Abstract
Background
Pregnant women are at greater risk of adverse outcomes, including mortality, as well as obstetrical complications resulting from COVID-19. However, pregnancy-specific changes that underlie such worsened outcomes remain unclear.
Methods
Plasma samples were collected from pregnant women and non-pregnant individuals (male and female) with (n = 72 pregnant, 52 non-pregnant) and without (n = 29 pregnant, 41 non-pregnant) COVID-19. COVID-19 patients were grouped as asymptomatic, mild, moderate, severe, or critically ill according to NIH classifications. Proteomic profiling of 7,288 analytes corresponding to 6,596 unique protein targets was performed using the SOMAmer platform.
Results
Herein, we profile the plasma proteome of pregnant and non-pregnant COVID-19 patients and controls and show alterations that display a dose-response relationship with disease severity; yet, such proteomic perturbations are dampened during pregnancy. In both pregnant and non-pregnant state, the proteome response induced by COVID-19 shows enrichment of mediators implicated in cytokine storm, endothelial dysfunction, and angiogenesis. Shared and pregnancy-specific proteomic changes are identified: pregnant women display a tailored response that may protect the conceptus from heightened inflammation, while non-pregnant individuals display a stronger response to repel infection. Furthermore, the plasma proteome can accurately identify COVID-19 patients, even when asymptomatic or with mild symptoms.
Conclusion
This study represents the most comprehensive characterization of the plasma proteome of pregnant and non-pregnant COVID-19 patients. Our findings emphasize the distinct immune modulation between the non-pregnant and pregnant states, providing insight into the pathogenesis of COVID-19 as well as a potential explanation for the more severe outcomes observed in pregnant women.
Plain language summary
Pregnant COVID-19 patients are at increased risk of experiencing complications and severe outcomes compared to the general population. However, the reasons for this heightened risk are still unclear. We measured the proteins present in the blood of pregnant and non-pregnant patients with COVID-19 and compared these to healthy individuals. We found that some COVID-19-associated proteins were present at lower levels in pregnant women, which could help to protect the fetus from harmful inflammation, the body’s natural response to infection. While some proteins affected by COVID-19 are shared between pregnant and non-pregnant patients, others were distinctly affected only in pregnant women, providing a potential explanation for the more severe outcomes in this group.
Similar content being viewed by others
Introduction
Coronavirus disease 2019 (COVID-19) represents an ongoing threat to people around the world1,2. As of October 2022, over 600 million people have been infected with SARS-CoV-2, the virus responsible for COVID-19, and the death toll has neared 6.5 million1. A growing body of evidence has indicated that pregnant women are at an increased risk of adverse outcomes resulting from COVID-19, ranging from greater rates of admission to the intensive care unit and need for mechanical ventilation to higher risk of death compared to non-pregnant women3,4,5. Moreover, pregnant women with COVID-19 have also been shown to experience more obstetrical complications such as preeclampsia6,7, preterm birth6,7, and stillbirth8. Thus, COVID-19 during pregnancy not only adversely affects the mother, but can also negatively affect short- and long-term quality of life for the offspring by causing respiratory distress syndrome, increasing neonatal intensive care unit admission, and impairing cognitive development, among others9,10,11,12,13,14,15,16,17. Hence, there is an urgent need to understand the pregnancy-driven biological pathways, including immune responses, underlying the increased susceptibility to severe COVID-19 and obstetrical disease.
Upon the onset of the COVID-19 pandemic, multiple investigations have sought to uncover the effects of SARS-CoV-2 infection on maternal, fetal/placental, and neonatal physiology, including the immune response18,19,20,21,22,23,24,25,26,27,28,29,30,31. Indeed, we and others have characterized the changes in systemic parameters such as cellular immune responses, virus-specific immunoglobulins, and inflammatory mediators in the maternal peripheral blood and/or cord blood to generate a profile of the maternal-fetal immune responses against SARS-CoV-2 infection25,32,33,34,35. In particular, comparative studies of pregnant and non-pregnant COVID-19 patients showed that pregnant women were more likely to experience a cytokine storm36 characterized by specific mediators37,38,39, together with elevated neutrophil counts37,38 and lymphopenia39, providing insight into the potential mechanisms underlying the increased susceptibility to severe COVID-19 during pregnancy. The recent utilization of longitudinal and multi-omics approaches has allowed for the identification of specific processes contributing to COVID-19 progression and severity in the general population40,41,42,43. Thus, to further understand the consequences of COVID-19 in pregnant women, the application of high-throughput omics platforms can facilitate the identification of relevant molecules and biological pathways implicated in this disease. Indeed, a recent study profiled over 1400 proteins in maternal peripheral blood and cord blood and indicated that pregnant women with severe COVID-19 display increased inflammatory and anti-viral signaling compared to asymptomatic pregnant women, while their offspring displayed elevated cytokines associated with T-cell responses and/or inflammasome activation44. However, the proteomic dysregulation that distinguishes pregnant from non-pregnant COVID-19 patients has not been elucidated.
Aptamer-based technologies that allow the identification and monitoring of over 1000 potential target proteins have been utilized to profile the human proteome during normal pregnancy and/or its complications in the maternal plasma45,46,47,48,49 and amniotic fluid50. Yet, the much-expanded version (4.1) of the SOMAScan platform, which allows measuring of over 7000 analytes, had not been utilized to study pathology in obstetrics.
In this study, we classify pregnant and non-pregnant women according to COVID-19 status and severity and perform proteomic profiling using the high-throughput SOMAScan platform to determine the differentially affected proteins. We show that COVID-19 drives changes in the plasma proteome in a dose-response relation with disease severity. Importantly, the response to COVID-19 is dampened during pregnancy, regardless of disease severity. Distinct and overlapping proteomic changes are identified in pregnant and non-pregnant COVID-19 patients: pregnant women display a tailored proteomic response, potentially to protect the conceptus from inflammation, while non-pregnant women display a stronger response that may help fight off infection. Moreover, the stereotypical proteomic response induced by COVID-19 in the pregnant and non-pregnant states shows enrichment of mediators implicated in a cytokine storm, endothelial dysfunction, and angiogenesis; yet, such a response is dampened during pregnancy. Finally, we apply machine learning to demonstrate that the plasma proteome can be used to discriminate COVID-19 patients from controls, even those who are asymptomatic or have mild symptoms.
Methods
Study design
The study involved profiling 7288 proteomic targets in plasma samples collected from pregnant women (n = 101) and from non-pregnant individuals (n = 93, including both men and women) of Hispanic ethnicity (Colombia). Hispanic refers to a person of South or Central American, or other Spanish culture or origin, regardless of race, as defined by the NIH (https://grants.nih.gov/grants/guide/notice-files/not-od-15-089.html). Pregnant patients were enrolled at admission to the labor and delivery unit or at the time of attending the clinic due to obstetrical indications or clinical deterioration warranting inpatient management. While the term “pregnant women” is used to describe this cohort, we note that we did not ask whether any of these patients identified as transgender or nonbinary. Variables considered to be indicative of clinical deterioration were the following: SOFA score at patient admission; presence of dyspnea; vital signs: heart rate >90, respiratory rate >20, median blood pressure; altered mental status; ICU admission; white blood cell count; D-dimer value; troponin values; OIT requirement; admission to in care diagnosis; admission to obstetric in care diagnosis; Glasgow coma scale; abnormal diagnostic images; abnormal electrocardiogram findings and echocardiogram; platelet values; hemoglobin values; serum lactate values; arterial blood gas values; serum electrolyte values; procalcitonin values; positive blood cultures; and clotting times. Moreover, for maternal clinical deterioration, we considered the following maternal (ICU admission; mechanical ventilation; non-severe preeclampsia; severe preeclampsia; gestational hypertension; eclampsia; pulmonary embolism; abruption; prelabor rupture of membranes; preterm labor; postpartum hemorrhage; maternal mortality; antenatal corticosteroid therapy) and perinatal (abnormal antepartum fetal monitoring; fetal growth restriction; acute respiratory distress syndrome; neonatal sepsis; IVH; prematurity; congenital malformation; NICU admission; perinatal mortality) outcomes.
All patients were screened for COVID-19 according to standard clinical care. Patients with suspected COVID-19 were triaged and admitted to the respiratory ward, where they underwent PCR testing to confirm the diagnosis of COVID-19. Depending on disease severity, patients were transported to the pulmonary ICU for treatment as well as clinical/paraclinical management. Of all controls (patients without COVID-19) enrolled in the study, we retained those who had samples collected within the same gestational age window as cases. Non-pregnant patients were enrolled at the time of admission for any medical indication, and all were tested for COVID-19 as described above. For non-pregnant women with COVID-19, diagnoses at admission were recorded as viral pneumonia, ARDS, septic shock, or other. For non-pregnant women without COVID-19, diagnoses at admission were recorded as coronary disease, kidney failure, or other.
All analyses accounted for the age and sex of patients as well as the presence of chronic hypertension or high-risk pathology. All patients provided written informed consent, and the use of biological specimens and clinical data for research purposes was approved by the Biomedical Research Ethics Committee of the Fundacion Valle del Lili (Protocol No. 1611), Cali, Colombia. Patients diagnosed with COVID-19 were grouped as asymptomatic, mild, moderate, severe, or critically ill according to NIH classification51. Blood samples were collected in tubes containing ethylenediaminetetraacetic acid (EDTA), and plasma was separated by centrifugation (1600 × g for 10 min at 4 °C). Plasma samples were immediately stored at −80 °C until proteomic analysis.
Plasma proteomics
Maternal plasma protein abundance was determined using the SOMAmer (Slow Off-rate Modified Aptamers) platform, which enables the multiplexed quantification of 7288 analytes corresponding to 6596 unique protein targets52,53,54. We present these results at the level of analytes, also referred to as proteins. Experiments were run in randomized batches by blinded laboratory personnel. Briefly, plasma samples were diluted and then incubated with the respective SOMAmer mixes pre-immobilized onto streptavidin-coated beads, which were then washed to remove unbound proteins and other matrix constituents. Bound proteins were tagged using an NHS-biotin reagent. After labeling, the beads were exposed to an anionic competitor solution that prevents non-specific interactions from reforming after disruption. Pure cognate-SOMAmer complexes and unbound SOMAmer reagents were released from the streptavidin beads using ultraviolet light. The photo-cleavage eluate was separated from the beads and then incubated with a second streptavidin-coated bead that binds the biotin-labeled proteins and the biotin-labeled protein-SOMAmer complexes. Subsequently, free SOMAmer reagents were removed by washing. Finally, protein-bound SOMAmer reagents were released from their cognate proteins using denaturing conditions. SOMAmer reagents were quantified by hybridization to custom DNA microarrays to detect the cyanine-3 signal52,53,54. Proteomics profiling was performed by Somalogic, Inc. (Boulder, CO, USA).
