Abstract
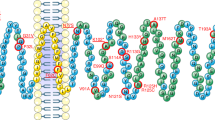
The glucagon-like peptide 1 receptor (GLP1R) is a major drug target with several agonists being prescribed in individuals with type 2 diabetes and obesity1,2. The impact of genetic variability of GLP1R on receptor function and its association with metabolic traits are unclear with conflicting reports. Here, we show an unexpected diversity of phenotypes ranging from defective cell surface expression to complete or pathway-specific gain of function (GoF) and loss of function (LoF), after performing a functional profiling of 60 GLP1R variants across four signalling pathways. The defective insulin secretion of GLP1R LoF variants is rescued by allosteric GLP1R ligands or high concentrations of exendin-4/semaglutide in INS-1 823/3 cells. Genetic association studies in 200,000 participants from the UK Biobank show that impaired GLP1R cell surface expression contributes to poor glucose control and increased adiposity with increased glycated haemoglobin A1c and body mass index. This study defines impaired GLP1R cell surface expression as a risk factor for traits associated with type 2 diabetes and obesity and provides potential treatment options for GLP1R LoF variant carriers.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by R.J. Links for databases are provided below.
Human genome (hg19/GRCh37): https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.13/
UK Biobank (application no. 67575): https://www.ukbiobank.ac.uk/
pVCF format (field no. 23156): https://biobank.ndph.ox.ac.uk/ukb/field.cgi?id=23156
ExAC browser: https://gnomad.broadinstitute.org/.
Source data are provided with this paper.
References
Muller, T. D. et al. Glucagon-like peptide 1 (GLP-1). Mol. Metab. 30, 72–130 (2019).
Graaf, C. et al. Glucagon-like peptide-1 and its class B G-protein-coupled receptors: a long march to therapeutic successes. Pharm. Rev. 68, 954–1013 (2016).
Zhang, Y. et al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. Nature 546, 248–253 (2017).
Koole, C. et al. Polymorphism and ligand dependent changes in human glucagon-like peptide-1 receptor (GLP-1R) function: allosteric rescue of loss of function mutation. Mol. Pharmacol. 80, 486–497 (2011).
Koole, C. et al. Minireview: signal bias, allosterism, and polymorphic variation at the GLP-1R: implications for drug discovery. Mol. Endocrinol. 27, 1234–1244 (2013).
Liu, T., Ji, R. L. & Tao, Y. X. Naturally occurring mutations in G-protein-coupled receptors associated with obesity and type 2 diabetes mellitus. Pharmacol. Ther. 234, 108044 (2022).
Katsonis, P. & Lichtarge, O. A formal perturbation equation between genotype and phenotype determines the evolutionary action of protein-coding variations on fitness. Genome Res 24, 2050–2058 (2014).
Lichtarge, O., Bourne, H. R. & Cohen, F. E. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 257, 342–358 (1996).
Wootten, D. et al. The extracellular surface of the GLP-1 receptor is a molecular trigger for biased agonism. Cell 165, 1632–1643 (2016).
Bonnefond, A. et al. Pathogenic variants in actionable MODY genes are associated with type 2 diabetes. Nat. Metab. 2, 1126–1134 (2020).
Naylor, J. et al. Use of CRISPR/Cas9-engineered INS-1 pancreatic beta cells to define the pharmacology of dual GIPR/GLP-1R agonists. Biochem. J. 473, 2881–2891 (2016).
Ast, J. et al. Super-resolution microscopy compatible fluorescent probes reveal endogenous glucagon-like peptide-1 receptor distribution and dynamics. Nat. Commun. 11, 467 (2020).
Bonnefond, A. et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat. Genet. 44, 297–301 (2012).
Karamitri, A. et al. Type 2 diabetes-associated variants of the MT(2) melatonin receptor affect distinct modes of signaling. Sci. Signal. 11, eaan6622 (2018).
Folon, L. et al. Contribution of heterozygous PCSK1 variants to obesity and implications for precision medicine: a case-control study. Lancet Diabetes Endocrinol. 11, 182–190 (2023).
