percentage. Values in parentheses indicate uncertain results. kb, kilobase (1,000 base pairs or nucleotides).
Genotyping
Genotyping was performed by TaqMan assays (Applied Biosystems, Foster City, California). Assay conditions for each variant can be found at http://snp500cancer.nci.nih.gov (26). For quality control purposes, a 10 percent random sample was assayed in duplicate with more than 99 percent concordance between replicates. Sample completion for the 16 assays was greater than 90 percent, and genotype failures were randomly distributed among all samples. Specifically, there was no significant difference in the distribution of dropouts between cases or controls or between the African-American or Caucasian populations. Deviations from Hardy-Weinberg equilibrium were determined in the control population, stratified by race. No substantial deviations were observed (27, 28).
Analytic methods
Because of the differences in linkage disequilibrium between SLC23A1 and SLC23A2, different analytic approaches were applied. As stated above, the strong association between variants within the SLC23A1 gene allowed us to estimate a few common haplotypes that accounted for greater than 90 percent of the common ancestral haplotypes within this gene. In order to estimate SLC23A1 haplotypes by use of unphased genotype data in unrelated subjects, a Bayesian algorithm, known as PHASE (version 2.1), was used (29, 30) (http://www.stat.washington.edu/stephens/software.html). We treated a haplotype as missing when phase probabilities were below 90 percent (i.e., when the program could not distinguish with a posterior probability of at least 90 percent that it had correctly assigned an allele to a specific chromosome). Similarly, we excluded subjects in whom more than 50 percent of the genotype data were missing. A case-control permutation test for haplotypes was conducted using PHASE (version 2.1). This software tests the null hypothesis that the overall distribution of haplotypes (i.e., frequencies) is similar in cases and controls versus the alternative that there are differences in the distribution of haplotypes. This may provide a more powerful approach than testing single haplotypes individually (30).
Linkage disequilibrium was estimated by use of Haploview (version 2.05) (31) (http://www.broad.mit.edu/personal/jcbarret/haploview/). In Haploview, linkage disequilibrium is measured as D′, which is a standardized measure ranging from 0 to 1, where 0 indicates randomness and 1 indicates complete nonrandomness, namely, that the two alleles always reside on the same ancestral chromosome.
We applied two different approaches for estimating the effects of genetic variants in SLC23A1 and SLC23A2 on the risk of spontaneous preterm birth. For SLC23A1, as stated above, we were able to estimate a few common haplotypes (five in African Americans, three in Caucasians) that accounted for the vast majority of variation across the gene and simultaneously accounted for all of the sites of genetic variation that we measured. We therefore applied conventional maximum likelihood methods for estimating odds ratios and 95 percent confidence intervals. Unconditional logistic regression, contrasting individuals heterozygous or homozygous for the index haplotype to individuals homozygous for the most common haplotype in the population, was applied (32). All models were adjusted for smoking and were stratified according to the maternal self-described ethnic background, Caucasian or African American.
We applied two analytic approaches to estimating the effect of SLC23A2 variants on the risk of spontaneous preterm delivery. To obtain odds ratio estimates for each SNP that simultaneously adjusted for the presence of all other measured polymorphisms within the gene, we implemented semi-Bayesian hierarchical regression analysis by use of the SAS GLIMMIX procedure (SAS Institute, Inc., Cary, North Carolina) (33–37). Hierarchical regression estimates have been shown in simulation studies to be more accurate and stable than those derived from conventional maximum likelihood models, especially when multiple variants are considered simultaneously and individual cell sizes are small (35). By shrinking unstable estimates toward the estimated prior means of the target parameters, the likelihood of observing large effects due to chance is reduced. Our first-stage logistic regression model regressed preterm case status on the individual SLC23A2 polymorphisms. Indicator variables were created to examine each polymorphism within genotype categories. All models were adjusted for smoking during pregnancy, maternal age, and maternal education and were stratified by maternal self-reported ethnicity. We explored two different second-stage linear models. Because we were interested in the possibility of an allele-dosing effect and had no a priori reason to suspect that specific polymorphisms shared a common functional effect (since polymorphisms were selected to represent the most common variation across the gene, and not because of their presumed effect on gene function), we examined a second-level model that assumed that the true effects of homozygotes (for any given polymorphism) and heterozygotes, respectively, were exchangeable. We also examined a second-stage model that regressed all genotype categories toward a common prior normal distribution (an intercept-only second-stage model) and found very similar effects. In both cases, residual variation unexplained by the second-stage covariates was captured in the independent random variable δ, which has a mean of 0 and variance τ2. We assumed with 95 percent certainty that the odds ratio for each genotype, after adjustment for the second-stage covariates, would fall within a 10-fold range (τ2 of 0.35). We also analyzed each variant genotype separately by use of conventional logistic regression. As in SLC23A1, all models were adjusted for smoking and stratified according to maternal self-described ethnic background, Caucasian or African American. All statistical analyses were performed using SAS, version 8, software (SAS Institute, Inc.).
RESULTS
Haplotypes in SLC23A1
Based on genotype data, there were three common haplotypes observed in the Caucasian group (table 1); pair-wise D′ values were between 0.85 and 1.00, indicating strong linkage disequilibrium within this block shown in figure 1 (38). In Caucasians, the three most common haplotypes represent more than 96 percent of the observed variation. The block structure in the African-American population was more complex (table 2) (figure 1). In the African-American control group, the five most common haplotypes amount to 95 percent of the observed variation. It is notable that the most common haplotype in the African-American population is the second most common one observed in Caucasian controls and vice versa (table 2). Reported haplotypes were inferred at greater than 95 percent probability.
SLC23A1 single-nucleotide polymorphisms and allele frequencies, Pregnancy, Infection, and Nutrition Study, 1995–2000
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A1-18 | rs10063949 | 5′/−584 | A/G | A = 0.24 | G = 0.33 |
| SLC23A1-04‡ | Pending | Ex8/3423 | A/G | G = 0.08 | No variation |
| SLC23A1-05‡ | Pending | Ex8/3441 | G/A | A = 0.09 | A = 0.03 |
| SLC23A1-09 | rs4257763 | In10/4784 | T/C | T = 0.14 | C = 0.35 |
| SLC23A1-11 | Pending | In11/5095 | A/G | G = 0.08 | No variation |
| SLC23A1-21 | rs6596473 | In13/8367 | C/G | C = 0.37 | G = 0.31 |
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A1-18 | rs10063949 | 5′/−584 | A/G | A = 0.24 | G = 0.33 |
| SLC23A1-04‡ | Pending | Ex8/3423 | A/G | G = 0.08 | No variation |
| SLC23A1-05‡ | Pending | Ex8/3441 | G/A | A = 0.09 | A = 0.03 |
| SLC23A1-09 | rs4257763 | In10/4784 | T/C | T = 0.14 | C = 0.35 |
| SLC23A1-11 | Pending | In11/5095 | A/G | G = 0.08 | No variation |
| SLC23A1-21 | rs6596473 | In13/8367 | C/G | C = 0.37 | G = 0.31 |
SNP500 ID, SNP500Cancer database identifier from the Cancer Genome Anatomy Project, where “SNP” represents single-nucleotide polymorphism (http://snp500cancer.nci.nih.gov/); dbSNP ID, single-nucleotide polymorphism database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP/index.html).