Statistics and reproducibility
Demographic and clinical characteristics
These data were summarized using numbers and percentages for categorical variables or medians and interquartile range (IQR) for continuous variables. Differences between cases and controls were assessed using Fisher’s exact test for categorical data and the Wilcoxon test for continuous data. All statistical tests were two-tailed and significance was inferred based on p < 0.05.
Principal component analysis
Protein abundances expressed as relative fluorescence units (RFU) were log2-transformed to improve normality. The function prcomp in the R statistical language and environment (www.r-project.org) was used to calculate principal components (PC). The top three PC were tested for associations with COVID-19 and pregnancy status using linear models with interaction terms. The dose-response relationship between a given PC and disease severity was assessed using a linear model in which the response variable was the PC and the explanatory variable was an ordered factor with six levels ordered in the sequence: Control, Asymptomatic, Mild, Moderate, Severe, and Critical. This analysis included also pregnancy status, age, and sex of participants as possible confounding variables. All statistical tests were two-tailed and significance was inferred based on p < 0.05. The top 2% of proteins with the largest absolute loading values on each of the first three principal components were interpreted via biological process enrichment analysis as described below in Gene ontology enrichment analysis.
Differential abundance analysis
The proteomic data preprocessing, including an adaptive normalization by maximum likelihood (ANML) step and a calibration step, were performed by SomaLogic, Inc. The goal of these steps was to make data comparable across samples by calculating plate-specific and analyte-specific scale factors. After log (base 2) transformation, data were compared between pooled COVID-19 cases and controls or compared separately between each disease severity group against controls. When analyzing data from pregnant women, maternal age, body mass index (BMI), and linear and quadratic terms of gestational age were included as co-variates. Analysis of data from non-pregnant subjects included adjustment for age, BMI, and sex of the participant. Models were fit using the limma package55,56 in R. Protein abundance was considered to have changed significantly with COVID-19 if the fold change was >1.25 and false discovery rate (FDR)57 adjusted p value (q value) was <0.1. Spearman correlation coefficients and significance p-values were calculated to determine the similarity of log2 fold changes in protein abundance obtained for different COVID-19 severity groups against controls, both within and between pregnant and non-pregnant subjects. Proteins with opposite dysregulation due to COVID-19 between pregnant and non-pregnant groups were defined as proteins being either a) significantly changed with COVID-19 in pregnant women (q < 0.1, fold change >1.25) but with opposite direction of change in non-pregnant individuals (p < 0.05), or b) significantly changed with COVID-19 in non-pregnant individuals (q < 0.1, fold change >1.25) but with opposite direction of change in pregnant women (p < 0.05).
Gene ontology enrichment analysis
Proteins were mapped using the Entrez gene database58 identifiers based on SomaLogic, Inc. protein annotation followed by Gene Ontology59. Biological processes over-represented among a given protein set were identified using Fisher’s exact tests. Gene ontology terms with three or more hits and an adjusted enrichment q value <0.1 were considered as significantly enriched. The MSigDB collection60 of curated canonical pathways (C2 collection) was also analyzed. Enrichment tests were performed using the GOStats package61 in Bioconductor enrichment analyses. Biological processes over-represented among a given protein set were identified using Fisher’s exact tests. Gene ontology terms with three or more hits and an adjusted enrichment q value <0.1 were considered as significantly enriched. The MSigDB collection60 of curated canonical pathways (C2 collection) was also analyzed. Enrichment tests were performed using the GOStats package61 in Bioconductor62.
Predictive model development
To assess the value of plasma proteomic data to discriminate between COVID-19 and controls, we have developed random forest models using up to 50 proteins. The proteins were selected based on their importance to the accuracy of the models using the randomForest function in R. The protein selection and random forest model fitting steps were evaluated using leave-one-out cross-validation (LOOCV), and receiver operating characteristic curves were derived using the pROC package in R.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Characteristics of the study population
Pregnant individuals
Plasma samples were collected from 101 pregnant women (23.2–39.3 weeks of gestation), including those diagnosed with COVID-19 at the time of admission (n = 72) and those who tested negative for SARS-CoV-2 during prenatal care visits (hereafter referred to as pregnant controls; n = 29) (Fig. 1a, b and Table 1). Parameters such as maternal age, BMI, parity, presence of labor at the time of sampling, frequency of chronic hypertension, and diagnosis of preeclampsia in the current pregnancy were comparable between the pregnant COVID-19 and control groups (Table 1). Gestational age at delivery was similar between groups; yet, sampling of COVID-19 cases was performed ~5 weeks earlier in gestation than in controls [median weeks (IQR) controls: 36.1 (32.6–37.5) vs. COVID-19: 31.3 (28.1–35.6), p < 0.05] (Table 1), which was considered in the data analysis. Among the pregnant COVID-19 cases, 6 (8%) were asymptomatic, 20 (28%) were mild, 13 (18%) were moderate, 12 (17%) were severe, and 21 (29%) were critically ill according to NIH classification51.
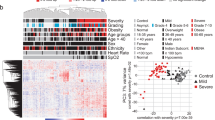
a Illustration of the study design showing the number of non-pregnant controls (n = 41; 22 male, 19 female), non-pregnant COVID-19 cases (n = 52; 22 male, 30 female) pregnant controls (n = 29), and pregnant COVID-19 cases (n = 72) from whom peripheral plasma samples were profiled. b Gestational age at sampling (gray circle) and at delivery (green triangle) for each pregnant control (upper panel) and case (lower panel). c Principal component (PC) plot of the plasma proteome of all study samples according to PC1 and PC2. Black = control, red = case. Circle = non-pregnant, triangle = pregnant. Increasing shape size corresponds to increasing COVID-19 severity. d PC plot representing the relationship between the plasma proteome of all study samples according to PC1 and PC3. e Violin plot representing the relationship between PC3 and COVID-19 severity among all study samples.
Non-pregnant individuals
Plasma samples were also collected from 93 non-pregnant individuals, which included 52 COVID-19 cases and 41 controls (Fig. 1a and Table 1). Among the non-pregnant COVID-19 cases, 1 (2%) was mild, 4 (8%) were moderate, 12 (23%) were severe, and 35 (67%) were critically ill.
COVID-19 drives shared and unique changes in the plasma proteome in pregnant and non-pregnant individuals that follow a dose response with disease severity
Over 7000 protein analytes were determined using the SOMAScan v4.1 platform in cases and controls to characterize the plasma proteome responses induced by COVID-19 according to its severity in pregnant and non-pregnant individuals (Fig. 1a). Uniform Manifold Approximation and Projection (UMAP) plots of the proteomic profiles illustrate that patients are clustered according to COVID-19 status and severity in both pregnant and non-pregnant (Supplementary Fig. 1a, b) individuals. It is worth mentioning that, in non-pregnant individuals, the plasma proteome was heavily modulated by COVID-19 status, regardless of sex (Supplementary Fig. 2). We performed an unsupervised projection of high-dimensional proteomic profiles of all controls and COVID-19 patients onto the first three principal components (PC), which can be understood as meta-proteins that are uncorrelated with each other (Fig. 1c, d). Notably, pregnancy status represented a source of variability in the proteome, as PC2 values (18% of variance explained) perfectly discriminated between pregnant and non-pregnant individuals (p < 0.001, Fig. 1c). The proteins with the top 2% highest loadings in PC2 were enriched for biological processes such as pregnancy, reproductive process, and gonadotropin secretion (q < 0.1, odds ratios >3 for all). Yet, the host response to COVID-19 represented the primary source of variability in the proteome, as PC1 and PC3 (PC1, 27% of variance explained; PC3, 6% of variance explained) were significantly different between COVID-19 cases and controls, regardless of pregnancy status (p < 0.001 for both, Fig. 1d). Proteins with the highest contribution to PC1 and PC3 were enriched for biological processes such as exocytic process (PC1), anti-viral innate immune response, antimicrobial humoral response, and positive regulation of type I interferon production (PC3) (q < 0.1, odds ratios >4 for all). The proteomic changes with COVID-19 were larger for non-pregnant than for pregnant women based on both PC1 and PC3 (interaction p < 0.005) (Fig. 1d), which is partly explained by the greater proportions of severe and critically ill cases in the non-pregnant than in the pregnant population. Moreover, we observed a dose-response relationship between PC3 and disease severity, regardless of pregnancy status (p < 0.001 for both linear and quadratic trends, Fig. 1e). Together, these data provide an overview of the plasma proteome in pregnant and non-pregnant individuals infected with SARS-CoV-2, and suggest dramatic changes with infection in a dose-response relationship with disease severity. In addition, these data hint that the host response to SARS-CoV-2 includes shared and unique processes between pregnant and non-pregnant individuals, which we further explore below.
The plasma proteome response to COVID-19 follows a dose-response relationship with disease severity in pregnant and non-pregnant individuals, yet such a response is dampened in pregnancy
Pregnant women have been reported to display heightened susceptibility to severe COVID-193,4,5. Therefore, we first explored the differential effects of COVID-19 on the maternal proteome compared to gestational age-matched control pregnancies according to disease severity (Fig. 2a). When comparing pregnant COVID-19 cases to controls after adjustment for maternal age, BMI, and gestational age at sampling, we identified 68, 81, 242, 144, and 1072 differentially abundant proteins in asymptomatic, mild, moderate, severe, and critically ill cases, respectively (Fig. 2b–f). Given that both disease severity and sample size may affect the number of differentially abundant proteins in specific groups, we next used the protein changes between critically ill patients and controls (1072 proteins) as a reference to compare with the changes observed in the less severe COVID-19 groups (Fig. 2g). The log2-transformed fold change of protein abundance between COVID-19 subgroups (i.e., asymptomatic, mild, moderate, and severe) and controls were more attenuated than those between critically ill patients and controls (regression slopes <1.0) (Fig. 2g). Yet, the magnitude of correlation and the correlation slope followed a dose-response relationship with disease severity, and even asymptomatic patients showed plasma proteomic changes that were significantly correlated to those observed in critically ill patients (r = 0.34 for Asymptomatic vs. Control; r = 0.72 for Mild vs. Control; r = 0.87 for Moderate vs. Control; r = 0.88 for Severe vs. Control; p < 0.001 for all) (Fig. 2g).
a Graphical representation showing the comparison of plasma proteomes between each classified subset of pregnant COVID-19 cases and controls. b Volcano plot showing the proteins modulated in asymptomatic COVID-19 cases compared to controls. Red = proteins with q < 0.1 and fold change > 1.25, green = proteins with q ≥ 0.1 and fold change >1.25, gray = proteins with q ≥ 0.1 and fold change ≤1.25, blue = proteins with q < 0.1 and fold change ≤1.25. c Volcano plot showing the proteins modulated in mild COVID-19 cases compared to controls. d Volcano plot showing the proteins modulated in moderate COVID-19 cases compared to controls. e Volcano plot showing the proteins modulated in severe COVID-19 cases compared to controls. f Volcano plot showing the proteins modulated in critical COVID-19 cases compared to controls. g Comparison of the magnitude of proteomic changes among pregnant COVID-19 case subsets, using the comparison between critical cases vs. controls as the reference. Spearman’s correlation and p-value are provided for the asymptomatic vs. control, mild vs. control, moderate vs. control, and severe vs. control contrasts compared to the reference. The proteins included in this analysis (gray dots) are those 1,072 identified as differentially abundant in the comparison between pregnant critically ill cases vs. controls.