Jones, B. et al. Targeting GLP-1 receptor trafficking to improve agonist efficacy. Nat. Commun. 9, 1602 (2018).
Kushner, R. F. et al. Semaglutide 2.4 mg for the treatment of obesity: key elements of the STEP trials 1 to 5. Obesity 28, 1050–1061 (2020).
Knudsen, L. B. et al. Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc. Natl Acad. Sci. USA 104, 937–942 (2007).
Koole, C. et al. Allosteric ligands of the glucagon-like peptide 1 receptor (GLP-1R) differentially modulate endogenous and exogenous peptide responses in a pathway-selective manner: implications for drug screening. Mol. Pharmacol. 78, 456–465 (2010).
Wootten, D. et al. Differential activation and modulation of the glucagon-like peptide-1 receptor by small molecule ligands. Mol. Pharmacol. 83, 822–834 (2013).
Rosenstock, J. et al. Efficacy and safety of a novel dual GIP and GLP-1 receptor agonist tirzepatide in patients with type 2 diabetes (SURPASS-1): a double-blind, randomised, phase 3 trial. Lancet 398, 143–155 (2021).
Malik, F. & Li, Z. Non-peptide agonists and positive allosteric modulators of glucagon-like peptide-1 receptors: alternative approaches for treatment of type 2 diabetes. Br. J. Pharmacol. 179, 511–525 (2022).
Bueno, A. B. et al. Structural insights into probe-dependent positive allosterism of the GLP-1 receptor. Nat. Chem. Biol. 16, 1105–1110 (2020).
Jones, B. The therapeutic potential of GLP-1 receptor biased agonism. Br. J. Pharmacol. 179, 492–510 (2022).
Sonoda, N. et al. Beta-arrestin-1 mediates glucagon-like peptide-1 signaling to insulin secretion in cultured pancreatic beta cells. Proc. Natl Acad. Sci. USA 105, 6614–6619 (2008).
Quoyer, J. et al. GLP-1 mediates antiapoptotic effect by phosphorylating Bad through a beta-arrestin 1-mediated ERK1/2 activation in pancreatic beta-cells. J. Biol. Chem. 285, 1989–2002 (2010).
Baron, M. et al. Loss-of-function mutations in MRAP2 are pathogenic in hyperphagic obesity with hyperglycemia and hypertension. Nat. Med. 25, 1733–1738 (2019).
Schonegge, A. M. et al. Evolutionary action and structural basis of the allosteric switch controlling β2AR functional selectivity. Nat. Commun. 8, 2169 (2017).
Lima-Fernandes, E. et al. Distinct functional outputs of PTEN signalling are controlled by dynamic association with beta-arrestins. EMBO J. 30, 2557–2568 (2011).
Berthault, C., Staels, W. & Scharfmann, R. Purification of pancreatic endocrine subsets reveals increased iron metabolism in beta cells. Mol. Metab. 42, 101060 (2020).
Guillaume, J. L. et al. The PDZ protein mupp1 promotes Gi coupling and signaling of the Mt1 melatonin receptor. J. Biol. Chem. 283, 16762–16771 (2008).
Chen, M. et al. Melatonin MT1 and MT2 receptor ERK signaling is differentially dependent on Gi/o and Gq/11 proteins. J. Pineal Res. 68, e12641 (2020).
Sun, J., Zheng, Y. & Hsu, L. A unified mixed-effects model for rare-variant association in sequencing studies. Genet. Epidemiol. 37, 334–344 (2013).
Galinsky, K. J., Loh, P. R., Mallick, S., Patterson, N. J. & Price, A. L. Population structure of UK Biobank and ancient eurasians reveals adaptation at genes influencing blood pressure. Am. J. Hum. Genet 99, 1130–1139 (2016).
Kenakin, T. New concepts in pharmacological efficacy at 7TM receptors: IUPHAR review 2. Br. J. Pharmacol. 168, 554–575 (2013).
Cecon, E., Oishi, A. & Jockers, R. Melatonin receptors: molecular pharmacology and signalling in the context of system bias. Br. J. Pharmacol. 175, 3263–3280 (2018).