“Ex” and “In” refer to exon and intron, respectively.
SLC23A1-04 and SLC23A1-05 are nonsynonymous (both valine to methionine at positions 258 and 264, respectively). All variants are in Hardy-Weinberg equilibrium.
SLC23A1 single-nucleotide polymorphisms and allele frequencies, Pregnancy, Infection, and Nutrition Study, 1995–2000
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A1-18 | rs10063949 | 5′/−584 | A/G | A = 0.24 | G = 0.33 |
| SLC23A1-04‡ | Pending | Ex8/3423 | A/G | G = 0.08 | No variation |
| SLC23A1-05‡ | Pending | Ex8/3441 | G/A | A = 0.09 | A = 0.03 |
| SLC23A1-09 | rs4257763 | In10/4784 | T/C | T = 0.14 | C = 0.35 |
| SLC23A1-11 | Pending | In11/5095 | A/G | G = 0.08 | No variation |
| SLC23A1-21 | rs6596473 | In13/8367 | C/G | C = 0.37 | G = 0.31 |
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A1-18 | rs10063949 | 5′/−584 | A/G | A = 0.24 | G = 0.33 |
| SLC23A1-04‡ | Pending | Ex8/3423 | A/G | G = 0.08 | No variation |
| SLC23A1-05‡ | Pending | Ex8/3441 | G/A | A = 0.09 | A = 0.03 |
| SLC23A1-09 | rs4257763 | In10/4784 | T/C | T = 0.14 | C = 0.35 |
| SLC23A1-11 | Pending | In11/5095 | A/G | G = 0.08 | No variation |
| SLC23A1-21 | rs6596473 | In13/8367 | C/G | C = 0.37 | G = 0.31 |
SNP500 ID, SNP500Cancer database identifier from the Cancer Genome Anatomy Project, where “SNP” represents single-nucleotide polymorphism (http://snp500cancer.nci.nih.gov/); dbSNP ID, single-nucleotide polymorphism database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP/index.html).
“Ex” and “In” refer to exon and intron, respectively.
SLC23A1-04 and SLC23A1-05 are nonsynonymous (both valine to methionine at positions 258 and 264, respectively). All variants are in Hardy-Weinberg equilibrium.
SLC23A1 inferred haplotypes,* Pregnancy, Infection, and Nutrition Study, 1995–2000
Haplotype . | Variants† . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|
| 1 | GAGCAG | 0.61 | 0.29 |
| 2 | AAGTAC | 0.14 | 0.64 |
| 3 | GAACAC | 0.09 | 0.03 |
| 4 | AGGCGC | 0.06 | Not observed |
| 5 | AAGCAC | 0.05 | Not observed |
Haplotype . | Variants† . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|
| 1 | GAGCAG | 0.61 | 0.29 |
| 2 | AAGTAC | 0.14 | 0.64 |
| 3 | GAACAC | 0.09 | 0.03 |
| 4 | AGGCGC | 0.06 | Not observed |
| 5 | AAGCAC | 0.05 | Not observed |
All loci inferred with more than 95% probability.
Underlined A's in haplotypes 1–3 show no variation in Caucasians.
SLC23A1 inferred haplotypes,* Pregnancy, Infection, and Nutrition Study, 1995–2000
Haplotype . | Variants† . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|
| 1 | GAGCAG | 0.61 | 0.29 |
| 2 | AAGTAC | 0.14 | 0.64 |
| 3 | GAACAC | 0.09 | 0.03 |
| 4 | AGGCGC | 0.06 | Not observed |
| 5 | AAGCAC | 0.05 | Not observed |
Haplotype . | Variants† . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|
| 1 | GAGCAG | 0.61 | 0.29 |
| 2 | AAGTAC | 0.14 | 0.64 |
| 3 | GAACAC | 0.09 | 0.03 |
| 4 | AGGCGC | 0.06 | Not observed |
| 5 | AAGCAC | 0.05 | Not observed |
All loci inferred with more than 95% probability.
Underlined A's in haplotypes 1–3 show no variation in Caucasians.
Genotype analysis of SLC23A2
The analysis of SLC23A2 was restricted to 10 common variants distributed across the gene. In the control populations, the range in frequency was from 4 percent to 46 percent (table 3). The distribution of SNP markers was insufficient to adequately capture the common block structures of SLC23A2 (figure 1) (23).
SLC23A2 single-nucleotide polymorphisms and allele frequencies, Pregnancy, Infection, and Nutrition Study, 1995–2000
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A2-31 | rs12479919 | In1/–67652 | G/A | A = 0.19 | A = 0.37 |
| SLC23A2-32 | rs2681118 | In1/–54795 | A/C | C = 0.20 | C = 0.15 |
| SLC23A2-08 | rs6139591 | In2/–38152 | C/T | T = 0.35 | T = 0.41 |
| SLC23A2-09 | rs2681116 | In2/–38124 | G/A | A = 0.34 | A = 0.46 |
| SLC23A2-33 | rs4813725 | In2/–4777 | G/A | A = 0.08 | A = 0.32 |
| SLC23A2-26 | rs1715365 | In3/14121 | G/A | A = 0.46 | A = 0.43 |
| SLC23A2-03 | rs1776964 | Ex6/32901‡ | C/T | T = 0.39 | T = 0.43 |
| SLC23A2-05 | rs4987219 | In8/48263 | G/C | G = 0.20 | C = 0.42 |
| SLC23A2-01§ | rs1110277 | Ex11/58527‡ | T/C | C = 0.15 | C = 0.31 |
| SLC23A2-02§ | Pending | 3′/78315 | C/T | T = 0.04 | T = 0.11 |
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A2-31 | rs12479919 | In1/–67652 | G/A | A = 0.19 | A = 0.37 |
| SLC23A2-32 | rs2681118 | In1/–54795 | A/C | C = 0.20 | C = 0.15 |
| SLC23A2-08 | rs6139591 | In2/–38152 | C/T | T = 0.35 | T = 0.41 |
| SLC23A2-09 | rs2681116 | In2/–38124 | G/A | A = 0.34 | A = 0.46 |
| SLC23A2-33 | rs4813725 | In2/–4777 | G/A | A = 0.08 | A = 0.32 |
| SLC23A2-26 | rs1715365 | In3/14121 | G/A | A = 0.46 | A = 0.43 |
| SLC23A2-03 | rs1776964 | Ex6/32901‡ | C/T | T = 0.39 | T = 0.43 |
| SLC23A2-05 | rs4987219 | In8/48263 | G/C | G = 0.20 | C = 0.42 |
| SLC23A2-01§ | rs1110277 | Ex11/58527‡ | T/C | C = 0.15 | C = 0.31 |
| SLC23A2-02§ | Pending | 3′/78315 | C/T | T = 0.04 | T = 0.11 |
SNP500 ID, SNP500Cancer database identifier from the Cancer Genome Anatomy Project, where “SNP” represents single-nucleotide polymorphism (http://snp500cancer.nci.nih.gov/); dbSNP ID, single-nucleotide polymorphism database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP/index.html).
“Ex” and “In” refer to exon and intron, respectively.
The single-nucleotide polymorphisms of exons 6 and 11 are synonymous.