We then performed the same analysis of differential protein abundance in non-pregnant patients (Fig. 3a), and identified 21, 1961, and 2966 differentially abundant proteins in moderate, severe, and critically ill cases, respectively (Fig. 3b–d), after adjusting for relevant covariates. Similar to the analysis in pregnant women, the log2-transformed fold changes of protein abundance between COVID-19 subgroups and controls were more attenuated than those found between critically ill cases and controls, and followed a dose-response with disease severity (r = 0.84 for Moderate vs. Controls; r = 0.94 for Severe vs. Controls; p < 0.001 for both) (Fig. 3e).
a Graphical representation showing the comparison of plasma proteomes between each classified subset of non-pregnant COVID-19 cases and controls. b Volcano plot showing the proteins modulated in moderate COVID-19 cases compared to controls. Red = proteins with q < 0.1 and fold change >1.25, green = proteins with q ≥ 0.1 and fold change >1.25, gray = proteins with q ≥ 0.1 and fold change ≤1.25, blue = proteins with q < 0.1 and fold change ≤1.25. c Volcano plot showing the proteins modulated in severe COVID-19 cases compared to controls. d Volcano plot showing the proteins modulated in critical COVID-19 cases compared to controls. e Comparison of the magnitude of proteomic changes among non-pregnant COVID-19 case subsets, using the comparison between critical cases vs. controls as the reference. Spearman’s correlation and p-value are provided for the moderate vs. control and severe vs. control contrasts compared to the reference. The proteins included in this analysis (gray dots) are those 2966 identified as differentially abundant in the comparison between non-pregnant critically ill cases vs. controls.
To contrast the magnitude of COVID-19-driven changes in the proteome between pregnant and non-pregnant patients, we then performed correlation analysis based on a core set of 486 proteins with significant and consistent changes in both pregnant and non-pregnant patients (Fig. 4a and Supplementary Data 1). These proteins with shared perturbation in pregnant and non-pregnant women with COVID-19 can be grouped into four main clusters enriched for distinct biological processes (Fig. 4b and Supplementary Data 2). Clusters 1 and 2 were predominantly increased in COVID-19 cases, regardless of sex or pregnancy status, while Clusters 3 and 4 were primarily decreased (Fig. 4b and Supplementary Data 2). By comparing the magnitude of changes between the pregnant and non-pregnant groups, we showed that the magnitude of changes for this set of core proteins was diminished during pregnancy for the same disease severity group, as indicated by the regression line slopes below 1.0 (Fig. 4c–e, p < 0.05 for all).
a Graphical representation showing the comparison of 486 plasma proteins that are modulated in both pregnant COVID-19 cases vs. controls and in non-pregnant COVID-19 cases vs. controls. b Heatmap representation of the 486 proteins with shared dysregulation between pregnant and non-pregnant COVID-19 patients. Clusters 1 and 2 include proteins with increased abundance while clusters 1 and 3 include proteins with decreased abundance in cases compared to controls. c Correlation between the magnitude of proteomic changes in pregnant moderate cases vs. controls and that in non-pregnant moderate cases vs. controls. Slope of the regression line (red line), Spearman’s correlation, and p-value are provided. Dotted blue line represents the parity line. d Correlation between the magnitude of proteomic changes in pregnant severe cases vs. controls and that in non-pregnant severe cases vs. controls. e Correlation between the magnitude of proteomic changes in pregnant critical cases vs. controls and that in non-pregnant critical cases vs. controls.
Together, these results demonstrate that there is a perturbation of the plasma proteome in both pregnant and non-pregnant women with COVID-19, and that the magnitude of such changes increases with COVID-19 severity. However, relative to the plasma proteome perturbations observed in non-pregnant individuals, the magnitude of changes with COVID-19 in the pregnant state is attenuated, suggesting a dampened response.
Shared and distinct changes in the plasma proteome of pregnant and non-pregnant women with COVID-19
We then sought to further unravel pregnancy-driven differences in the plasma proteomic response to COVID-19 as well as changes that are shared between pregnant and non-pregnant states. First, we identified all proteins that were differentially abundant with COVID-19, which resulted in 708 differentially abundant proteins for pregnant women (Fig. 5a and Supplementary Data 3). Labor status and maternal deterioration did not confound the COVID-19-related differences observed in the proteome, as demonstrated by sensitivity analysis (Supplementary Data 3). Similarly, we identified 2605 significant proteins with COVID-19 for non-pregnant individuals (Fig. 5b and Supplementary Data 1). From these two lists, we could identify the abovementioned 486 proteins that were significantly affected by COVID-19 in both pregnant and non-pregnant groups and had a similar direction of change (Supplementary Data 1).
a Volcano plot showing the proteins modulated in all pregnant COVID-19 cases compared to controls. Red = proteins with q < 0.1 and fold change >1.25, green = proteins with q ≥ 0.1 and fold change >1.25, gray = proteins with q ≥ 0.1 and fold change ≤1.25, blue = proteins with q < 0.1 and fold change ≤1.25. b Volcano plot showing the proteins modulated in all non-pregnant COVID-19 cases compared to controls. c Venn diagram showing the overlap of biological processes enriched among proteins modulated by COVID-19 between pregnant and non-pregnant cases compared to controls. d Bar plot showing the odds ratios for top biological processes enriched among proteins modulated by COVID-19 in pregnant cases compared to controls. Asterisk indicates the odds ratio calculated as infinite. e Bar plot showing the odds ratios for top biological processes enriched among proteins modulated by COVID-19 in non-pregnant cases compared to controls. f Bar plot showing the odds ratios for top biological processes enriched among proteins modulated by COVID-19 in both pregnant and non-pregnant cases compared to controls. g Venn diagram showing the overlap of C2 pathways enriched among proteins modulated by COVID-19 in pregnant and non-pregnant cases compared to controls. h Bar plot showing the odds ratios for top C2 pathways enriched among proteins modulated by COVID-19 in pregnant cases compared to controls. i Bar plot showing the odds ratios for top C2 pathways enriched among proteins modulated by COVID-19 in non-pregnant cases compared to controls. j Bar plot showing the odds ratios for top C2 pathways enriched among proteins modulated by COVID-19 in both pregnant and non-pregnant cases compared to controls.
Next, we explored the biological processes that were enriched among the entire set of differentially abundant proteins for pregnant (708 proteins) and non-pregnant (2605 proteins) COVID-19 patients to characterize the differences in host response (Fig. 5c–f). As expected, enriched biological processes in pregnant women with COVID-19 were fewer than those in non-pregnant patients, given the dampened protein response (Fig. 5c and Supplementary Data 4–6). Consistent with such an observation, pregnant COVID-19 patients showed enrichment of processes related to extracellular matrix, defense response, and immune response (Fig. 5d), whereas those enriched in non-pregnant individuals included protein localization and transport, peptide biosynthesis, and translation (Fig. 5e and Supplementary Data 4, 5). Shared processes were characterized by cell adhesion and immune responses as well as response to wounding and blood coagulation (Fig. 5f). We summarized the biological processes perturbed with COVID-19 in pregnant and non-pregnant patients according to disease severity (Supplementary Data 6).
In addition to biological processes, we also evaluated the enrichment of pathways and gene sets derived from the C2 collection of the MSigDB database, which includes canonical pathways and experimental gene sets such as those associated with disease and viral infection (Fig. 5g and Supplementary Data 7–9). Similar to biological processes, pathways enriched in pregnant women with COVID-19 included terms related to extracellular matrix; yet, pathways associated with viral infection or anti-viral defenses were also observed (Fig. 5h and Supplementary Data 7). Enriched pathways in non-pregnant COVID-19 patients included terms related to platelet activation, VEGF, and PDGF (Fig. 5i), while shared pathways included virus- and cancer-related terms (Fig. 5j and Supplementary Data 8). The shared C2 pathways perturbed with COVID-19 between pregnant and non-pregnant patients were also summarized while considering disease severity (Supplementary Data 9).
Together, these data further demonstrate that, although there is a set of common responses to COVID-19 in both pregnant and non-pregnant state, pregnancy-specific changes are detectable in the maternal circulation. These data put forward evidence for a working hypothesis that pregnancy tailors the immune response against pathogens29,63,64 threatening the successful completion of gestation.
