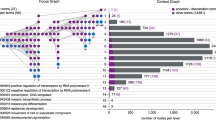
Abstract
Gene expression is regulated by a large variety of mechanisms. Previous studies attempting to model the quantitative relationships between gene expression levels and regulatory mechanisms have considered only one or a few mechanisms at a time, which cannot provide a full picture of the complex interactions among different mechanisms. This was partially due to the heterogeneity of the mechanisms, which involve different types of biological objects and data representations, making it hard to study them in a unified way. Here, we describe a flexible framework that can integrate very different types of data for studying their joint effects on gene expression. In this framework, domain knowledge is represented by metapaths, while the manifestations of their effects in actual data are summarized by an embedding of the biological objects in a latent space. We demonstrate the use of our framework in integrating several diverse types of data that are related to gene expression in different ways, including DNA contacts in three-dimensional genome architecture, protein–protein interactions, genomic neighbourhoods and broad chromatin accessibility domains. The modelling results reveal that these several types of data are able to model gene expression fairly well individually, but even better when integrated.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Example data for testing the source code are available at https://doi.org/10.24433/CO.1518993.v143. The public data used for producing the results in this study and the embeddings produced from the five cell lines can be downloaded from http://yiplab.cse.cuhk.edu.hk/geek/.
Code availability
Source codes are available at https://doi.org/10.24433/CO.1518993.v143.
References
Lodish, H. et al. Molecular Cell Biology 8th edn (W. H. Freeman and Company, 2016).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Tang, J., Qu, M. & Mei, Q. PTE: predictive text embedding through large-scale heterogeneous text networks. In Proceedings of ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 1165–1174 (ACM, 2015).
Sun, Y., Han, J., Yan, X., Yu, P. S. & Wu, T. PathSim: meta path-based top-k similarity search in heterogeneous information networks. Proc. VLDB Endowment 4, 992–1003 (2011).
Mikolov, T., Chen, K., Corrado, G. & Dean, J. Efficient estimation of word representations in vector space. Preprint at https://arxiv.org/abs/1301.3781 (2013).
Mikolov, T., Sutskever, I., Chen, K., Corrado, G. S. & Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of Advances in Neural Information Processing Systems (NeurIPS) 3111–3119 (NIPS, 2013).
Grover, A. & Leskovec, J. node2vec: scalable feature learning for networks. In Proceedings of ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 855–864 (ACM, 2016).
Zitnik, M. & Leskovec, J. Predicting multicellular function through multi-layer tissue networks. Bioinformatics 33, i190–i198 (2017).
Zeng, W., Wu, M. & Jiang, R. Prediction of enhancer–promoter interactions via natural language processing. BMC Genomics 19, 84 (2018).
Dong, Y., Chawla, N. V. & Swami, A. metapath2vec: scalable representation learning for heterogeneous networks. In Proceedings of ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 135–144 (ACM, 2017).
Faruqui, M. et al. Retrofitting word vectors to semantic lexicons. In Proceedings of Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (NAACL-HLT) 1606–1615 (Association for Computational Linguistics, 2015).
Vulić, I. & Mrkšić, N. Specialising word vectors for lexical entailment. In Proceedings of Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (NAACL-HLT) 1134–1145 (Association for Computational Linguistics, 2018).
Xu, C. et al. Rc-net: a general framework for incorporating knowledge into word representations. In Proceedings of International Conference on Information and Knowledge Management (CIKM) 1219–1228 (ACM, 2014).
Yu, M. & Dredze, M. Improving lexical embeddings with semantic knowledge. In Annual Meeting of the Association for Computational Linguistics (ACL) (Short Papers) 545–550 (ACL, 2014).
Marcotte, E. M., Pellegrini, M., Thompson, M. J., Yeates, T. O. & Eisenberg, D. A combined algorithm for genome-wide prediction of protein function. Nature 402, 83–86 (1999).
Michalak, P. Coexpression coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 91, 243–248 (2008).
Hu, X., Shi, C. H. & Yip, K. Y. A novel method for discovering local spatial clusters of genomic regions with functional relationships from DNA contact maps. Bioinformatics 32, i111–i120 (2016).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In Proceedings of ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, 2016).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. Preprint at https://arxiv.org/abs/1609.02907 (2017).