In African Americans, the Hardy-Weinberg equilibrium deviation for SLC23A2-01 was χ2 = 6.71 (p = 0.01) and for SLC23A2-14 was χ2 = 4.59 (p = 0.03). In Caucasians, the Hardy-Weinberg equilibrium deviation for SLC23A2-02 was χ2 = 5.85 (p = 0.02). All other variants were in Hardy-Weinberg equilibrium for both populations.
SLC23A2 single-nucleotide polymorphisms and allele frequencies, Pregnancy, Infection, and Nutrition Study, 1995–2000
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A2-31 | rs12479919 | In1/–67652 | G/A | A = 0.19 | A = 0.37 |
| SLC23A2-32 | rs2681118 | In1/–54795 | A/C | C = 0.20 | C = 0.15 |
| SLC23A2-08 | rs6139591 | In2/–38152 | C/T | T = 0.35 | T = 0.41 |
| SLC23A2-09 | rs2681116 | In2/–38124 | G/A | A = 0.34 | A = 0.46 |
| SLC23A2-33 | rs4813725 | In2/–4777 | G/A | A = 0.08 | A = 0.32 |
| SLC23A2-26 | rs1715365 | In3/14121 | G/A | A = 0.46 | A = 0.43 |
| SLC23A2-03 | rs1776964 | Ex6/32901‡ | C/T | T = 0.39 | T = 0.43 |
| SLC23A2-05 | rs4987219 | In8/48263 | G/C | G = 0.20 | C = 0.42 |
| SLC23A2-01§ | rs1110277 | Ex11/58527‡ | T/C | C = 0.15 | C = 0.31 |
| SLC23A2-02§ | Pending | 3′/78315 | C/T | T = 0.04 | T = 0.11 |
SNP500 ID* . | dbSNP ID* . | Position† . | Allele . | African-American frequencies . | Caucasian frequencies . |
|---|---|---|---|---|---|
| SLC23A2-31 | rs12479919 | In1/–67652 | G/A | A = 0.19 | A = 0.37 |
| SLC23A2-32 | rs2681118 | In1/–54795 | A/C | C = 0.20 | C = 0.15 |
| SLC23A2-08 | rs6139591 | In2/–38152 | C/T | T = 0.35 | T = 0.41 |
| SLC23A2-09 | rs2681116 | In2/–38124 | G/A | A = 0.34 | A = 0.46 |
| SLC23A2-33 | rs4813725 | In2/–4777 | G/A | A = 0.08 | A = 0.32 |
| SLC23A2-26 | rs1715365 | In3/14121 | G/A | A = 0.46 | A = 0.43 |
| SLC23A2-03 | rs1776964 | Ex6/32901‡ | C/T | T = 0.39 | T = 0.43 |
| SLC23A2-05 | rs4987219 | In8/48263 | G/C | G = 0.20 | C = 0.42 |
| SLC23A2-01§ | rs1110277 | Ex11/58527‡ | T/C | C = 0.15 | C = 0.31 |
| SLC23A2-02§ | Pending | 3′/78315 | C/T | T = 0.04 | T = 0.11 |
SNP500 ID, SNP500Cancer database identifier from the Cancer Genome Anatomy Project, where “SNP” represents single-nucleotide polymorphism (http://snp500cancer.nci.nih.gov/); dbSNP ID, single-nucleotide polymorphism database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/SNP/index.html).
“Ex” and “In” refer to exon and intron, respectively.
The single-nucleotide polymorphisms of exons 6 and 11 are synonymous.
In African Americans, the Hardy-Weinberg equilibrium deviation for SLC23A2-01 was χ2 = 6.71 (p = 0.01) and for SLC23A2-14 was χ2 = 4.59 (p = 0.03). In Caucasians, the Hardy-Weinberg equilibrium deviation for SLC23A2-02 was χ2 = 5.85 (p = 0.02). All other variants were in Hardy-Weinberg equilibrium for both populations.
Preterm birth and genetic variation in SLC23A1 and SLC23A2
The case-control permutation test did not detect any differences in the haplotype frequency distributions between cases and controls for SLC23A1 in African-American or Caucasian populations. In an analysis stratified by ethnic group, associations between common haplotypes in SLC23A1 and preterm outcomes were observed (table 4). In the Caucasian population, individuals heterozygous for haplotype 1 (GAGCAG) had a reduced risk for spontaneous preterm birth compared with individuals homozygous for haplotype 2 (AAGTAC) (odds ratio (OR) = 0.5, 95 percent confidence interval (CI): 0.3, 0.9). Individuals homozygous for haplotype 1 had a reduction in risk when compared with individuals homozygous for haplotype 2 (OR = 0.7, 95 percent CI: 0.2, 1.9), but the estimate was imprecise. In the African-American population, there was no consistent pattern of association between the five common haplotypes in SLC23A1 and spontaneous preterm birth (table 4).
SLC23A1 haplotypes and risk for spontaneous preterm birth, Pregnancy, Infection, and Nutrition Study, 1995–2000
Haplotype* . | Cases (no./ haplotype) . | Controls (no./ haplotype) . | Logistic regression† . | . | |
|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | |
| African Americans | |||||
| 1/2 | 28/15 | 96/63 | 0.9 | 0.4, 1.9 | |
| 1/3 | 28/7 | 96/40 | 0.7 | 0.3, 1.8 | |
| 1/4 | 28/7 | 96/25 | 1.1 | 0.4, 3.1 | |
| 1/5 | 28/11 | 96/20 | 2.3 | 0.9, 5.6 | |
| Caucasians | |||||
| 2/1 | 37/26 | 138/168 | |||
| X/1‡ | /18 | /140 | 0.5 | 0.3, 0.9 | |
| 1/1‡ | /8 | /28 | 0.7 | 0.2, 1.9 | |
| 2/3 | 37/6 | 138/18 | 0.6 | 0.0, 0.9 | |
Haplotype* . | Cases (no./ haplotype) . | Controls (no./ haplotype) . | Logistic regression† . | . | |
|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | |
| African Americans | |||||
| 1/2 | 28/15 | 96/63 | 0.9 | 0.4, 1.9 | |
| 1/3 | 28/7 | 96/40 | 0.7 | 0.3, 1.8 | |
| 1/4 | 28/7 | 96/25 | 1.1 | 0.4, 3.1 | |
| 1/5 | 28/11 | 96/20 | 2.3 | 0.9, 5.6 | |
| Caucasians | |||||
| 2/1 | 37/26 | 138/168 | |||
| X/1‡ | /18 | /140 | 0.5 | 0.3, 0.9 | |
| 1/1‡ | /8 | /28 | 0.7 | 0.2, 1.9 | |
| 2/3 | 37/6 | 138/18 | 0.6 | 0.0, 0.9 | |
Haplotype 1: GAGCAG; 2: AAGTAC; 3: GAACAC; 4: AGGCGC; and 5: AAGCAC. The most common haplotype in African Americans (haplotype 1) or Caucasians (haplotype 2) was used as the referent for analysis.
Adjusted for smoking.
In Caucasians, heterozygous (X/1) and homozygous (1/1) individuals of haplotype 1 were contrasted to the referent group.