COVID-19 drives distinct and shared angiogenic and inflammatory proteomic changes in pregnant and non-pregnant individuals
Given our finding that pregnancy modifies the proteomic response to COVID-19, we further investigated whether any proteins were dysregulated with COVID-19 in opposite directions between pregnant and non-pregnant patients (see Methods). This analysis identified a core set of 33 proteins with opposing direction of change (Fig. 6a) and included proteins related to angiogenesis and wound healing as well as alarmins, cytokines, and growth factors (Supplementary Data 10). Proteins that decreased with COVID-19 in pregnancy but were increased in non-pregnant cases included vascular endothelial growth factor receptor 1 (VEGF-sR1 or sFLT-1) and angiotensinogen (AGT); yet, this could potentially be explained by their already elevated baseline among pregnant patients (Fig. 6b, c and Supplementary Data 10). Consistent with these findings, proteins that underwent pregnancy-specific regulation with COVID-19 were enriched for biological processes and pathways related to vasodilation, angiogenesis, and regulation of inflammatory response (Supplementary Data 11). A previous report indicated that COVID-19 during pregnancy is characterized by a profile of proteomic factors that is distinct from but overlaps with that observed in preeclampsia6, an obstetric syndrome characterized by intravascular inflammation65. Therefore, we further evaluated changes in angiogenic or endothelial factors between pregnant and non-pregnant COVID-19 patients. Several factors such as soluble TNF receptor II (TNFRSF1B) and von Willebrand factor (VWF) were found to increase with COVID-19 regardless of pregnancy status (Fig. 6d, e). Notably, neutrophil elastase (ELANE), a neutrophil degranulation factor66 as well as a component of neutrophil extracellular traps (NETs)67, was elevated in both pregnant and non-pregnant COVID-19 cases (Fig. 6f), as was histone H3.1 (H3C1), another NET component (Fig. 6g). These results provide insight into the unique biological processes in pregnant and non-pregnant individuals: while non-pregnant individuals exhibit increased abundance of angiogenic and inflammatory proteins in the circulation, the proteome of pregnant women hints at a systemic inflammatory response and no increase in anti-angiogenic sFLT-1, which is already elevated in the pregnant state.
a Representative diagram illustrating the comparison between pregnant and non-pregnant COVID-19 cases and controls for specific proteins associated with angiogenesis, endothelial dysfunction, and intravascular inflammation. A core set of 33 proteins that are significantly modulated with COVID-19 in opposite directions between pregnant and non-pregnant patients. Note the negative slope and correlation coefficient. b–g Violin plots showing the modulation of b sFLT-1, c AGT, d TNFRSF1B, e VWF, f ELANE, and g H3C1 levels with COVID-19 severity in non-pregnant and pregnant cases and controls. Black = control, gray = asymptomatic, blue = mild, yellow = moderate, red = severe, and brown = critical. RFU = relative fluorescence units.
Pregnant women with COVID-19 display a dampened systemic cytokine response
COVID-19 is characterized by a cytokine storm, components of which can display a dose-response with disease severity41. Therefore, we next focused on the protein expression changes of specific inflammatory mediators (Fig. 7a). The classical inflammatory cytokines IL-6, IL-1β, and IL-18 were increased in COVID-19 cases compared to controls for both pregnant and non-pregnant patients; yet, the latter two did not reach significance in pregnant women (IL-1β, p = 0.074; IL-18, p = 0.052), likely due to the dampened proteomic response (Fig. 7b–d). Similarly, TNF and IL-17A were upregulated with COVID-19 in non-pregnant patients and only showed a slight tendency to increase during pregnancy (Fig. 7e, f). The alarmin IL-1α was found to be downregulated only in pregnant COVID-19 cases, although a tendency towards the same reduction was observed in non-pregnant patients (Fig. 7g). By contrast, IFNγ was reduced with COVID-19 in non-pregnant individuals but not pregnant patients (Fig. 7h). The anti-inflammatory cytokine IL-10 was downregulated in pregnant and non-pregnant COVID-19 cases (Fig. 7i), whereas TGFβ1 was upregulated in both groups (Fig. 7j). Several chemokines were also found to exhibit differential regulation with COVID-19 in the pregnant and non-pregnant states: CXCL10 and CCL22 were consistently increased or diminished, respectively, in both non-pregnant and pregnant cases; yet, CCL1 was reduced and CXCL13 was increased only in non-pregnant COVID-19 patients, although data from pregnant patients showed similar tendencies (Fig. 7k–n). These findings suggest that COVID-19 induces a cytokine storm in the circulation of both pregnant and non-pregnant individuals; yet, pregnant women display a dampened soluble immune response.
a Representative diagram illustrating the evaluation and comparison of specific cytokines in the circulation of pregnant and non-pregnant COVID-19 cases and controls. b–n Violin plots showing the modulation of b IL-6, c IL-1β, d IL-18, e TNF, f IL-17A, g IL-1α, h IFNγ, i IL-10, j TGFβ1, k CCL1, l CCL22, m CXCL13, and n CXCL10 levels with COVID-19 severity in non-pregnant and pregnant cases and controls. Black = control, gray = asymptomatic, blue = mild, yellow = moderate, red = severe, and brown = critical. RFU = relative fluorescence units.
The plasma proteome can discriminate COVID-19 cases from uninfected controls, even when mild or asymptomatic
Last, we evaluated the ability of the proteomic profiles to discriminate between COVID-19 cases and controls, regardless of pregnancy status. For this purpose, we developed random forest models that included up to 50 proteins and evaluated their accuracy via leave-one-out cross-validation (LOOCV). The resulting proteomics model was able to accurately discriminate COVID-19 cases from controls, in the absence of any other inputs (Fig. 8a). The area under the Receiver Operating Characteristic curve (AUC) was 0.978 for the full analysis set, 0.974 for pregnant women, and 0.985 for non-pregnant individuals (Fig. 8a). The relative importance of the proteomic predictors in the random forest model is displayed in Fig. 8b and includes several of the proteins with differential abundance as reported in Supplementary Data 1 and 3. When classification models were derived separately based on disease severity, the accuracy to distinguish most severe cases (severe or critical COVID-19) from controls was higher (AUC = 0.99) than the one obtained for discriminating between controls and moderate cases (AUC = 0.94) (Fig. 8c). Of interest, similarly high accuracy was obtained also for distinguishing asymptomatic or mild cases from uninfected controls (AUC = 0.95) (Fig. 8c). ISG15, MX1, ZBP1 and IFNL1 were the top four proteins most contributing to the accuracy of random forest models for discriminating all COVID-19 cases from controls, and these proteins were also among the top ones for prediction of severe and critical COVID-19 (Fig. 8d), moderate COVID-19 (Supplementary Fig. 3), and for mild or asymptomatic cases (Supplementary Fig. 4). Together, these data suggest that a shared proteomic signature can discriminate between COVID-19 patients and healthy individuals regardless of pregnancy status, and that disease severity is a driver of classification accuracy.
a Receiver operating characteristic (ROC) curves for discrimination of all COVID-19 cases from all controls (black curve), only pregnant COVID-19 cases from pregnant controls (red curve), and only non-pregnant COVID-19 cases from non-pregnant controls (blue curve). b Bar plot displaying the relative importance of the top 50 proteomic predictors for identifying all COVID-19 cases. c ROC curves for discrimination of severe/critical cases from controls (red curve), moderate cases from controls (yellow curve), and asymptomatic/mild cases from controls (blue curve). Data from both pregnant and non-pregnant cases are included in this analysis. d Bar plot displaying the relative importance of the top 50 proteomic predictors for distinguishing severe/critical COVID-19 cases from controls.
Discussion
In this study, we utilized the SOMAScan v4.1 platform to profile over 7000 protein targets in the peripheral blood of pregnant women and non-pregnant individuals diagnosed with COVID-19, and found that this disease drives changes in their plasma proteomes in a dose-response relation with disease severity. Importantly, we showed that the response to COVID-19 is dampened during pregnancy, regardless of disease severity. Distinct and overlapping proteomic changes were identified in pregnant and non-pregnant COVID-19 patients: pregnant women displayed a tailored proteomic response, potentially to protect the conceptus from the deleterious effects of inflammation, while non-pregnant women displayed a stronger response to fight off infection. The stereotypical proteomic response induced by COVID-19 in the pregnant and non-pregnant state showed enrichment of mediators implicated in cytokine storm, endothelial dysfunction, and angiogenesis, yet was dampened during pregnancy. Last, we utilized machine learning to demonstrate that the plasma proteome can be used to discriminate COVID-19 patients from controls, even those who were asymptomatic or had mild symptoms.
The proteomic dysregulations after COVID-19 revealed in our current study are suggestive of a dampened systemic immune response in pregnant women compared to non-pregnant individuals, both in terms of the number of proteins affected and magnitude of changes for proteins implicated in the pregnant and non-pregnant states. This phenomenon could be secondary to physiological changes that occur during pregnancy, such as the reversible thymic involution68,69,70 that impacts T-cell development71,72, or could be a primary outcome intended to prevent aberrant immune activation that could threaten pregnancy73,74. Immune suppression was originally considered to be a requirement for successful pregnancy, given the immunological puzzle of the mother displaying tolerance towards the semi-allograft fetus for 40 weeks75. Rather than complete inertness or unresponsiveness, as proposed by Peter Medawar75, pregnancy has since been shown to be a state of selective immune tolerance76,77,78,79,80,81,82,83,84,85,86,87, mediated by homeostatic cells such as regulatory T cells (Tregs)76,77,78,79,80,81,84,85,87,88,89,90,91,92,93,94,95,96,97 and macrophages92,98,99,100,101,102,103,104,105,106. This concept is further supported by studies of women with autoimmune diseases such as systemic lupus erythematosus (SLE), in whom such pregnancy-specific immune adaptations can fail to occur107,108, resulting in pregnancy complications108,109. Pertinent to our current findings, it was suggested that the suppression of key immune pathways such as IFN could underlie the higher risk of severe viral infection during pregnancy108. Given the high energy demands of fighting off infection, it is possible that the diversion of maternal resources to effectively clear the virus while avoiding a potentially harmful inflammatory response may result in a transient period of fetal “neglect”, which may contribute to the observed worsened outcomes in pregnant COVID-19 patients. Indeed, past and present viral pandemics have provided a large body of evidence showing that specific viruses, such as pandemic influenza viruses, Dengue virus, and coronaviruses, can result in disproportionately high rates of adverse outcomes in pregnant women110.
Peripheral T and B cells show decreased numbers, greater activation-induced proliferation, and altered phenotypes during pregnancy64,111, and such alterations can be further exacerbated by the lymphopenia that characterizes viral infections such as SARS-CoV-235,41,42,112. Moreover, given the demonstrated relationship between pathological maternal T-cell activation and pregnancy complications such as preterm labor73,74, it is imperative that maternal adaptive immunity remain under strict control until normal parturition at term113,114,115,116. Consistently, we recently undertook an ex vivo evaluation of peripheral cellular immune responses against SARS-CoV-2 particles and proteins in pregnant and non-pregnant women29. We demonstrated a pregnancy-specific reduction of unswitched memory-like and transitional-like B-cell subsets29, which is in line with a prior study showing that such reduction of B cells is associated with COVID-19 severity117. Thus, given such deficiencies in peripheral adaptive immunity, pregnant women infected with SARS-CoV-2 may rely more heavily on monocytes, which are also potent contributors to anti-viral host defense118. Consistently, monocytes undergo substantial expansion and differentiation in patients with severe COVID-19119,120,121, and we have shown that monocytes from pregnant women appear to undergo accelerated transition and activation in response to SARS-CoV-2 exposure29, which is in line with a previous report37.