Acknowledgements
We thank J. Naylor (Cambridge, United Kingdom) for kindly providing INS-1 832/3 (GLP1R KO) cells and M. Scott for giving advice for the selection of β-arr1/β-arr2 selective siRNA sequences. This work was supported by the Fondation de la Recherche Médicale (Equipe FRM DEQ20130326503 to R.J.), Agence Nationale de la Recherche (ANR-2011-BSV1-012-01 ‘MLT2D’, ANR-2011-META ‘MELA-BETES, ANR-21-CE18-0023 ‘alloGLP1R’) to R.J., Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS) and the ‘Who am I?’ laboratory of excellence no. ANR-11-LABX-0071 funded by the French Government through its ‘Investments for the Future’ program operated by The French National Research Agency under grant no. ANR-11-IDEX-0005-01 to R.J. This work was supported by grants from the European Union’s Horizon Europe Research and Innovation Programme under grant agreement 101080465 to A.B., P.F., R.J., J.D., the Ministry of Science and Technology (grant no. 2018YFA0507003 and 2021ZD0203302 to J.L.) and the National Natural Science Foundation of China (NSFC; grant numbers 81720108031, 81872945 and 31721002 to J.L.). This research was conducted using the UK Biobank application no. 67575 (A.B.). This study was further funded by the French National Research Agency (ANR-10-LABX-46 (European Genomics Institute for Diabetes)) to A.B. and P.F., the French National Research Agency (ANR-10-EQPX-07-01 (LIGAN-PM)) to A.B. and P.F., the European Research Council (ERC GEPIDIAB – 294785 to P.F.; ERC Reg-Seq – 715575 to A.B.), ‘France Génomique’ consortium (ANR-10-INBS-009) and the National Center for Precision Diabetic Medicine – PreciDIAB to A.B. and P.F., which is jointly supported by the French National Agency for Research (ANR-18-IBHU-0001), by the European Union (FEDER), by the Hauts-de-France Regional Council and by the European Metropolis of Lille (MEL). O.L. gratefully acknowledges support from the National Institutes of Health (GM066099). W.G. was supported by a doctoral fellowship from the Chinese Scholarship Council (China).
Author information
Authors and Affiliations
Contributions
W.G., L.L., R.S., A.B., J.L. and R.J. conceived and designed experiments. Z.F. and G.H. made constructions of variant plasmids and L.H. made constructions of the non-tagged WT GLP1R plasmid. W.G., L.L., F.G., E.C., M.O. and A.H. performed biochemical experiments. E.H. and O.L. did the clustering analysis. P.K. performed variant impact score analyses. A.B. and P.F. conceived and designed the next-generation sequencing study. A.B., M.D. and M.B. performed the genetic analyses. G.C., M.M., A.B., P.F. and B.B. contributed DNA samples from participants. W.G., L.L., A.B., O.L., J.D. and R.J. analysed data. A.B., P.F., J.L. and R.J. obtained the funding. W.G., A.B. and R.J. wrote the manuscript that was edited and/or approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Jonathan Campbell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Isabella Samuelson, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Evolutionary action analysis of GLP1R variants.
a, Evolutionary Action (EA) scores were calculated for the 132 indicated GLP1R variants from the general database. EA scores range from 0 to 100, with a score of 0 predicted as benign and a score of 100 predicted as highly impactful or detrimental to protein function. Variants selected for functional profiling are highlighted in bold. b, EA scores were calculated for 26 indicated GLP1R variants obtained from the RaDiO study.
Extended Data Fig. 2 Expression analysis of WT and mutant GLP1R monitored by ELISA and LUXendin binding.