SLC23A1 haplotypes and risk for spontaneous preterm birth, Pregnancy, Infection, and Nutrition Study, 1995–2000
Haplotype* . | Cases (no./ haplotype) . | Controls (no./ haplotype) . | Logistic regression† . | . | |
|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | |
| African Americans | |||||
| 1/2 | 28/15 | 96/63 | 0.9 | 0.4, 1.9 | |
| 1/3 | 28/7 | 96/40 | 0.7 | 0.3, 1.8 | |
| 1/4 | 28/7 | 96/25 | 1.1 | 0.4, 3.1 | |
| 1/5 | 28/11 | 96/20 | 2.3 | 0.9, 5.6 | |
| Caucasians | |||||
| 2/1 | 37/26 | 138/168 | |||
| X/1‡ | /18 | /140 | 0.5 | 0.3, 0.9 | |
| 1/1‡ | /8 | /28 | 0.7 | 0.2, 1.9 | |
| 2/3 | 37/6 | 138/18 | 0.6 | 0.0, 0.9 | |
Haplotype* . | Cases (no./ haplotype) . | Controls (no./ haplotype) . | Logistic regression† . | . | |
|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | |
| African Americans | |||||
| 1/2 | 28/15 | 96/63 | 0.9 | 0.4, 1.9 | |
| 1/3 | 28/7 | 96/40 | 0.7 | 0.3, 1.8 | |
| 1/4 | 28/7 | 96/25 | 1.1 | 0.4, 3.1 | |
| 1/5 | 28/11 | 96/20 | 2.3 | 0.9, 5.6 | |
| Caucasians | |||||
| 2/1 | 37/26 | 138/168 | |||
| X/1‡ | /18 | /140 | 0.5 | 0.3, 0.9 | |
| 1/1‡ | /8 | /28 | 0.7 | 0.2, 1.9 | |
| 2/3 | 37/6 | 138/18 | 0.6 | 0.0, 0.9 | |
Haplotype 1: GAGCAG; 2: AAGTAC; 3: GAACAC; 4: AGGCGC; and 5: AAGCAC. The most common haplotype in African Americans (haplotype 1) or Caucasians (haplotype 2) was used as the referent for analysis.
Adjusted for smoking.
In Caucasians, heterozygous (X/1) and homozygous (1/1) individuals of haplotype 1 were contrasted to the referent group.
The analysis of each SNP used to construct the SLC23A1 haplotypes was unremarkable. Both standard contingency tables and hierarchical regression analyses were performed and did not demonstrate significant findings overall or when stratified by self-described ethnic group (data not shown).
In the analysis of 10 SNPs across SLC23A2 in Caucasian subjects, two were individually associated with an increased risk for preterm birth in conventional logistic regression models (table 5). Individuals heterozygous for the T allele of SLC23A2-08 had an odds ratio of 1.7 (95 percent CI: 0.9, 3.3). Individuals homozygous for the T allele have an odds ratio of 2.7 (95 percent CI: 1.2, 6.3). Hierarchical regression results were very similar. In individuals heterozygous for the minor allele A of SLC23A2-09, an increased risk for preterm birth was also observed (OR = 1.9, 95 percent CI: 1.0, 3.6), although not in subjects homozygous for the minor A allele; hierarchical regression did not reveal an effect for homozygotes (OR = 1.1, 95 percent CI: 0.5, 2.3). Individuals homozygous for the T allele in SLC23A2-03 had a decreased risk of spontaneous preterm birth (OR = 0.3, 95 percent CI: 0.1, 0.8), although this was not confirmed in hierarchical regression analysis (OR = 0.7, 95 percent CI: 0.3, 1.6).
SLC23A2 variants and risk for spontaneous preterm birth, Pregnancy, Infection, and Nutrition Study, 1995–2000*
Variant . | Cases (no./SNP†) . | Controls (no./SNP) . | Logistic regression‡ . | . | Semi-Bayesian hierarchical regression . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | ||||||
| African Americans | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 42/18 | 146/63 | 1.0 | 0.5, 1.9 | 0.8 | 0.4, 1.7 | ||||||
| GG/AA | /3 | /9 | 1.3 | 0.3, 5.3 | 1.3 | 0.5, 3.5 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 33/17 | 148/52 | 1.4 | 0.7, 2.8 | 1.3 | 0.6, 2.8 | ||||||
| AA/CC | /5 | /6 | 4.0 | 1.1, 14.1 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 28/25 | 72/108 | 0.8 | 0.4, 1.4 | 1.0 | 0.5, 2.1 | ||||||
| CC/TT | /2 | /35 | 0.2 | 0.0, 0.8 | 0.5 | 0.2, 1.3 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 22/20 | 67/103 | 0.6 | 0.3, 1.2 | 0.6 | 0.3, 1.2 | ||||||
| GG/AA | /7 | /33 | 0.8 | 0.3, 2.0 | 1.0 | 0.4, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 38/19 | 129/79 | 0.8 | 0.4, 1.6 | 1.1 | 0.5, 2.4 | ||||||
| GG/AA | /3 | /18 | 0.4 | 0.1, 1.7 | 0.9 | 0.3, 2.4 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 14/27 | 66/97 | 1.4 | 0.6, 3.0 | 1.0 | 0.5, 2.3 | ||||||
| GG/AA | /11 | /51 | 1.1 | 0.5, 2.8 | 1.1 | 0.5, 2.5 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 21/29 | 77/105 | 0.8 | 0.4, 1.6 | 0.7 | 0.4, 1.5 | ||||||
| CC/TT | /9 | /35 | 0.9 | 0.4, 2.2 | 1.0 | 0.4, 2.4 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 7/28 | 36/93 | 1.4 | 0.5, 3.9 | 1.3 | 0.6, 2.9 | ||||||
| GG/CC | /17 | /80 | 1.2 | 0.4, 3.4 | 1.1 | 0.5, 2.7 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 34/16 | 129/71 | 0.8 | 0.4, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| TT/CC | /4 | /6 | 2.4 | 0.6, 9.0 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 52/8 | 187/36 | 0.8 | 0.3, 1.8 | 0.7 | 0.3, 1.5 | ||||||
| CC/TT | /2 | /1 | 8.0 | 0.7, 94.5 | 1.0 | 0.3, 3.2 | ||||||
| Caucasians | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 25/31 | 125/141 | 1.3 | 0.7, 2.3 | 1.5 | 0.8, 2.8 | ||||||
| GG/AA | /7 | /43 | 1.0 | 0.4, 2.6 | 1.1 | 0.5, 2.4 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 41/20 | 197/92 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| AA/CC | /2 | /11 | 0.5 | 0.1, 3.7 | 1.0 | 0.4, 3.0 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 16/37 | 115/151 | 1.7 | 0.9, 3.3 | 1.9 | 1.0, 3.6 | ||||||