Notably, we found that the cytokine profile of peripheral leukocytes was also impacted by pregnancy, as the release of IFN-β and IL-8 in response to SARS-CoV-2 was diminished compared to non-pregnant women29. The abovementioned studies, together with our current results, point to a specific dampening of the maternal proteomic response to COVID-19 to protect the fetus from heightened inflammation that could jeopardize pregnancy. This may not be the only mechanism protecting the fetus, as the placenta has also been shown to play a critical role in anti-SARS-CoV-2 defenses35,122. The incidence of vertical transmission of SARS-CoV-2 has been shown to be rare, which may be due in part to the minimal co-expression of the canonical viral cell entry mediators ACE2 and TMPRSS2 in this organ18. Moreover, the placenta exhibits strong anti-viral properties123,124, and in women with COVID-19 the placental anti-viral response was shown to include the activation of leukocytes such as T cells, NK cells, and macrophages together with elevated expression of genes related to immune and cytokine signaling, even in the absence of detectable placental infection35,122. Thus, the diminished maternal systemic response to SARS-CoV-2 infection may be partially offset by the protective functions of the placenta, thereby preventing a cytokine storm that could damage the fetus while still ensuring a barrier to prevent viral transmission.
Herein, we found that pregnant and non-pregnant patients infected with SARS-CoV-2 exhibit a perturbed proteomic profile characterized by the enhanced release of cytokines and other mediators associated with inflammation, endothelial dysfunction, and angiogenesis. A hallmark of severe COVID-19 is the systemic inflammatory response that includes the exacerbated release of pro-inflammatory immune mediators, termed a cytokine storm125,126,127,128,129. Multiple cytokines involved in this response have been proposed as biomarkers of severity and prognosis for COVID-19130. Indeed, the peripheral blood concentration of cytokines, including IL-6, is highly correlated with mortality in patients with COVID-19130,131, hinting at a key role for IL-6 in the pathophysiology of severe disease. In fact, it has been proposed that IL-6 acts as an amplifier of the inflammatory response triggered by SARS-CoV-2 by activating the NF-κB and STAT3 pathways in non-immune cells such as the vascular endothelium132. This concept is in line with the clinical findings showing that the cytokine storm can lead to generalized endothelial dysfunction127,133, as was initially suspected early in the pandemic given the rapid emergence of cardiovascular complications in COVID-19 patients134,135. The vascular endothelium is an organ with multiple endocrine, paracrine, and autocrine functions, which are required for vascular homeostasis and regulation of vascular tone136,137. Therefore, any disruption in these functions can induce vasoconstriction that can progress to ischemia, inflammation, edema, and culminate in a pro-coagulant state138.
In addition to the indirect induction of endothelial dysfunction due to the host inflammatory response139,140, SARS-CoV-2 can also directly interact with the vascular endothelium, as evidenced by viral inclusion structures observed in vascular endothelial cells at multiple body sites in deceased COVID-19 patients140,141. SARS-CoV-2 binds to the ACE2 receptor to enter cells, which can impair the activity of the enzyme ACE2 to neutralize angiotensin vasopressors133,142. Such impaired ACE2 activity can activate the kallikrein-bradykinin pathway that results in increased vascular permeability133,143. Moreover, the activation of innate immune cells induces the release of toxic mediators such as reactive oxygen species (ROS) and vasoactive substances that can lead to inter-endothelial gaps, thereby further enhancing endothelial permeability133. The activation of endothelial cells leads to the production of multiple pro-coagulant factors, such as P-selectin, fibrinogen, and Von Willebrand factor (VWF), which initiates the coagulation cascade133,139. These processes can also lead to platelet aggregation and the release of other factors such as VEGF, which upregulates the endothelial cell production of tissue factor, the primary stimulator of the coagulation cascade133,144, ultimately leading to a pro-thrombotic state. Consistently, herein we showed that, while non-pregnant patients with COVID-19 exhibit angiogenic and inflammatory circulatory profiles, the proteome of pregnant women is characterized by a systemic inflammation without dysregulating the anti-angiogenic factor sFLT-1, which is already elevated in pregnant controls.
Initial investigations of pregnant women infected with SARS-CoV-2 had revealed the development of a preeclampsia-like syndrome145,146. Later evidence supported COVID-19 as a risk factor for preeclampsia7,147 and indicated a dose-response relationship with disease severity6; however, the mechanisms and causality of such an association are still poorly understood65,147,148. Our current findings revealed that some proteins implicated in inflammatory and angiogenic processes were perturbed in patients with COVID-19, regardless of pregnancy status; yet, specific proteins were only modified by SARS-CoV-2 infection in pregnancy. As preeclampsia is a primarily systemic endothelial-inflammatory obstetrical disease65,149,150,151,152,153, our findings support the fact that some perturbed pathways may be shared between COVID-19 and preeclampsia. A previous study compared circulating biomarkers in pregnant women with COVID-19 and those of women with preeclampsia, and demonstrated that these conditions display distinct biomarker profiles154. The partial overlap between these two disease states may be driven by the placental inflammatory response induced by maternal SARS-CoV-2 infection, even in asymptomatic pregnant women35,122. Such inflammation can affect the fetus even in the absence of vertical transmission35; therefore, it is imperative to follow and evaluate these infants for eventual adverse outcomes, as suggested by recent evidence demonstrating neurodevelopmental sequelae at one year of life in children exposed to SARS-CoV-2 in utero16,155.
The establishment of biomarkers that allow for the classification and monitoring of COVID-19 outcomes is essential to guide patient management, particularly during pregnancy. In the current study, we demonstrated that the systemic proteome can be utilized to distinguish COVID-19 patients and controls, in the absence of any other patient risk factors. Of importance, the plasma proteome was able to discriminate asymptomatic cases and those with mild symptoms from controls with high accuracy. The top proteomic contributors in our discriminatory model included interferon-induced GTP-binding protein Mx1 (MX1 or MXA), interferon-stimulated gene 15 (ISG15), Z-DNA-binding protein 1 (ZBP1), and interferon lambda 1 (IFNL1), also referred to as IL-29. Given their responsiveness to IFN signaling, ISG15 and its downstream targets, including MX1156, have been implicated as key participants during SARS-CoV-2 infection by using in vitro and in vivo models157,158,159 as well as samples from COVID-19 patients160,161,162. Similarly, the induction of IFNL1 by SARS-CoV-2 infection was previously reported in vitro163. Analysis of serum samples derived from pregnant women with COVID-19 using a different proteomic platform indicated the specific upregulation of IFNL1 and its receptor, IFNLR1, in severe/critical patients compared to mild/moderate or asymptomatic44, which conflicts with earlier findings that IFN responses are delayed and impaired in non-pregnant COVID-19 patients164. Here, we observed consistent upregulation of IFNL1 in pregnant and non-pregnant COVID-19 patients compared to controls; yet, consistent with the abovementioned study44, this interferon most strongly contributed to the discrimination of severe/critical cases from controls, suggesting it is most greatly impacted in such patients. ZBP1 is part of the innate immune response165 that is induced by IFN signaling166,167,168 and mediates PANoptosis (pyroptosis, necroptosis, and apoptosis) as part of host defense against pathogens such as influenza169. Importantly, ZBP1 has also been shown to be activated by SARS-CoV-2 infection166,170,171,172. Given its function as an inducer of PANoptosis, the IFN-mediated upregulation of ZBP1 represents an obstacle to the proposed use of IFNs as a potential therapy for COVID-19173. Further studies exploring such therapy may consider the inhibition of ZBP1 in conjunction with IFN therapy to help prevent the associated tissue damage and lethality173.
Our current findings are in line with a prior multi-omics investigation that evaluated 1400 plasma proteins together with single-cell immune features for the classification of non-pregnant COVID-19 patients174. In the latter study, such integrated modeling showed value for the distinction of mild, moderate, and severe COVID-19 cases, and identified specific immune features that showed dose-response changes with disease174. The use of specific inflammatory mediators in the circulation to characterize COVID-19 was evaluated since the onset of the pandemic, with elevated levels of cytokines (such as IL-6), chemokines, and interferons being reported in cases of severe COVID-19175,176 and high systemic levels of IL-6, IL-8, and TNF at the time of hospitalization showing use as biomarkers of disease severity and mortality130. In-depth investigations have used longitudinal profiling of COVID-19 patients to identify multiple immune signatures that correlated with different disease trajectories41, or utilized proteomic determinations and machine learning to identify 11 host proteins and biomarker combinations that could distinguish and predict COVID-19 outcomes177. Interestingly, the presence of neutralizing immunoglobulin G (IgG) autoantibodies against type I interferons has also been shown to represent a likely indicator of severe disease in COVID-19 patients, given that such autoantibodies were absent in most of the individuals with asymptomatic or mild SARS-CoV-2 infection178. Together with our current data, these observations point to the value of identifying specific proteomic changes that can serve as biomarkers of COVID-19 severity, particularly during the vulnerable period of pregnancy. It is worth mentioning that the proteomic approach used herein was previously validated by our group179, indicating that our determinations are robust and can serve for future studies using targeted assays.
The current study has some limitations. Information regarding the interval from infection or symptom onset to sampling collection was not available, and thus any potential differences in the kinetics of the proteomic response to COVID-19 between pregnant and non-pregnant patients could not be considered. In addition, while the non-pregnant controls included in the study were matched by age and comorbidity with non-pregnant cases, we acknowledge that the inclusion of a healthy control group representing a non-perturbed proteomic state would be ideal. It should also be noted that specifically profiling the proteome to diagnose COVID-19 may not be practical; yet, the signature of COVID-19 described herein could be used to generate risk scores for potential COVID-19 at the time of sample collection in studies focused on other outcomes. Indeed, blood proteomic Soma Signal tests have already been proposed as patient-specific health indicators for cardiovascular events180, liver fat, kidney health, percentage body fat, cardiopulmonary fitness, and diabetes, among others181. The cardiovascular events risk score was shown to be informative of subsequent COVID-19 severity and mortality180, supporting the implementation of the COVID-19 signature proposed herein as a signal test to provide a more comprehensive characterization of the health status of subjects participating in studies with other primary endpoints.