a-e, FACS-sorting of intact mouse ß-cells expressing endogenous GLP1R (a), of INS-1 823/3 (Glp1r KO) cells expressing SNAP-flag-tagged WT GLP1R (b), or of HEK293T cells expressing SNAP-flag-tagged WT GLP1R (c), after fluorescent LUXendin binding (100 nM). Specificity LUXendin binding was determined in the presence of an excess of Ex-4 (10 μM) in ß-cells (a) and by mock transfection in INS-1 823/3 (Glp1r KO) and HEK293T cells (b,c). Functional WT-GLP1R expression was monitored in parallel by determining Ex-4-induced cAMP production (a-c). d, Quantification of LUXendin binding of panels a-c (see Methods for more details). e, Quantification of LUXendin binding to non-tagged vs. SNAP-flag-tagged WT GLP1R in INS-1 823/3 (Glp1r KO) and HEK293T cells. a-e, The figure is representative of 3 independent experiments each performed in one replicate. cAMP for a, n = 3; b, n = 5; c, n = 3. n represents biologically independent experiments. f-h, Surface (Sur) and total (To) expression of SNAP-flag-tagged GLP1R in HEK293T and INS-1 823/3 (Glp1r KO) cells determined by ELISA. (f) Expression of WT and mutant GLP1R in INS-1 823/3 (Glp1r KO) cells. Surface expression is shown at X-axis, total receptor expression as color gradient and the Sur/To ratio as size of the bubble. Statistical significance of differences (compared with WT GLP1R) was determined by one-way analysis of variance and Dunnett’s post-test. (g-h) Comparison of the total expression of mutants in HEK293T and INS-1 823/3 (Glp1r KO) cells. P value from a two-way analysis of variance and Sidak’s multiple comparisons test is indicated. All values are shown as means ± s.e.m. Ex-4, Exendin-4; MFI, Mean Fluorescent Intensity. See also Fig. 1c,d for complete data set.
Extended Data Fig. 3 Signaling pathways activated by WT GLP1R in HEK293T cells.
a-f, Ex-4 concentration-response curves for cAMP accumulation (a), Ca2+ mobilization (b), β-arr2 recruitment (d) and ERK activation (f) of SNAP-flag-tagged WT GLP1R. Kinetics of β-arr2 recruitment (c) and ERK activation (e). a-d,f, Representative curves are shown; More than three replicates were conducted in signaling profiling experiments. e, n = 3. g-j, comparison of signaling of non-tagged vs. SNAP-flag-tagged WT GLP1R. g,j, n = 3; h,i, n = 4. k-n, At 5 minutes, the ERK1/2 activation was fully blocked by the PKA inhibitor H89 (1 h preincubation) (k) but not by β-arr1/2 silencing (72 hours prior to Ex-4) (i) indicating that the Gs/cAMP/PKA pathway is the predominant input pathway at 5 minutes of Ex-4 (100 nM) stimulation in HEK293T cells. Representative Western blots showing knockdown of β-arr1/2, and similar Flag-GLP1R expression levels in samples (m). Quantification of panel m (n). k-n, n = 3. o, Effect of the Gq/11 protein inhibitor YM-254890 (30 min preincubation) on Ex-4 (100 nM) stimulated Ca2+ mobilization. o, n = 3. All values are means ± s.e.m. Statistical significance of differences was determined by one-way analysis of variance and Dunnett’s post-test and two-tailed unpaired t-test (n). Ex-4, Exendin-4; Sema, Semaglutide; β-arr1/2, β-arrestin1/2; pERK, phosphorylation of ERK, Ctrl, control; AU, Arbitrary Unit.
Extended Data Fig. 4 Correlations between signaling parameters of cAMP production and cell surface expression and comparison of GLP-1 vs. Ex-4 in HEK293T cells.
a, Ex-4 concentration-response curves for cAMP accumulation at different quantities of cell surface expressed WT GLP1R. 100% refers to transfection of 50 ng WT GLP1R plasmid. b,c, Correlation of Emax (b) and logEC50 (c) of cAMP accumulation with cell surface expression of WT GLP1R. Inset: 0 to 15% cell surface expression range. a,c, n = 3; b, n values are shown in source data for Extended Data Fig. 4; d, Surface expression of WT and mutant GLP1R. Ctrl-1 = 50 ng and Ctrl-2 = 1 ng of WT GLP1R plasmid to match the low expression of some mutants. e,f, Ex-4 and GLP-1 concentration-response curves of cAMP accumulation at Ctrl-1 and Ctrl-2 WT GLP1R conditions. Inset: response at Ctrl-2 condition. d-f. n = 3. g, pEC50 values of panels e-f. h-k, Ex-4 (h) and GLP-1 (i) concentration-response curves of mutant GLP1R compared to WT GLP1R at Ctrl-2 conditions. p.H180Y, p.N320Y, p.G361R and p.I400R are complete loss-of-function (LoF) mutants for this pathway when stimulated with Ex-4 (h) or GLP-1 (i). p.H173P and p.R176R show a residual response and were classified as severely defective with similar results for Ex-4 (j) and GLP-1 (k) confirming the physiological relevance of this result h-k. n = 3. All values are means ± s.e.m. Ctrl, control; Exp, Expression; Ex-4, Exendin-4.