| CC/TT | /14 | /38 | 2.7 | 1.2, 6.3 | 2.3 | 1.0, 5.0 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 17/39 | 105/133 | 1.9 | 1.0, 3.6 | 1.5 | 0.8, 2.7 | ||||||
| GG/AA | /10 | /50 | 1.2 | 0.5, 2.9 | 1.1 | 0.5, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 41/18 | 208/89 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 2.0 | ||||||
| GG/AA | /3 | /14 | 0.7 | 0.2, 3.3 | 1.1 | 0.4, 3.0 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 20/31 | 98/135 | 1.1 | 0.6, 2.0 | 1.2 | 0.6, 2.3 | ||||||
| GG/AA | /11 | /50 | 1.0 | 0.4, 2.3 | 1.0 | 0.5, 2.3 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 31/29 | 112/135 | 0.7 | 0.4, 1.3 | 0.9 | 0.5, 1.7 | ||||||
| CC/TT | /7 | /61 | 0.3 | 0.1, 0.8 | 0.7 | 0.3, 1.6 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 16/21 | 66/123 | 0.8 | 0.4, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| GG/CC | /30 | /110 | 1.2 | 0.6, 2.5 | 1.4 | 0.7, 2.9 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 41/23 | 167/112 | 0.9 | 0.5, 1.7 | 1.0 | 0.6, 1.8 | ||||||
| TT/CC | /3 | /26 | 0.6 | 0.2, 2.0 | 0.9 | 0.4, 2.2 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 56/7 | 266/44 | 0.7 | 0.3, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| CC/TT | /1 | /3 | 1.5 | 0.2, 14.6 | 1.3 | 0.4, 3.9 | ||||||
Variant . | Cases (no./SNP†) . | Controls (no./SNP) . | Logistic regression‡ . | . | Semi-Bayesian hierarchical regression . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | ||||||
| African Americans | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 42/18 | 146/63 | 1.0 | 0.5, 1.9 | 0.8 | 0.4, 1.7 | ||||||
| GG/AA | /3 | /9 | 1.3 | 0.3, 5.3 | 1.3 | 0.5, 3.5 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 33/17 | 148/52 | 1.4 | 0.7, 2.8 | 1.3 | 0.6, 2.8 | ||||||
| AA/CC | /5 | /6 | 4.0 | 1.1, 14.1 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 28/25 | 72/108 | 0.8 | 0.4, 1.4 | 1.0 | 0.5, 2.1 | ||||||
| CC/TT | /2 | /35 | 0.2 | 0.0, 0.8 | 0.5 | 0.2, 1.3 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 22/20 | 67/103 | 0.6 | 0.3, 1.2 | 0.6 | 0.3, 1.2 | ||||||
| GG/AA | /7 | /33 | 0.8 | 0.3, 2.0 | 1.0 | 0.4, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 38/19 | 129/79 | 0.8 | 0.4, 1.6 | 1.1 | 0.5, 2.4 | ||||||
| GG/AA | /3 | /18 | 0.4 | 0.1, 1.7 | 0.9 | 0.3, 2.4 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 14/27 | 66/97 | 1.4 | 0.6, 3.0 | 1.0 | 0.5, 2.3 | ||||||
| GG/AA | /11 | /51 | 1.1 | 0.5, 2.8 | 1.1 | 0.5, 2.5 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 21/29 | 77/105 | 0.8 | 0.4, 1.6 | 0.7 | 0.4, 1.5 | ||||||
| CC/TT | /9 | /35 | 0.9 | 0.4, 2.2 | 1.0 | 0.4, 2.4 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 7/28 | 36/93 | 1.4 | 0.5, 3.9 | 1.3 | 0.6, 2.9 | ||||||
| GG/CC | /17 | /80 | 1.2 | 0.4, 3.4 | 1.1 | 0.5, 2.7 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 34/16 | 129/71 | 0.8 | 0.4, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| TT/CC | /4 | /6 | 2.4 | 0.6, 9.0 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 52/8 | 187/36 | 0.8 | 0.3, 1.8 | 0.7 | 0.3, 1.5 | ||||||
| CC/TT | /2 | /1 | 8.0 | 0.7, 94.5 | 1.0 | 0.3, 3.2 | ||||||
| Caucasians | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 25/31 | 125/141 | 1.3 | 0.7, 2.3 | 1.5 | 0.8, 2.8 | ||||||
| GG/AA | /7 | /43 | 1.0 | 0.4, 2.6 | 1.1 | 0.5, 2.4 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 41/20 | 197/92 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| AA/CC | /2 | /11 | 0.5 | 0.1, 3.7 | 1.0 | 0.4, 3.0 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 16/37 | 115/151 | 1.7 | 0.9, 3.3 | 1.9 | 1.0, 3.6 | ||||||
| CC/TT | /14 | /38 | 2.7 | 1.2, 6.3 | 2.3 | 1.0, 5.0 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 17/39 | 105/133 | 1.9 | 1.0, 3.6 | 1.5 | 0.8, 2.7 | ||||||
| GG/AA | /10 | /50 | 1.2 | 0.5, 2.9 | 1.1 | 0.5, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 41/18 | 208/89 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 2.0 | ||||||
| GG/AA | /3 | /14 | 0.7 | 0.2, 3.3 | 1.1 | 0.4, 3.0 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 20/31 | 98/135 | 1.1 | 0.6, 2.0 | 1.2 | 0.6, 2.3 | ||||||
| GG/AA | /11 | /50 | 1.0 | 0.4, 2.3 | 1.0 | 0.5, 2.3 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 31/29 | 112/135 | 0.7 | 0.4, 1.3 | 0.9 | 0.5, 1.7 | ||||||
| CC/TT | /7 | /61 | 0.3 | 0.1, 0.8 | 0.7 | 0.3, 1.6 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 16/21 | 66/123 | 0.8 | 0.4, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| GG/CC | /30 | /110 | 1.2 | 0.6, 2.5 | 1.4 | 0.7, 2.9 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 41/23 | 167/112 | 0.9 | 0.5, 1.7 | 1.0 | 0.6, 1.8 | ||||||
| TT/CC | /3 | /26 | 0.6 | 0.2, 2.0 | 0.9 | 0.4, 2.2 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 56/7 | 266/44 | 0.7 | 0.3, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| CC/TT | /1 | /3 | 1.5 | 0.2, 14.6 | 1.3 | 0.4, 3.9 | ||||||
Heterozygous or homozygous variant alleles were contrasted to the homozygous wild-type allele.
SNP, single-nucleotide polymorphism.
Adjusted for smoking.