Collectively, the study herein represents the most comprehensive characterization of the plasma proteome of pregnant and non-pregnant individuals diagnosed with COVID-19. The findings reported herein emphasize the distinct immune modulation between the non-pregnant and pregnant states, providing insight into the pathogenesis of COVID-19 as well as a potential explanation for the more severe outcomes observed in pregnant women. Importantly, the unique proteomic profiles observed in pregnant women suggest that the preeclampsia-like disease in this population may differ in pathogenesis from the canonical pathways implicated in the obstetrical syndrome of preeclampsia. Yet, further investigation is required to decipher the unique molecular mechanisms whereby SARS-CoV-2 infection induces a maternal cytokine storm and, more importantly, its effects on the offspring.
Data availability
The majority of the data generated in this study are included in the manuscript and/or in the Supplementary Materials. Proteomic data generated in this study are available at the Gene Expression Omnibus (accession number GSE207015). All software and R packages used herein are detailed in the Methods.
References
World Health Organization. COVID-19 Weekly Epidemiological Update <https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports> (2022).
Centers for Disease Control. COVID-19 Data from the National Center for Health Statistics, <https://www.cdc.gov/nchs/covid19/index.htm> (2021).
Zambrano, L. D. et al. Update: characteristics of symptomatic women of reproductive age with laboratory-confirmed SARS-CoV-2 Infection by pregnancy status - United States, January 22-October 3, 2020. MMWR Morb. Mort. Wkly. Rep. 69, 1641–1647 (2020).
Jamieson, D. J. & Rasmussen, S. A. An update on coronavirus disease 2019 (COVID-19) and pregnancy. Am. J. Obstet. Gynecol. 226, 177–186 (2021).
Lokken, E. M. et al. Disease severity, pregnancy outcomes, and maternal deaths among pregnant patients with severe acute respiratory syndrome coronavirus 2 infection in Washington State. Am. J. Obstet. Gynecol. 225, 77 e71–77.e14 (2021).
Lai, J. et al. SARS-CoV-2 and the subsequent development of preeclampsia and preterm birth: evidence of a dose-response relationship supporting causality. Am. J. Obstet. Gynecol. 225, 689–693.e681 (2021).
Conde-Agudelo, A. & Romero, R. SARS-CoV-2 infection during pregnancy and risk of preeclampsia: a systematic review and meta-analysis. Am. J. Obstet. Gynecol. 226, 68–89.e63 (2022).
DeSisto, C. L. et al. Risk for stillbirth among women with and without COVID-19 at delivery hospitalization — United States, March 2020–september 2021. MMWR Morb. Mort. Wkly. Rep. 70, 1640–1645 (2021).
Wang, Y. et al. Impact of Covid-19 in pregnancy on mother’s psychological status and infant’s neurobehavioral development: a longitudinal cohort study in China. BMC Med. 18, 347 (2020).
Deoni, S. C., Beauchemin, J., Volpe, A. & V, D. S. Impact of the COVID-19 Pandemic on Early Child Cognitive Development: Initial Findings in a Longitudinal Observational Study of Child Health. medRxiv, https://doi.org/10.1101/2021.08.10.21261846 (2021).
Huang, P. et al. Association Between the COVID-19 Pandemic and Infant Neurodevelopment: A Comparison Before and During COVID-19. Front. Pediatr. 9, 662165 (2021).
Norman, M. et al. Association of maternal SARS-CoV-2 infection in pregnancy with neonatal outcomes. JAMA 325, 2076–2086 (2021).
Villar, J. et al. Maternal and neonatal morbidity and mortality among pregnant women with and without COVID-19 infection: the INTERCOVID multinational cohort study. JAMA Pediatr 175, 817–826 (2021).
Shook, L. L., Sullivan, E. L., Lo, J. O., Perlis, R. H. & Edlow, A. G. COVID-19 in pregnancy: implications for fetal brain development. Trends Mol Med 28, 319–330 (2022).
Shuffrey, L. C. et al. Association of birth during the COVID-19 pandemic with neurodevelopmental status at 6 months in infants with and without in utero exposure to maternal SARS-CoV-2 infection. JAMA Pediatr. e215563, (2022).
Edlow, A. G., Castro, V. M., Shook, L. L., Kaimal, A. J. & Perlis, R. H. Neurodevelopmental outcomes at 1 year in infants of mothers who tested positive for SARS-CoV-2 during pregnancy. JAMA Netw Open 5, e2215787 (2022).
Ayed, M. et al. Neurodevelopmental outcomes of infants born to mothers with SARS-CoV-2 infections during pregnancy: a national prospective study in Kuwait. BMC Pediatr 22, 319 (2022).
Pique-Regi, R. et al. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife 9, https://doi.org/10.7554/eLife.58716 (2020).
Shanes, E. D. et al. Placental pathology in COVID-19. Am J Clin Pathol 154, 23–32 (2020).
Vivanti, A. J. et al. Transplacental transmission of SARS-CoV-2 infection. Nat Commun 11, 3572 (2020).
Zelop, C. M. & Bonney, E. A. COVID-19 in pregnancy: possible mechanisms not to be discounted. J. Matern. Fetal Neonatal. Med. 35, 3016–3019 (2020).
Sharps, M. C. et al. A structured review of placental morphology and histopathological lesions associated with SARS-CoV-2 infection. Placenta 101, 13–29 (2020).
Bordt, E. A. et al. Maternal SARS-CoV-2 infection elicits sexually dimorphic placental immune responses. Sci. Transl. Med. 13, eabi7428 (2021).
Ovies, C., Semmes, E. C. & Coyne, C. B. Pregnancy influences immune responses to SARS-CoV-2. Sci. Transl. Med. 13, eabm2070 (2021).
Atyeo, C. et al. Compromised SARS-CoV-2-specific placental antibody transfer. Cell 184, 628–642.e610 (2021).
Valdespino-Vázquez, M. Y. et al. Fetal and placental infection with SARS-CoV-2 in early pregnancy. J. Med. Virol. 93, 4480–4487 (2021).
Verma, S. et al. SARS-CoV-2 colonization of maternal and fetal cells of the human placenta promotes alteration of local renin-angiotensin system. Med (N Y) 2, 575–590.e575 (2021).
Shook, L. L. et al. SARS-CoV-2 placentitis associated with B.1.617.2 (Delta) variant and fetal distress or demise. J Infect Dis 225, 754–758 (2022).
Gomez-Lopez, N. et al. Distinct cellular immune responses to SARS-CoV-2 in pregnant women. J. Immunol. 208, 1857–1872 (2022).
Mithal, L. B. et al. Low-level SARS-CoV-2 viremia coincident with COVID placentitis and stillbirth. Placenta 121, 79–81 (2022).
Argueta, L. B. et al. Inflammatory responses in the placenta upon SARS-CoV-2 infection late in pregnancy. iScience 25, 104223 (2022).
Fenizia, C. et al. Analysis of SARS-CoV-2 vertical transmission during pregnancy. Nat. Commun. 11, 5128 (2020).
Edlow, A. G. et al. Assessment of maternal and neonatal SARS-CoV-2 viral load, transplacental antibody transfer, and placental pathology in pregnancies during the COVID-19 pandemic. JAMA Netw. Open. 3, e2030455 (2020).
Flannery, D. D. et al. Assessment of maternal and neonatal cord blood SARS-CoV-2 antibodies and placental transfer ratios. JAMA Pediatr. 175, 594–600 (2021).
Garcia-Flores, V. et al. Maternal-fetal immune responses in pregnant women infected with SARS-CoV-2. Nat. Commun. 13, 320 (2022).
Muthuka, J. K., Kiptoo, M., Oluoch, K. & Nyamai, E. An association of pregnancy with coronavirus cytokine storm: systematic review and meta-analysis. JMIR Pediatr. Parent. https://doi.org/10.2196/31579 (2022).
De Biasi, S. et al. Endogenous control of inflammation characterizes pregnant women with asymptomatic or paucisymptomatic SARS-CoV-2 infection. Nat. Commun. 12, 4677 (2021).
Chen, G. et al. Immune response to COVID-19 during pregnancy. Front Immunol 12, 675476 (2021).
Chen, G. et al. Differential immune responses in pregnant patients recovered from COVID-19. Signal Transduct. Target Ther. 6, 289 (2021).
Meckiff, B. J. et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell 183, 1340–1353.e1316 (2020).
Lucas, C. et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020).
Bernardes, J. P. et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity 53, 1296–1314.e1299 (2020).
Lipman, D., Safo, S. E. & Chekouo, T. Multi-omic analysis reveals enriched pathways associated with COVID-19 and COVID-19 severity. PLoS One 17, e0267047 (2022).
Foo, S. S. et al. The systemic inflammatory landscape of COVID-19 in pregnancy: extensive serum proteomic profiling of mother-infant dyads with in utero SARS-CoV-2. Cell. Rep. Med. 2, 100453 (2021).
Romero, R. et al. The maternal plasma proteome changes as a function of gestational age in normal pregnancy: a longitudinal study. Am. J. Obstet. Gynecol. 217, 67 e61–67.e21 (2017).
Erez, O. et al. The prediction of late-onset preeclampsia: results from a longitudinal proteomics study. PLoS One 12, e0181468 (2017).
Tarca, A. L. et al. The prediction of early preeclampsia: results from a longitudinal proteomics study. PLoS One 14, e0217273 (2019).
Ghaemi, M. S. et al. Proteomic signatures predict preeclampsia in individual cohorts but not across cohorts - implications for clinical biomarker studies. J. Matern. Fetal. Neonatal. Med. 35, 5621–5628 (2021).
Tarca, A. L. et al. Crowdsourcing assessment of maternal blood multi-omics for predicting gestational age and preterm birth. Cell Rep. Med. 2, 100323 (2021).
Bhatti, G. et al. The amniotic fluid proteome changes with gestational age in normal pregnancy: a cross-sectional study. Sci. Rep. 12, 601 (2022).
Health, N. I. o. Clinical Spectrum of SARS-CoV-2 Infection, <https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/>.
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5, e15004 (2010).
Davies, D. R. et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA 109, 19971–19976 (2012).
SomaLogic. SOMAmer Technical notes: http://www.somalogic.com/somalogic/media/Assets/PDFs/SSM-017-Rev-3-SOMAmer-Technical-Note-3-7-15.pdf.
Smyth, G. K. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor (eds R. Gentleman et al.) 397–420 (Springer, 2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. B 57, 289–300 (1995).
Maglott, D., Ostell, J., Pruitt, K. D. & Tatusova, T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 33, D54–D58 (2005).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Liberzon, A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015).
Falcon, S. & Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007).
Gentleman, R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5, R80 (2004).
Farias-Jofre, M. et al. Pregnancy tailors endotoxin-induced monocyte and neutrophil responses in the maternal circulation. Inflamm. Res. 71, 653–668 (2022).