Extended Data Fig. 5 Correlations between signaling parameters of Ca2+ mobilization, ERK activation, β-arr2 recruitment and cell surface expression in HEK293T cells.
a-i, Different amounts of WT GLP1R were expressed in HEK293T cells and Ex-4 concentration-response curves were generated for Ca2+ mobilization (a), ERK1/2 activation (d), and β-arr2 recruitment (g) and calculated Emax (b, e, h) and logEC50 (c, f, i) values correlated with surface expression. These correlation curves allowed us to determine the signaling parameters (Emax and EC50) of receptor mutants at matched WT GLP1R expression levels. 100% surface expression refers to transfection of 50 ng of WT GLP1R plasmid. a-d,f, n = 3; e, g-i, n values are shown in source data for Extended Data Fig. 5. All values are means ± s.e.m. Exp, Expression; Ctrl, control; Ex-4, Exendin-4.
Extended Data Fig. 6 Affinity of Ex-4 for WT and mutant GLP1R with modified EC50 values in functional assays determined in TAG-LITE® GLP1 receptor competition binding experiments.
All values are expressed as means ± s.e.m of at least three independent experiments. ND refers to ‘no detectable binding’. The cumulative pIC50 = 8.13 ± 0.06 for the WT GLP1R. The data were analyzed by comparing independent fits with a global fit that share the selected parameter (#).
Extended Data Fig. 7 Correlations between experimentally determined phenotypic score and different predictive scores.
We calculated the predicted scores by using 5 different scoring algorithms including EA (a), REVEL (b), CADD (c), MutationAssessor (d), SIFT (e), and PolyPhen2 (f), and correlated them with the experimentally obtained phenotypic scores.
Extended Data Fig. 8 Association between rare deleterious GLP1R variants and adiposity and glucose control, in the UK Biobank.
The rare variants were analyzed as single clusters using the MiST method adjusted for age, sex and ancestry. The significance threshold of p-value after Bonferroni correction is p = 2.3×10−3. β-arr2, β-arrestin2; BMI, Body Mass Index; HbA1c, glycated hemoglobin A1C; SE, Standard Error.
Extended Data Fig. 9 cAMP production and insulin secretion activated by WT and mutant GLP1R in INS-1 823/3 (Glp1r KO) cells.
a,b, Semaglutide concentration-response curves for cAMP accumulation (a) and insulin secretion (b) induced by SNAP-flag-tagged WT GLP1R. a, n = 3; b, n = 4. (c-i) Ex-4 response in cells expressing mutants with (c-f), defective β-arr2 recruitment and (g-i), gain-of-function phenotype. Responses are normalized to glucose-induced insulin secretion in the absence of Ex-4. Responses are normalized to glucose-induced insulin secretion in the absence of Ex-4. c-f, n = 4; g,h, n = 6; i, n = 5. All values are means ± s.e.m. Statistical significance of differences (compared with control) was determined by one-way analysis of variance and Dunnett’s post-test. Ex-4, Exendin-4; G-ctrl, Glucose control, GIP, Glucose-dependent insulinotropic polypeptide.
Supplementary information
Supplementary Information
Supplementary Tables 1, 2 and 4, Figs. 1–9 and data analysis.
Supplementary Table 3
Supplementary Table 3a–d.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, W., Liu, L., Huh, E. et al. Human GLP1R variants affecting GLP1R cell surface expression are associated with impaired glucose control and increased adiposity. Nat Metab 5, 1673–1684 (2023). https://doi.org/10.1038/s42255-023-00889-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-023-00889-6