SLC23A2 variants and risk for spontaneous preterm birth, Pregnancy, Infection, and Nutrition Study, 1995–2000*
Variant . | Cases (no./SNP†) . | Controls (no./SNP) . | Logistic regression‡ . | . | Semi-Bayesian hierarchical regression . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | ||||||
| African Americans | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 42/18 | 146/63 | 1.0 | 0.5, 1.9 | 0.8 | 0.4, 1.7 | ||||||
| GG/AA | /3 | /9 | 1.3 | 0.3, 5.3 | 1.3 | 0.5, 3.5 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 33/17 | 148/52 | 1.4 | 0.7, 2.8 | 1.3 | 0.6, 2.8 | ||||||
| AA/CC | /5 | /6 | 4.0 | 1.1, 14.1 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 28/25 | 72/108 | 0.8 | 0.4, 1.4 | 1.0 | 0.5, 2.1 | ||||||
| CC/TT | /2 | /35 | 0.2 | 0.0, 0.8 | 0.5 | 0.2, 1.3 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 22/20 | 67/103 | 0.6 | 0.3, 1.2 | 0.6 | 0.3, 1.2 | ||||||
| GG/AA | /7 | /33 | 0.8 | 0.3, 2.0 | 1.0 | 0.4, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 38/19 | 129/79 | 0.8 | 0.4, 1.6 | 1.1 | 0.5, 2.4 | ||||||
| GG/AA | /3 | /18 | 0.4 | 0.1, 1.7 | 0.9 | 0.3, 2.4 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 14/27 | 66/97 | 1.4 | 0.6, 3.0 | 1.0 | 0.5, 2.3 | ||||||
| GG/AA | /11 | /51 | 1.1 | 0.5, 2.8 | 1.1 | 0.5, 2.5 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 21/29 | 77/105 | 0.8 | 0.4, 1.6 | 0.7 | 0.4, 1.5 | ||||||
| CC/TT | /9 | /35 | 0.9 | 0.4, 2.2 | 1.0 | 0.4, 2.4 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 7/28 | 36/93 | 1.4 | 0.5, 3.9 | 1.3 | 0.6, 2.9 | ||||||
| GG/CC | /17 | /80 | 1.2 | 0.4, 3.4 | 1.1 | 0.5, 2.7 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 34/16 | 129/71 | 0.8 | 0.4, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| TT/CC | /4 | /6 | 2.4 | 0.6, 9.0 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 52/8 | 187/36 | 0.8 | 0.3, 1.8 | 0.7 | 0.3, 1.5 | ||||||
| CC/TT | /2 | /1 | 8.0 | 0.7, 94.5 | 1.0 | 0.3, 3.2 | ||||||
| Caucasians | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 25/31 | 125/141 | 1.3 | 0.7, 2.3 | 1.5 | 0.8, 2.8 | ||||||
| GG/AA | /7 | /43 | 1.0 | 0.4, 2.6 | 1.1 | 0.5, 2.4 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 41/20 | 197/92 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| AA/CC | /2 | /11 | 0.5 | 0.1, 3.7 | 1.0 | 0.4, 3.0 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 16/37 | 115/151 | 1.7 | 0.9, 3.3 | 1.9 | 1.0, 3.6 | ||||||
| CC/TT | /14 | /38 | 2.7 | 1.2, 6.3 | 2.3 | 1.0, 5.0 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 17/39 | 105/133 | 1.9 | 1.0, 3.6 | 1.5 | 0.8, 2.7 | ||||||
| GG/AA | /10 | /50 | 1.2 | 0.5, 2.9 | 1.1 | 0.5, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 41/18 | 208/89 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 2.0 | ||||||
| GG/AA | /3 | /14 | 0.7 | 0.2, 3.3 | 1.1 | 0.4, 3.0 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 20/31 | 98/135 | 1.1 | 0.6, 2.0 | 1.2 | 0.6, 2.3 | ||||||
| GG/AA | /11 | /50 | 1.0 | 0.4, 2.3 | 1.0 | 0.5, 2.3 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 31/29 | 112/135 | 0.7 | 0.4, 1.3 | 0.9 | 0.5, 1.7 | ||||||
| CC/TT | /7 | /61 | 0.3 | 0.1, 0.8 | 0.7 | 0.3, 1.6 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 16/21 | 66/123 | 0.8 | 0.4, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| GG/CC | /30 | /110 | 1.2 | 0.6, 2.5 | 1.4 | 0.7, 2.9 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 41/23 | 167/112 | 0.9 | 0.5, 1.7 | 1.0 | 0.6, 1.8 | ||||||
| TT/CC | /3 | /26 | 0.6 | 0.2, 2.0 | 0.9 | 0.4, 2.2 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 56/7 | 266/44 | 0.7 | 0.3, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| CC/TT | /1 | /3 | 1.5 | 0.2, 14.6 | 1.3 | 0.4, 3.9 | ||||||
Variant . | Cases (no./SNP†) . | Controls (no./SNP) . | Logistic regression‡ . | . | Semi-Bayesian hierarchical regression . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | Odds ratio . | 95% confidence interval . | Odds ratio . | 95% confidence interval . | ||||||
| African Americans | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 42/18 | 146/63 | 1.0 | 0.5, 1.9 | 0.8 | 0.4, 1.7 | ||||||
| GG/AA | /3 | /9 | 1.3 | 0.3, 5.3 | 1.3 | 0.5, 3.5 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 33/17 | 148/52 | 1.4 | 0.7, 2.8 | 1.3 | 0.6, 2.8 | ||||||
| AA/CC | /5 | /6 | 4.0 | 1.1, 14.1 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 28/25 | 72/108 | 0.8 | 0.4, 1.4 | 1.0 | 0.5, 2.1 | ||||||
| CC/TT | /2 | /35 | 0.2 | 0.0, 0.8 | 0.5 | 0.2, 1.3 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 22/20 | 67/103 | 0.6 | 0.3, 1.2 | 0.6 | 0.3, 1.2 | ||||||
| GG/AA | /7 | /33 | 0.8 | 0.3, 2.0 | 1.0 | 0.4, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 38/19 | 129/79 | 0.8 | 0.4, 1.6 | 1.1 | 0.5, 2.4 | ||||||
| GG/AA | /3 | /18 | 0.4 | 0.1, 1.7 | 0.9 | 0.3, 2.4 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 14/27 | 66/97 | 1.4 | 0.6, 3.0 | 1.0 | 0.5, 2.3 | ||||||
| GG/AA | /11 | /51 | 1.1 | 0.5, 2.8 | 1.1 | 0.5, 2.5 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 21/29 | 77/105 | 0.8 | 0.4, 1.6 | 0.7 | 0.4, 1.5 | ||||||
| CC/TT | /9 | /35 | 0.9 | 0.4, 2.2 | 1.0 | 0.4, 2.4 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 7/28 | 36/93 | 1.4 | 0.5, 3.9 | 1.3 | 0.6, 2.9 | ||||||
| GG/CC | /17 | /80 | 1.2 | 0.4, 3.4 | 1.1 | 0.5, 2.7 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 34/16 | 129/71 | 0.8 | 0.4, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| TT/CC | /4 | /6 | 2.4 | 0.6, 9.0 | 1.1 | 0.4, 3.1 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 52/8 | 187/36 | 0.8 | 0.3, 1.8 | 0.7 | 0.3, 1.5 | ||||||
| CC/TT | /2 | /1 | 8.0 | 0.7, 94.5 | 1.0 | 0.3, 3.2 | ||||||
| Caucasians | ||||||||||||
| SLC23A2-31 | ||||||||||||
| GG/AG | 25/31 | 125/141 | 1.3 | 0.7, 2.3 | 1.5 | 0.8, 2.8 | ||||||
| GG/AA | /7 | /43 | 1.0 | 0.4, 2.6 | 1.1 | 0.5, 2.4 | ||||||
| SLC23A2-32 | ||||||||||||
| AA/AC | 41/20 | 197/92 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| AA/CC | /2 | /11 | 0.5 | 0.1, 3.7 | 1.0 | 0.4, 3.0 | ||||||
| SLC23A2-08 | ||||||||||||
| CC/CT | 16/37 | 115/151 | 1.7 | 0.9, 3.3 | 1.9 | 1.0, 3.6 | ||||||
| CC/TT | /14 | /38 | 2.7 | 1.2, 6.3 | 2.3 | 1.0, 5.0 | ||||||
| SLC23A2-09 | ||||||||||||
| GG/GA | 17/39 | 105/133 | 1.9 | 1.0, 3.6 | 1.5 | 0.8, 2.7 | ||||||
| GG/AA | /10 | /50 | 1.2 | 0.5, 2.9 | 1.1 | 0.5, 2.3 | ||||||
| SLC23A2-33 | ||||||||||||
| GG/GA | 41/18 | 208/89 | 0.9 | 0.5, 1.7 | 1.0 | 0.5, 2.0 | ||||||
| GG/AA | /3 | /14 | 0.7 | 0.2, 3.3 | 1.1 | 0.4, 3.0 | ||||||
| SLC23A2-26 | ||||||||||||
| GG/GA | 20/31 | 98/135 | 1.1 | 0.6, 2.0 | 1.2 | 0.6, 2.3 | ||||||
| GG/AA | /11 | /50 | 1.0 | 0.4, 2.3 | 1.0 | 0.5, 2.3 | ||||||
| SLC23A2-03 | ||||||||||||
| CC/CT | 31/29 | 112/135 | 0.7 | 0.4, 1.3 | 0.9 | 0.5, 1.7 | ||||||