Demery-Poulos, C. et al. Pregnancy imparts distinct systemic adaptive immune function. Am. J. Reprod. Immunol. 88, e13606 (2022).
Jung, E. et al. The etiology of preeclampsia. Am. J. Obstet. Gynecol. 226, S844–S866 (2022).
Amulic, B., Cazalet, C., Hayes, G. L., Metzler, K. D. & Zychlinsky, A. Neutrophil function: from mechanisms to disease. Annu. Rev. Immunol. 30, 459–489 (2012).
Papayannopoulos, V., Metzler, K. D., Hakkim, A. & Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell. Biol. 191, 677–691 (2010).
Chambers, S. P. & Clarke, A. G. Measurement of thymus weight, lumbar node weight and progesterone levels in syngeneically pregnant, allogeneically pregnant, and pseudopregnant mice. J. Reprod. Fertil. 55, 309–315 (1979).
Shinomiya, N. et al. Thymic depletion in pregnancy: kinetics of thymocytes and immunologic capacities of the hosts. J. Clin. Lab. Immunol. 34, 11–22 (1991).
Clarke, A. G. & Kendall, M. D. The thymus in pregnancy: the interplay of neural, endocrine and immune influences. Immunol. Today 15, 545–551 (1994).
Laan, M., Haljasorg, U., Kisand, K., Salumets, A. & Peterson, P. Pregnancy-induced thymic involution is associated with suppression of chemokines essential for T-lymphoid progenitor homing. Eur. J. Immunol. 46, 2008–2017 (2016).
Zoller, A. L., Schnell, F. J. & Kersh, G. J. Murine pregnancy leads to reduced proliferation of maternal thymocytes and decreased thymic emigration. Immunology 121, 207–215 (2007).
Gomez-Lopez, N. et al. In vivo T-cell activation by a monoclonal αCD3ε antibody induces preterm labor and birth. Am. J. Reprod. Immunol. 76, 386–390 (2016).
Arenas-Hernandez, M. et al. Effector and activated T cells induce preterm labor and birth that is prevented by treatment with progesterone. J. Immunol. 202, 2585–2608 (2019).
Medawar, P. B. Some immunological and endocrinological problems raised by the evolution of viviparity in vertebrates. Symp. Soc. Exp. Biol. 7, 320–328 (1953).
Chaouat, G., Voisin, G. A., Escalier, D. & Robert, P. Facilitation reaction (enhancing antibodies and suppressor cells) and rejection reaction (sensitized cells) from the mother to the paternal antigens of the conceptus. Clin. Exp. Immunol. 35, 13–24 (1979).
Bonney, E. A. & Onyekwuluje, J. The H-Y response in mid-gestation and long after delivery in mice primed before pregnancy. Immunol. Invest. 32, 71–81 (2003).
Zenclussen, A. C. et al. Abnormal T-cell reactivity against paternal antigens in spontaneous abortion: adoptive transfer of pregnancy-induced CD4+CD25+ T regulatory cells prevents fetal rejection in a murine abortion model. Am. J. Pathol. 166, 811–822 (2005).
Robertson, S. A., Guerin, L. R., Moldenhauer, L. M. & Hayball, J. D. Activating T regulatory cells for tolerance in early pregnancy - the contribution of seminal fluid. J. Reprod. Immunol. 83, 109–116 (2009).
Kahn, D. A. & Baltimore, D. Pregnancy induces a fetal antigen-specific maternal T regulatory cell response that contributes to tolerance. Proc. Natl. Acad. Sci. USA 107, 9299–9304 (2010).
Shima, T. et al. Regulatory T cells are necessary for implantation and maintenance of early pregnancy but not late pregnancy in allogeneic mice. J. Reprod. Immunol. 85, 121–129 (2010).
Zenclussen, M. L. et al. The persistence of paternal antigens in the maternal body is involved in regulatory T-cell expansion and fetal-maternal tolerance in murine pregnancy. Am. J. Reprod. Immunol. 63, 200–208 (2010).
Dimova, T. et al. Maternal Foxp3 expressing CD4+ CD25+ and CD4+ CD25- regulatory T-cell populations are enriched in human early normal pregnancy decidua: a phenotypic study of paired decidual and peripheral blood samples. Am. J. Reprod. Immunol. 66, 44–56 (2011).
Rowe, J. H., Ertelt, J. M., Xin, L. & Way, S. S. Pregnancy imprints regulatory memory that sustains anergy to fetal antigen. Nature 490, 102–106 (2012).
Samstein, R. M., Josefowicz, S. Z., Arvey, A., Treuting, P. M. & Rudensky, A. Y. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell 150, 29–38 (2012).
Ramhorst, R. et al. Modulation and recruitment of inducible regulatory T cells by first trimester trophoblast cells. Am. J. Reprod. Immunol. 67, 17–27 (2012).
Shima, T. et al. Paternal antigen-specific proliferating regulatory T cells are increased in uterine-draining lymph nodes just before implantation and in pregnant uterus just after implantation by seminal plasma-priming in allogeneic mouse pregnancy. J. Reprod. Immunol. 108, 72–82 (2015).
Aluvihare, V. R., Kallikourdis, M. & Betz, A. G. Regulatory T cells mediate maternal tolerance to the fetus. Nat Immunol 5, 266–271 (2004).
Sasaki, Y. et al. Decidual and peripheral blood CD4+CD25+ regulatory T cells in early pregnancy subjects and spontaneous abortion cases. Mol. Hum. Reprod. 10, 347–353 (2004).
Heikkinen, J., Möttönen, M., Alanen, A. & Lassila, O. Phenotypic characterization of regulatory T cells in the human decidua. Clin. Exp. Immunol. 136, 373–378 (2004).
Jiang, T. T. et al. Regulatory T cells: new keys for further unlocking the enigma of fetal tolerance and pregnancy complications. J Immunol. 192, 4949–4956 (2014).
Svensson-Arvelund, J. et al. The human fetal placenta promotes tolerance against the semiallogeneic fetus by inducing regulatory T cells and homeostatic M2 macrophages. J. Immunol. 194, 1534–1544 (2015).
Bonney, E. A. Immune regulation in pregnancy: a matter of perspective? Obstet. Gynecol. Clin. North Am. 43, 679–698 (2016).
Tsuda, S. et al. Clonally expanded decidual effector regulatory T cells increase in late gestation of normal pregnancy, but not in preeclampsia, in humans. Front Immunol 9, 1934 (2018).
Salvany-Celades, M. et al. Three types of functional regulatory T cells control T cell responses at the human maternal-fetal interface. Cell Rep. 27, 2537–2547.e2535 (2019).
Gomez-Lopez, N. et al. Regulatory T cells play a role in a subset of idiopathic preterm labor/birth and adverse neonatal outcomes. Cell Rep. 32, 107874 (2020).
Zhang, D., Lin, Y., Li, Y., Zhao, D. & Du, M. Mesenchymal stem cells enhance Treg immunosuppressive function at the fetal-maternal interface. J. Reprod. Immunol. 148, 103366 (2021).
Hunt, J. S., Manning, L. S. & Wood, G. W. Macrophages in murine uterus are immunosuppressive. Cell Immunol. 85, 499–510 (1984).
Gustafsson, C. et al. Gene expression profiling of human decidual macrophages: evidence for immunosuppressive phenotype. PLoS One 3, e2078 (2008).
Nagamatsu, T. & Schust, D. J. The immunomodulatory roles of macrophages at the maternal-fetal interface. Reprod. Sci. 17, 209–218 (2010).
Svensson, J. et al. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J. Immunol. 187, 3671–3682 (2011).
Houser, B. L., Tilburgs, T., Hill, J., Nicotra, M. L. & Strominger, J. L. Two unique human decidual macrophage populations. J. Immunol. 186, 2633–2642 (2011).
Svensson-Arvelund, J. & Ernerudh, J. The role of macrophages in promoting and maintaining homeostasis at the fetal-maternal interface. Am. J. Reprod. Immunol. 74, 100–109 (2015).
Xu, Y. et al. An M1-like macrophage polarization in decidual tissue during spontaneous preterm labor that is attenuated by rosiglitazone treatment. J. Immunol. 196, 2476–2491 (2016).
Chambers, M. et al. Macrophage plasticity in reproduction and environmental influences on their function. Front. Immunol. 11, 607328 (2020).
Gomez-Lopez, N. et al. Macrophages exert homeostatic actions in pregnancy to protect against preterm birth and fetal inflammatory injury. JCI Insight. 6, https://doi.org/10.1172/jci.insight.146089 (2021).
Doria, A. et al. Effect of pregnancy on serum cytokines in SLE patients. Arthritis Res. Ther. 14, R66 (2012).
Hong, S. et al. Longitudinal profiling of human blood transcriptome in healthy and lupus pregnancy. J. Exp. Med. 216, 1154–1169 (2019).
Bundhun, P. K., Soogund, M. Z. & Huang, F. Impact of systemic lupus erythematosus on maternal and fetal outcomes following pregnancy: a meta-analysis of studies published between years 2001-2016. J. Autoimmun. 79, 17–27 (2017).
Cornish, E. F., Filipovic, I., Asenius, F., Williams, D. J. & McDonnell, T. Innate immune responses to acute viral infection during pregnancy. Front. Immunol. 11, 572567 (2020).
Kraus, T. A. et al. Characterizing the pregnancy immune phenotype: results of the viral immunity and pregnancy (VIP) study. J Clin. Immunol. 32, 300–311 (2012).
Song, J. W. et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 11, 3410 (2020).
Aghaeepour, N. et al. An immune clock of human pregnancy. Sci. Immunol. 2, https://doi.org/10.1126/sciimmunol.aan2946 (2017).
Gomez-Lopez, N. et al. The cellular transcriptome in the maternal circulation during normal pregnancy: a longitudinal study. Front. Immunol. 10, 2863 (2019).
Han, X. et al. Differential dynamics of the maternal immune system in healthy pregnancy and preeclampsia. Front. Immunol. 10, 1305 (2019).
Tarca, A. L. et al. Targeted expression profiling by RNA-Seq improves detection of cellular dynamics during pregnancy and identifies a role for T cells in term parturition. Sci. Rep. 9, 848 (2019).
Sosa-Hernandez, V. A. et al. B cell subsets as severity-associated signatures in COVID-19 patients. Front. Immunol. 11, 611004 (2020).