Tang, Z. et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163, 1611–1627 (2015).
Schmitt, A. D. et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 (2016).
Sima, J. et al. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell 176, 816–830 (2019).
Ma, J. & Duan, Z. Replication timing becomes intertwined with 3D genome organization. Cell 176, 681–684 (2019).
Ernst, J. & Kellis, M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 33, 364–376 (2015).
Artetxe, M., Labaka, G., Lopez-Gazpio, I. & Agirre, E. Uncovering divergent linguistic information in word embeddings with lessons for intrinsic and extrinsic evaluation. In Proceedings of Conference on Computational Natural Language Learning (CoNLL) 282–291 (Association for Computational Linguistics, 2018).
Kiela, D., Hill, F. & Clark, S. Specializing word embeddings for similarity or relatedness. In Proceedings of Conference on Empirical Methods in Natural Language Processing (EMNLP) 2044–2048 (Association for Computational Linguistics, 2015).
Chatr-Aryamontri, A. et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 45, D369–D379 (2017).
Harrow, J. et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 22, 1760–1774 (2012).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 (2012).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Ay, F., Bailey, T. L. & Noble, W. S. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 24, 999–1011 (2014).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. Theory Exp. 2008, P10008 (2008).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong.Nucleic Acids Res 47, D330–D338 (2018).
Klopfenstein, D. et al. GOATOOLS: a Python library for gene ontology analyses. Sci. Rep. 8, 10872 (2018).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Wang, Y. et al. The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 19, 151 (2018).
The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome.Nature 489, 57–74 (2012).
Cao, Q. et al. GEEK (Gene Expression Embedding frameworK) Demo (GM12878, Chromosome 1) (CodeOcean, 2020); https://doi.org/10.24433/CO.1518993.v1
Acknowledgements
This work was supported by Hong Kong Research Grants Council General Research Funds 14170217 (C.C., D.L. and K.Y.Y.), 14203119 (A.S.L.C. and K.Y.Y.), 14200817, 15200715 and 15204116 (E.L.), Collaborative Research Funds 4045-18WF (A.S.L.C. and K.Y.Y.), 4054-16G (T.-L.L. and K.Y.Y.) and C4057-18EF (K.Y.Y.), Area of Excellence AoE/P-404/18 (E.L.), Theme-based Research Scheme T12C-714/14-R (K.Y.Y.), the Hong Kong Innovation and Technology Commission Innovative and Technology Fund ITS/310/18 (E.L.) and the Hong Kong Epigenomics Project (EpiHK). K.Y.Y. was also supported by CUHK Young Researcher Award, Outstanding Fellowship and Seed Funding for Strategic Areas.
Author information
Authors and Affiliations
Contributions
K.Y.Y. conceived the study. Q.C. and Z.Z. designed and implemented GEEK. Q.C., Z.Z., A.X.F., Q.W. and K.Y.Y. performed data analyses. Q.C., A.X.F., T.-L.L., E.L., A.S.L.C., C.C., D.L. and K.Y.Y. interpreted the results. K.Y.Y. and Q.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary text (including algorithms 1 and 2), Table 1 and Figs. 1–23.
Supplementary Table 2
Functional enrichment analysis of gene clusters based on their embeddings. Each row represents the genes in a cluster that are annotated with the same functional term. The different columns specify the embedding setting, enrichment results and gene symbols, with their full descriptions provided in the file.
Rights and permissions
About this article
Cite this article
Cao, Q., Zhang, Z., Fu, A.X. et al. A unified framework for integrative study of heterogeneous gene regulatory mechanisms. Nat Mach Intell 2, 447–456 (2020). https://doi.org/10.1038/s42256-020-0205-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-0205-2
This article is cited by
-
Reusability report: Capturing properties of biological objects and their relationships using graph neural networks
Nature Machine Intelligence (2022)
-
Reusability report: Compressing regulatory networks to vectors for interpreting gene expression and genetic variants
Nature Machine Intelligence (2021)