| CC/TT | /7 | /61 | 0.3 | 0.1, 0.8 | 0.7 | 0.3, 1.6 | ||||||
| SLC23A2-05 | ||||||||||||
| GG/GC | 16/21 | 66/123 | 0.8 | 0.4, 1.7 | 1.0 | 0.5, 1.9 | ||||||
| GG/CC | /30 | /110 | 1.2 | 0.6, 2.5 | 1.4 | 0.7, 2.9 | ||||||
| SLC23A2-01 | ||||||||||||
| TT/TC | 41/23 | 167/112 | 0.9 | 0.5, 1.7 | 1.0 | 0.6, 1.8 | ||||||
| TT/CC | /3 | /26 | 0.6 | 0.2, 2.0 | 0.9 | 0.4, 2.2 | ||||||
| SLC23A2-02 | ||||||||||||
| CC/CT | 56/7 | 266/44 | 0.7 | 0.3, 1.6 | 0.8 | 0.4, 1.7 | ||||||
| CC/TT | /1 | /3 | 1.5 | 0.2, 14.6 | 1.3 | 0.4, 3.9 | ||||||
Heterozygous or homozygous variant alleles were contrasted to the homozygous wild-type allele.
SNP, single-nucleotide polymorphism.
Adjusted for smoking.
In an analysis of the 10 SLC23A2 SNPs in African-American subjects, the data are less conclusive (table 5). Two SNPs, SLC23A2-08 and SLC23A2-32, were associated with risk for preterm birth in conventional logistic regression models but not by hierarchical analyses. Specifically, subjects homozygous for the minor C allele in SLC23A2-32 carried an increased risk for preterm birth (OR = 4.0, 95 percent CI: 1.1, 14.1), but in hierarchical regression analysis the finding was not confirmed (OR = 1.1, 95 percent CI: 0.4, 3.1). Interestingly, African-American subjects who were homozygous for the T allele of SLC23A2-08 had a decreased risk (OR = 0.2, 95 percent CI: 0.0, 0.8), but in hierarchical analysis no effect was observed (OR = 0.5, 95 percent CI: 0.2, 1.3).
We also examined the interaction among vitamin C intake, vitamin C transporter polymorphisms, and risk of preterm birth in this population. The results, however, were very unstable because of sparse numbers and severely limited interpretation.
DISCUSSION
In this study, we report on our analysis of common SNPs and haplotypes in the sodium-dependent vitamin C transporters, SLC23A1 and SLC23A2, in relation to outcomes of preterm birth. Specifically, our findings indicate that common genetic variants within the larger, ubiquitously expressed vitamin C transporter, SLC23A2, could be associated with spontaneous preterm delivery. This study was designed to examine the major transport genes for vitamin C uptake in a nested case-control study drawn from a well-characterized, prospective pregnancy cohort. Since there is a substantial size difference between the two genes, SLC23A1 and SLC23A2, we chose complementary approaches for analyses of SNPs in a case-control association study, namely, haplotype analysis for the former and individual genotype analysis for the latter. The substantially different linkage disequilibrium patterns across these two genes required us to adopt separate approaches. However, with the deposition of new SNPs from the International HapMap Project (http://www.hapmap.org), it is likely that the linkage disequilibrium structures across SLC23A2 will be available at a later time for analysis (39), especially in follow-up of notable findings in this study.
In our analysis, we stratified according to the two major ethnic groups enrolled in the Pregnancy, Infection, and Nutrition Study, because of the observed differences between allele frequencies and extent of differences in linkage disequilibrium between African-American and Caucasian populations. These differences are consistent with the reports of others (40–42). Differences in linkage disequilibrium patterns could partially explain the differences in risk by ethnic groups, which could also be influenced by differences in common genetic variants in additional genes not tested in this study. Although the biologic basis for this observed difference is unknown, it is possible that additional variants could be in linkage disequilibrium, either within or neighboring the gene, that could account for the observation. In this regard, further studies are needed to confirm our results and to investigate the biologic implications of variation in SLC23A2 and vitamin C transport. Still, our genetic association study provides preliminary evidence that differences in the major sodium-dependent transporters for vitamin C could affect pregnancy and its outcome.
In our analysis, we report that three individual SNPs were associated with preterm birth in Caucasian individuals, but when further analyzed by hierarchical regression, only one, namely, SLC23A2-08, was strongly associated. Hierarchical analysis was performed to address the issues related to small cell numbers for select analyses and also to account for linkage disequilibrium and the inherent associations that arise between neighboring SNPs. Individuals heterozygous for the T allele of SLC23A2-08 showed some elevation in risk; individuals homozygous for the T allele were observed to have a nearly threefold elevated risk of preterm delivery. We consider this finding robust, as it was apparent in both conventional logistic regression analysis and semi-Bayesian hierarchical regression. In Caucasian subjects, the results suggest that there could be a gene-dosage effect; namely, the effect in homozygous subjects exceeds that observed in heterozygous subjects. The analysis in the African-American population is more complex, mainly because of the more complex pattern of linkage disequilibrium across the SNPs surveyed. These findings are consistent with the observations of surveys of genes comparing populations (40–42). Because of incomplete analysis of the linkage disequilibrium pattern across SLC23A2, our preliminary analysis has been restricted to high-frequency SNPs distributed across the gene. Further analysis of SLC23A2 in African Americans will most likely require additional haplotype-tagging SNPs in a second study.
The strongest association between preterm birth and SLC23A2 was observed for an intron 2 SNP (known as SLC23A2-08, rs6139591). This SNP lies in the 5′ end of this large gene and in a region with flanking conserved sequence between mouse and human, suggestive of possible functional consequences. At this time, the functional importance of this SNP is not known, but since it lies in an early intron, in this case the second one, it is plausible that it resides in a region responsible for regulation of the gene. For example, variants in intron 1 or intron 2 have been shown to alter expression of genes (43). Follow-up studies are needed to fully characterize the detailed haplotype structure, as well as the functional consequences of the variant. It is possible that this common SNP is in linkage disequilibrium with one or more causal SNPs in this gene. Nonetheless, it will be important to investigate the biologic basis of our observation, namely, the effect of genetic variation in the ubiquitous vitamin C transporter, SLC23A2.