Nikitina, E., Larionova, I., Choinzonov, E. & Kzhyshkowska, J. Monocytes and macrophages as viral targets and reservoirs. Int. J. Mol. Sci. 19, https://doi.org/10.3390/ijms19092821 (2018).
Wen, W. et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 6, 31 (2020).
Chilunda, V. et al. Transcriptional changes in CD16+ monocytes may contribute to the pathogenesis of COVID-19. Front. Immunol. 12, 665773 (2021).
Zhang, D. et al. Frontline science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc. Biol. 109, 13–22 (2021).
Lu-Culligan, A. et al. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med (N Y) 2, 591–610.e510 (2021).
Delorme-Axford, E., Sadovsky, Y. & Coyne, C. B. The Placenta as a Barrier to Viral Infections. Annu. Rev. Virol. 1, 133–146 (2014).
Megli, C. J. & Coyne, C. B. Infections at the maternal-fetal interface: an overview of pathogenesis and defence. Nat. Rev. Microbiol. 20, 67–82 (2022).
Mehta, P. et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet 395, 1033–1034 (2020).
Moore, J. B. & June, C. H. Cytokine release syndrome in severe COVID-19. Science 368, 473–474 (2020).
Pedersen, S. F. & Ho, Y. C. SARS-CoV-2: a storm is raging. J. Clin. Invest. 130, 2202–2205 (2020).
Tay, M. Z., Poh, C. M., Rénia, L., MacAry, P. A. & Ng, L. F. P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 (2020).
Hu, B., Huang, S. & Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 93, 250–256 (2021).
Del Valle, D. M. et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020).
Santa Cruz, A. et al. Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 12, 613422 (2021).
Hojyo, S. et al. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 40, 37 (2020).
Teuwen, L. A., Geldhof, V., Pasut, A. & Carmeliet, P. COVID-19: the vasculature unleashed. Nat. Rev. Immunol. 20, 389–391 (2020).
Guo, T. et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol. 5, 811–818 (2020).
Zheng, Y. Y., Ma, Y. T., Zhang, J. Y. & Xie, X. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 17, 259–260 (2020).
Vane, J. R., Anggard, E. E. & Botting, R. M. Regulatory functions of the vascular endothelium. N. Engl. J. Med. 323, 27–36 (1990).
Flammer, A. J. et al. The assessment of endothelial function: from research into clinical practice. Circulation 126, 753–767 (2012).
Celermajer, D. S. Endothelial dysfunction: does it matter? Is it reversible. J. Am. Coll. Cardiol. 30, 325–333 (1997).
Evans, P. C. et al. Endothelial dysfunction in COVID-19: a position paper of the ESC Working Group for atherosclerosis and vascular biology, and the ESC Council of basic cardiovascular science. Cardiovasc. Res. 116, 2177–2184 (2020).
Varga, Z. et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418 (2020).
Ackermann, M. et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 383, 120–128 (2020).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease Inhibitor. Cell 181, 271–280 e278 (2020).
Nagashima, S. et al. COVID-19 and lung mast cells: the Kallikrein-Kinin activation pathway. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms23031714 (2022).
Giesen, P. L. et al. Blood-borne tissue factor: another view of thrombosis. Proc. Natl. Acad. Sci. USA 96, 2311–2315 (1999).
Mendoza, M. et al. Pre-eclampsia-like syndrome induced by severe COVID-19: a prospective observational study. BJOG 127, 1374–1380 (2020).
Rosenbloom, J. I., Raghuraman, N., Carter, E. B. & Kelly, J. C. Coronavirus disease 2019 infection and hypertensive disorders of pregnancy. Am. J. Obstet. Gynecol. 224, 623–624 (2021).
Khalil, A., Samara, A., Chowdhury, T. & O’Brien, P. Does COVID-19 cause pre-eclampsia? Ultrasound Obstet. Gynecol. 59, 146–152 (2022).
Conde-Agudelo, A. & Romero, R. Mechanisms that may underlie a causal association between SARS-COV-2 infection and preeclampsia. Am. J. Obstet. Gynecol. 226, 280–281 (2022).
Redman, C. W. Immunological aspects of pre-eclampsia. Baillieres Clin. Obstet. Gynaecol. 6, 601–615 (1992).
Borzychowski, A. M., Sargent, I. L. & Redman, C. W. Inflammation and pre-eclampsia. Semin. Fetal. Neonatal. Med. 11, 309–316 (2006).
Erez, O. et al. Preeclampsia and eclampsia: the conceptual evolution of a syndrome. Am. J. Obstet. Gynecol. 226, S786–s803 (2022).
Miller, D. et al. Cellular immune responses in the pathophysiology of preeclampsia. J. Leukoc. Biol. 111, 237–260 (2022).
Staff, A. C. et al. Failure of physiological transformation and spiral artery atherosis: their roles in preeclampsia. Am. J. Obstet. Gynecol. 226, S895–s906 (2022).
Palomo, M. et al. Differences and similarities in endothelial and angiogenic profiles of preeclampsia and COVID-19 in pregnancy. Am. J. Obstet. Gynecol. https://doi.org/10.1016/j.ajog.2022.03.048 (2022).
Shook, L. L., Fourman, L. T. & Edlow, A. G. Immune responses to SARS-CoV-2 in pregnancy: implications for the health of the next generation. J. Immunol. 209, 1465–1473 (2022).
Zhao, C., Denison, C., Huibregtse, J. M., Gygi, S. & Krug, R. M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. USA 102, 10200–10205 (2005).
Munnur, D. et al. Altered ISGylation drives aberrant macrophage-dependent immune responses during SARS-CoV-2 infection. Nat. Immunol. 22, 1416–1427 (2021).
Liu, G. et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 6, 467–478 (2021).
Schroeder, S. et al. Interferon antagonism by SARS-CoV-2: a functional study using reverse genetics. Lancet Microbe 2, e210–e218 (2021).
Toro, A. et al. Pin-pointing the key hubs in the IFN-gamma pathway responding to SARS-CoV-2 infection. Viruses 14, https://doi.org/10.3390/v14102180 (2022).
Bizzotto, J. et al. SARS-CoV-2 infection boosts MX1 antiviral effector in COVID-19 patients. iScience 23, 101585 (2020).
Ghoreshi, Z. A. et al. Evaluation of MX1 gene promoter methylation in different severities of COVID-19 considering patient gender. Clin. Lab. 68, https://doi.org/10.7754/Clin.Lab.2022.220104 (2022).
Cheng, L. C. et al. Novel signaling pathways regulate SARS-CoV and SARS-CoV-2 infectious disease. Medicine (Baltimore) 100, e24321 (2021).
Galani, I. E. et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol 22, 32–40 (2021).
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007).
Kuriakose, T. et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1, https://doi.org/10.1126/sciimmunol.aag2045 (2016).
Pham, T. H., Kwon, K. M., Kim, Y. E., Kim, K. K. & Ahn, J. H. DNA sensing-independent inhibition of herpes simplex virus 1 replication by DAI/ZBP1. J. Virol. 87, 3076–3086 (2013).
Upton, J. W., Kaiser, W. J. & Mocarski, E. S. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 11, 290–297 (2012).
Zhang, T. et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180, 1115–1129.e1113 (2020).
Rothan, H. A. et al. SARS-CoV-2 infects primary neurons from human ACE2 expressing mice and upregulates genes involved in the inflammatory and necroptotic pathways. Pathogens 11, https://doi.org/10.3390/pathogens11020257 (2022).
Peng, R. et al. Human ZBP1 induces cell death-independent inflammatory signaling via RIPK3 and RIPK1. EMBO Rep. 23, e55839, (2022).
Basavaraju, S., Mishra, S., Jindal, R. & Kesavardhana, S. Emerging role of ZBP1 in Z-RNA sensing, influenza virus-induced cell death, and pulmonary inflammation. mBio 13, e0040122 (2022).
Karki, R. et al. ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Sci. Immunol. 7, eabo6294 (2022).
Feyaerts, D. et al. Integrated plasma proteomic and single-cell immune signaling network signatures demarcate mild, moderate, and severe COVID-19. bioRxiv https://doi.org/10.1101/2021.02.09.430269 (2021).
Blanco-Melo, D. et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181, 1036–1045.e1039 (2020).
Laing, A. G. et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 26, 1623–1635 (2020).
Shu, T. et al. Plasma proteomics identify biomarkers and pathogenesis of COVID-19. Immunity 53, 1108–1122.e1105 (2020).
Bastard, P. et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, https://doi.org/10.1126/science.abd4585 (2020).
Tarca, A. L. et al. Human plasma proteome during normal pregnancy. J. Proteome Res. https://doi.org/10.1021/acs.jproteome.2c00391 (2022).
Williams, S. A. et al. A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci. Transl. Med. 14, eabj9625 (2022).
Williams, S. A. et al. Plasma protein patterns as comprehensive indicators of health. Nat. Med. 25, 1851–1857 (2019).
Acknowledgements
This research was supported by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, US Department of Health and Human Services (NICHD/NIH/DHHS) under Contract no. HHSN275201300006C (R.R.). This research was also supported by the Wayne State University Perinatal Initiative in Maternal, Perinatal, and Child Health (N.G.-L. and A.L.T.) and the Clinical Research Center, Department of Pathology and Laboratory Medicine, High Obstetric Complexity Unit, and Intensive Care Unit of the Fundacion Valle del Lili (M.F.E.). R.R. has contributed to this work as part of his official duties as an employee of the United States Federal Government. Figures include elements created using BioRender.com.
Author information
Authors and Affiliations
Contributions
N.G.-L., R.R., and A.L.T.: designed and supervised the study, analyzed data, provided intellectual input, and wrote the paper. M.F.E.: conceived, designed, and supervised the study, analyzed data and provided intellectual input. J.A.C., M.P.E., and L.L.A. D.N.: provided human samples used in the study, analyzed and recorded data. D.M., J.G., and M.A.-H.: analyzed data, provided intellectual input, and wrote the paper. D.M.G.: analyze data and provided intellectual input. G.B. and B.D.: analyzed data and provided intellectual input. M.A.Z., I.R., P.A.F., and L.P.: provided human samples used in the study and recorded data. T.C., E.J., V.G.-F., M.S., F.G., M.B., and N.G.T.: provided intellectual input.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Nelly Amenyogbe and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gomez-Lopez, N., Romero, R., Escobar, M.F. et al. Pregnancy-specific responses to COVID-19 revealed by high-throughput proteomics of human plasma. Commun Med 3, 48 (2023). https://doi.org/10.1038/s43856-023-00268-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-023-00268-y