It is notable that this is the first study of the vitamin C transporters, SLC23A1, and SLC23A2 in preterm birth outcomes and, thus, several findings of elevated risk, which were imprecise and possibly due to chance, should nonetheless be considered for future follow-up studies. We observed that, in the SLC23A2-09 SNP, there is an effect for heterozygous individuals. However, analysis of the homozygous carriers was not informative, probably because of small numbers. For SLC23A2-03, there was a decreased risk for preterm delivery for the homozygous T individuals in the Caucasian population.
Recently, Siega-Riz et al. (10) reported an association between dietary intake of vitamin C and premature rupture of membranes within the Pregnancy, Infection, and Nutrition Study cohort. In light of this study, our findings provide new data to link vitamin C transport mechanisms, albeit indirectly, to preterm birth. Our findings provide further evidence in support of the role of vitamin C in pregnancy. It is theoretically possible, though unlikely, that an additional component in fruits and vegetables, not vitamin C, could be related to the observed effect of the SLC23A2 transporter. Also, we did not measure vitamin C directly in study subjects. In follow-up studies, measurement of vitamin C should be considered, particularly in an effort to correlate levels with common genetic variants. Furthermore, DNA samples from the newborn were not collected and available for genotyping in the Pregnancy, Infection, and Nutrition Study. An analysis relating newborn genotypes to preterm birth would clearly be of interest. However, on the basis of the proposed mechanism of vitamin C in preterm birth (membrane tensile strength), we believe that investigation should address the role of maternal genotypes in premature outcomes. It is notable that there are possible associations with both transporters, which differ in their expression and perhaps contribution to vitamin C transport. In Caucasian subjects only, we observed a common haplotype, which resulted in a decrease in risk for preterm delivery. Though this was not seen in African-American subjects, our results are robust; the likelihood of a false positive finding is small because of the size of our study (44). We also observed an increased risk for common SNPs in SLC23A2, in particular, a high-minor-allel-frequency SNP in intron 2, which is ubiquitous in its expression pattern and, perhaps, regulates transport to all cells.
The exploratory analysis of estimated dietary vitamin C intake and transporter genotypes yielded imprecise results, possibly because of small numbers, and no obvious pattern was observed. In conclusion, our study represents the first step in investigating the contribution of common genetic variation in the sodium-dependent vitamin C transporters to pregnancy-related outcomes. This study establishes a foundation for investigating the basis for the intersection between genetic risk and vitamin C intake.
Conflict of interest: none declared.
References
Martin JA, Hamilton BE, Sutton PD, et al. Births: final data for 2002.
Wideman GL, Baird GH, Bolding OT. Ascorbic acid deficiency and premature rupture of fetal membranes.
Levine M, Rumsey SC, Daruwala R, et al. Criteria and recommendations for vitamin C intake.
Guajardo L, Beharry KD, Modanlou HD, et al. Ascorbic acid concentrations in umbilical cord veins and arteries of preterm and term newborns.
Odendaal HJ, Popov I, Schoeman J, et al. Preterm labour—is bacterial vaginosis involved?
Steyn PS, Odendaal HJ, Schoeman J, et al. A randomised, double-blind placebo-controlled trial of ascorbic acid supplementation for the prevention of preterm labour.
Barrett BM, Sowell A, Gunter E, et al. Potential role of ascorbic acid and beta-carotene in the prevention of preterm rupture of fetal membranes.
Casanueva E, Magana L, Pfeffer F, et al. Incidence of premature rupture of membranes in pregnant women with low leukocyte levels of vitamin C.
Hadley CB, Main DM, Gabbe SG. Risk factors for preterm premature rupture of the fetal membranes.
Siega-Riz AM, Promislow JH, Savitz DA, et al. Vitamin C intake and the risk of preterm delivery.
Wang H, Dutta B, Huang W, et al. Human Na(+)-dependent vitamin C transporter 1 (hSVCT1): primary structure, functional characteristics and evidence for a non-functional splice variant.
Stratakis CA, Taymans SE, Daruwala R, et al. Mapping of the human genes (SLC23A2 and SLC23A1) coding for vitamin C transporters 1 and 2 (SVCT1 and SVCT2) to 5q23 and 20p12, respectively. (Electronic letter).
Tsukaguchi H, Tokui T, Mackenzie B, et al. A family of mammalian Na+-dependent L-ascorbic acid transporters.
Wang Y, Russo TA, Kwon O, et al. Ascorbate recycling in human neutrophils: induction by bacteria.
Washko PW, Wang Y, Levine M. Ascorbic acid recycling in human neutrophils.
Rumsey SC, Kwon O, Xu GW, et al. Glucose transporter isoforms GLUT1 and GLUT3 transport dehydroascorbic acid.
Rumsey SC, Welch RW, Garraffo HM, et al. Specificity of ascorbate analogs for ascorbate transport. Synthesis and detection of [125I]6-deoxy-6-iodo-L-ascorbic acid and characterization of its ascorbate-specific transport properties.
Rumsey SC, Daruwala R, Al-Hasani H, et al. Dehydroascorbic acid transport by GLUT4 in Xenopus oocytes and isolated rat adipocytes.
Vera JC, Rivas CI, Fischbarg J, et al. Mammalian facilitative hexose transporters mediate the transport of dehydroascorbic acid.
Corpe CP, Lee JH, Kwon O, et al. 6-Bromo-6-deoxy-L-ascorbic acid: an ascorbate analog specific for Na+-dependent vitamin C transporter but not glucose transporter pathways.
Dhariwal KR, Hartzell WO, Levine M. Ascorbic acid and dehydroascorbic acid measurements in human plasma and serum.
Sotiriou S, Gispert S, Cheng J, et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival.
Eck P, Erichsen HC, Taylor JG, et al. Comparison of the genomic structure and variation in the two human sodium-dependent vitamin C transporters, SLC23A1 and SLC23A2.
Savitz DA, Dole N, Williams J, et al. Determinants of participation in an epidemiological study of preterm delivery.
Savitz DA, Terry JW Jr, Dole N, et al. Comparison of pregnancy dating by last menstrual period, ultrasound scanning, and their combination.
Packer BR, Yeager M, Staats B, et al. SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes.
Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy-Weinberg equilibrium.
Salanti G, Amountza G, Ntzani EE, et al. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power.
Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data.
Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data.
Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps.
Greenland S. Multilevel modeling and model averaging.
Greenland S, Poole C. Empirical-Bayes and semi-Bayes approaches to occupational and environmental hazard surveillance.
Witte JS, Greenland S, Kim LL, et al. Multilevel modeling in epidemiology with GLIMMIX.
Witte JS, Greenland S, Haile RW, et al. Hierarchical regression analysis applied to a study of multiple dietary exposures and breast cancer.
Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome.
Reich DE, Cargill M, Bolk S, et al. Linkage disequilibrium in the human genome.
Bonnen PE, Wang PJ, Kimmel M, et al. Haplotype and linkage disequilibrium architecture for human cancer-associated genes.
Carlson CS, Eberle MA, Rieder MJ, et al. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans.
Kellis M, Patterson N, Endrizzi M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements.