Abstract
Hepatocellular carcinoma (HCC) is a fatal tumor which is usually diagnosed at advanced stage. Molecular targeted drugs were used recently to treat HCC, however, due to serious side effects, mainly cardiotoxicity and emergence of resistance, there is demanding to explore new chemotherapeutics. 10 novel thiazoloquinoxaline derivatives coupled with different sulfonamide moieties 4(a–j) were designed and synthesized fulfilling pharmacophoric features of VEGFR-2 inhibition. Structures of all new compounds were verified via spectral and microanalytical data. After carrying in-vitro VEGFR-2 assay for compounds 4(a–j); sulfapyridine and sulfamethoxazole derivatives 4d and 4f showed potential inhibitory effect [61.04 and 83.35 nM], respectively, comparable to standard sorafenib [51.41 nM]. Both were then further evaluated for their cytocidal activity against HepG2 cell-line and against myocardium cells using H9C2 cell-line. As a result, only sulfapyridine derivative 4d exhibited a significant inhibition of HepG2 cells viability [IC50 = 4.31 μM]. Furthermore, it showed relatively lower cytotoxic impact against normal H9C2 myocardium cells [IC50, 33.47 μM] compared to that of sorafenib [IC50, 98.07 μM]. In-vivo study was carried out to determine myocardium safety of compound 4d on irradiated mice (8 Gy). In-vivo results of sulfapyridine derivative 4d showed normal cardiac enzyme function (CK) and serum catalase activity with significant reductions in LDH, cardiac TNF-α and caspase-9 levels, alongside with its efficacy in suppressing the expression of hepatic VEGF. In conclusion, sulfapyridine derivative 4d could be considered a promising candidate as VEGFR-2 inhibitor with less myocardium side effect.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC), establishes more than 90% of liver’s primary tumor. Recently, HCC constitutes the 5th most common cancer globally. HCC is the 2nd leading cause of cancer-specific mortality in men, after lung cancer and it is usually diagnosed at advanced stage1. Chemotherapeutics, either systemic or local, along with surgical and/or radiological intervention is a tactic used to increase HCC patients’ survival rate2,3. However, and due to toxic side effects on many organs, the clinical use of traditional chemotherapeutic drugs as 5-fluorouracil, doxorubicin and cisplatin, are significantly being restricted as they owe poor specificity4.
Recently, molecular targeted drugs show higher specificity on tumor tissues with better therapeutic effects against HCC5,6. Sorafenib, a multi kinase inhibitor, was approved from FDA as a first-line molecular targeted drug for the treatment of advanced HCC since 20077,8. Sorafenib blocked tyrosine kinases receptors; Platelet Derived Growth Factor receptor (PDGFR-β), and Vascular Endothelial Growth Factor Receptor (VEGFR) in endothelial cells, which further inactivate Mitogen Activated Protein Kinase/ Extracellular signal-regulated Kinases (MAPK/ERK) pathways involved in genes transcription that promotes angiogenesis, leading finally to tumor shrinkage, it also inhibits translations of pro-survivors Mcl-1 and Bcl-2, inducing apoptosis in various cancer cells types9,10,11. In spite of sorafenib potency in HCC treatment, primary or acquired resistance develops in many patients12. It exerts some disadvantageous side effects due to normal tissues’ non-specific uptake, as well as its narrow therapeutic index which limit its future clinical application13. Moreover, FDA approved sorafenib derivatives were explored hoping to overcome all the above-mentioned challenges, but there are still obstacles and encounters to overcome, such as the moderate clinical efficacy, mechanism-related toxicities and the incidence of clinical resistance.
More importantly, the ability of sorafenib to suppress more than 15 types of tyrosine kinases could negatively induce cardiovascular complications due to enzymatic and mitochondrial functions suppression14,15,16. The primary contributing factor to these compounds' undesirable cardiovascular output is the suppression of VEGFR expressed on endothelial and smooth muscle vascular cells, as they exhibited anti-proliferative, anti-angiogenic, and apoptotic manner by acting on the MAPK/ERK pathway and producing reactive oxygen species (ROS). This could further lead to unfavorable clinical outcomes like tissue injury, vascular dysfunction, hypertension, arrhythmia, myocarditis and even myocardial infarction which could limit the treatment protocol of cancer patient and even threatened their lives17,18,19,20. These complications could be diagnosed by measuring of creatine kinase (CK), an early marker of cardiovascular diseases, which is responsible for cardiac hemodynamics via upregulation of myocardium contractility, vascular resistance and cardiac output21. Abundance release of serum lactate dehydrogenase enzyme (LDH), which is mostly found in heart and skeletal muscles tissues, is also associated with myocardium injury22.
The cardiovascular risks associated with targeted therapies are not the only risk facing cancer patients, as the inclusion of radiotherapy in the treatment protocol of HCC together with TKIs targeted therapies is well established for better therapeutic outcomes23,24. Exposure to radiation therapy is considered another risk for vascular and myocardium tissue damage with high incidence of cardiomyopathy, valvular disease, atherosclerosis and even myocardial infarction25,26. Radiation induced endothelial cells dysfunction due to oxidative damage of DNA and production of pro-inflammatory mediators such as TNF-α, IL-1, IL-6, IL-8. Also, dysregulations of lipid metabolism and changes in calcium homeostasis are frequently recognized with radiotherapy27. Moreover, radiation induced apoptosis to cardiomyocytes and cardio fibroblasts in a dose dependent matter, while apoptosis of cardiac endothelial cells occurred only after exposure to moderate to high doses of radiation28,29.
Exploring novel HCC candidates with high selectivity is often desperately required. Quinoxaline has been proven to be a key moiety in drug design and exploration. It was reported to exert wide range of biological activities30. Owing to versatility of derivatives and chemical easiness, quinoxalines are considered promising attractive targets for selective anticancer development. Recently Khandan and co-workers replaced sorafenib’s N-methylpicolinamide group with quinoxalinedione for synthesis of sorafenib derivatives with potential cytotoxic effec 31.
Sulfonamide hybrids were reported for their potential anticancer efficacy with minimal toxicities32. Sayed and co-workers coupled sulfonamide with hydrazone for anticancer activity via VEGFR-2 inhibition33. A novel series of diazepam bearing sulfonamide moieties were reported as VEGFR-2 inhibitors34.
Seeking for improving lives quality in hepatic cancer patients, this study aims at exploring novel thiazoloquinoxaline derivatives endowed with sulfonamide moieties based upon the main pharmacophoric features for inhibition of VEGFR-2, with less adverse actions on cardiomyocytes.
Rational and design
Molecular targeted therapies which inhibit tyrosine kinases were a breakthrough in treatment of HCC 35. FDA approved many drugs downregulating VEGFRs and act as angiogenesis inhibitors. Pazopanib, was developed as sorafenib derivative in 2011 for treating HCC and it showed promising effectiveness and safety profile in its clinical trials before its approval in 201236,37. After then, Santoti and co-workers reported gastrointestinal toxicities for pazopanib38. Other toxicities of pazopanib were then reported as depigmentation phenomena, proteinuria, hepatotoxicity, hypothyroidism, as well as, hypertension, thrombosis and cardiac dysfunction39. Despite the reported progress in HCC treatment modalities, there is always an urgent need for HCC drug development with new scaffolds in order to obtain new candidates of better therapeutic index and to combat the continuous emergence of cancer cells’ acquired resistance40.
Quinoxalines are promising scaffolds for exploring new VEGFR-2 inhibitors and selective anticancer drugs 41. Sulfonyl hybrids were reported for anticancer activity through VEGFR-2 inhibition with minimal toxicities32,42.
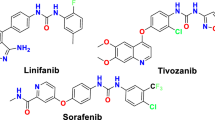
From all the above mentioned and in continuation to our efforts in exploring new candidates against HCC and as VEGFR-2 inhibitors43, herein we designed and synthesized a novel series of thiazoloquinoxaline derivatives coupled with sulfonamide moieties following the four pharmacophoric features required for occupying VEGFR-2 active binding site44. Pazopanib was taken as an example along with some bioisosteric modifications (Fig. 1). 1- Quinoxaline ring in our designed structures acts as the heteroaromatic moiety for occupying the hinge region, expecting to offer more binding interactions than pyridine ring of sorafenib and as a bioisoster to the indazole ring of pazopanib. This comes in accordance with Yu and co-workers45, who reported that substitution of sorafenib pyridine ring and urea contributed in more kinase targeting effect 2- Thiazole ring is the central aromatic linker in our designed compounds, to occupy the gate keeper region, where Chen and co-workers46 reported that 1,3-substituted ring display superior activity in VEGFR-2 inhibition, in addition to offering of more regioselectivity and stability. 3-Thiazole ring along with central imine, supposed as HBD-HBA pharmacophore, aiming to form H- bonds for inhibitory activity against VEGFR-2, thus resembling the amino pyrimidine ring of pazopanib. 4- The terminal sulfonyl phenyl ring is nominated to occupy the allosteric lipophilic pocket.
Results and discussion
Chemistry
Different sulfonamide substituents were used in our study as bioisostere to the sulfonyl terminal of pazopanib in order to investigate their interactions and effect on orientation within active binding site of VEGFR-2, which consequently may result in more residue interactions and probably more stabilization of newly synthesized compounds.
The novel target compounds were synthesized according to the pathways described in Scheme 1. A series of different substituted sulfonamides 1 (a–j) were reacted with ammoniumthiocyanate to yield thioureidobenzenesulfonamide derivatives 2 (a–j)47,48, respectively. To couple quinoxaline moiety with sulfonamide moieties, compounds 2 (a– j) were further reacted with 2,3-dichloroquinoxaline (3) to give the novel target N-(substituted)-4-(thiazolo[4,5-b]quinoxalin-2(3H)-ylideneamino)-benzenesulfonamide derivatives 4 (a–j) respectively, in considerable yields.
All the new compounds obtained were characterized and confirmed by spectral and microanalytical data.
Structure of the novel targeted N-(substituted)-4-(thiazolo[4,5-b]quinoxalin-2(3H)-ylideneamino)benzenesulfonamide derivatives 4 (a–j) were characterized by the disappearance of the NH2 and C=S bands of the thiourea in all their respective IR spectra and also the disappearance of the NH2 signals in their 1H NMR spectra exchangeable with D2O. Furthermore, 13C NMR spectra of compounds 4 (a–j) displayed characteristic signals of N=C–S at range 135.5–137.4 ppm. Signals of C=N also appeared at range 160.1–162.2 ppm. Signals of N=C–NH were shown at range 162.1–164.1 ppm. All data obtained confirmed incorporation of NH2 and C=S of thiourea intermediates 2 (a–j) in cyclization upon reaction with 2,3-dichloroquinoxaline (3) to form the novel targeted thiazoloquinoxaline derivatives 4 (a–j). While by more observation of their spectra, they showed the appearance of some significant signals of the sulfonamide series which had been used. For example, 1H-NMR spectrum of compound (4a) displayed singlet signal at 2.43 ppm, exchangeable with D2O, for NH2. 1H-NMR spectrum of compound (4b) showed the presence of singlet signal at 2.10 ppm attributed to CH3 group, while it appeared as the most shielded C at 21.2 ppm in its 13C NMR spectrum. 1H-NMR spectrum of compound (4c) exhibited singlet signals at 8.18, 8.34 ppm exchangeable with D2O for NH, NH2, respectively. while its 13C- NMR spectrum showed a downfield signal at 158.4 ppm attributed to C guanidine (NH=C–NH2) formed. 1H-NMR spectrum of compound (4d) displayed a triplet and two doublet signals at 6.71, 6.79, 7.85 ppm respectively for the CHs-pyridine ring. While 13C NMR spectrum of compound (4d) showed signals 136.4–148.4 pm for CHs pyridine ring and a signal at 152.7 ppm attributed (NH-C-pyridine). 1H-NMR spectrum of compound (4e) revealed triplet and doublet signals at 6.84 and 8.64 ppm, respectively ascribed for aromatic hydrogens of pyrimidine ring. 13C-NMR spectrum of compound (4e) displayed signals at 117.5, 157.1 ppm for CH-pyrimidine while the C-pyrimidine ring appeared at 168.5 ppm. 1H-NMR spectrum of compound (4f.) showed singlet signal at 6.12 ppm attributed to CH of isoxazole ring, while C of isoxazole CH appeared at 95.4 ppm in 13C-NMR spectrum. 1H-NMR spectrum of compound (4 g) displayed two doublet signals at 6.28, 6.86 ppm for 2 CH thiazole ring introduced. 13C-NMR spectrum of compound (4g) showed signals at 112.1, 136.3 ppm ascribed for 2CH thiazole ring. 1H-NMR spectrum of compound (4h) exhibited an upfield signal at 2.43 ppm of the methyl group. In addition to two doublet signals appeared at 6.83 and 7.97 ppm for the hydrogens of 2CH-pyrimidine ring while their Cs were displayed at 156.3, 169.2 in its respective 13C-NMR spectrum. 1H-NMR spectra of compounds (4i) and (4j) revealed upfield singlet signals at 2.33 ppm (2 CH3) and 3.75, 3.87 ppm (2 OCH3) groups, of sulfamethazine and sulfadimethoxine, respectively, while their Cs appeared at 24.0 ppm and 54.4 ppm, respectively at their corresponding 13C-NMR spectra.
In addition, the mass spectra and microanalytical data of compounds 4 (a–j) were in agreement with their postulated structures.
Biological evaluation
In-vitro assay
VEGF receptors are highly expressed in many tumor types and their prolonged presence indorsed vascular network, promoting tumor growth and metastases. VEGFR2 is the type II of tyrosine kinase transmembrane receptor and it is the principal angiogenic signaling mediator in tumors49. First, we carried in-vitro assay to compare VEGFR-2 suppression activities among the FDA approved VEGFR-2 inhibitor (Sorafenib) and our newly synthesized compounds 4 (a–j). Results showed that IC50 of sulfapyridine derivative 4d and sulfamethoxazole derivative 4f, was as low as the reference drug, sorafenib (Table. 1).
Accordingly, the two promising compounds 4d and 4f along with sorafenib had been further investigated for their cytotoxic anticancer activity against human HepG2 cell line. Results showed that sorafenib as well as sulfapyridine derivative 4d significantly decreased cell viabilities and achieved IC50 at concentrations of 2.97 μM and 4.31 μM, respectively (Table 2). The cytotoxic activity of sorafenib referred mainly to its ability to suppress the VEGFR-2, exerting its anti-angiogenic and apoptotic effects 50,51. Moreover, the suppressing potency of compound 4d on HepG2 cells proliferation (4.31 nM) was in line with earlier research, demonstrated the cytotoxic effects of sulfonamide derivatives, particularly sulfapyridine 52,53.
Based on the previous results, a further comparable screening between compound 4d and sorafenib on cardiomyocytes viability was carried out on H9C2 cell culture; results of compound 4d showed 2.93 times less cytotoxic than sorafenib (Fig. 2). Sorafenib interferes with the electron transport system by creating ROS, which led to ATP depletion, consequently causing proteins damage in the mitochondrial membrane, leading to an irreversible loss of membrane potential. This was reported to be the primary cause of sorafenib cardiotoxicity 16. Moreover, cardiac toxicity of sorafenib could be attributed also to depletion of taurine which plays a major cellular protective role as an anti-oxidant, stabilizing proteins factor and neuro-modulating agent and its lack promoted surging of NF-Kβ, P53 and P38 leading to systolic and diastolic abnormalities and even myocardial infarction 54,55.
Acute oral toxicity test
The in-vivo acute toxicity signs associated with compound 4d were also assessed. No mortalities, weight loss, or behavioral alterations were seen in mice treated with 200, 500, 800, or 1000 mg/kg of compound 4d up to14 days. Overall, compound 4d displayed a high level of safety up to 1000 mg/kg.
In-vivo assay
Cardiovascular dysfunction is a significant risk connected with exposure to γ-radiation, particularly for cancer patients who may also be given tyrosine kinase inhibitors concurrently23,24. Radiotherapy could exert many adverse cardiac reactions such as myocardial infarction, hypertension, vascular dysfunction, apoptosis and even necrosis of normal heart tissue25,56.
Owing to this risk, a demand to new targeted therapies with less cardiotoxic effect was the goal of this study. In order to evaluate the cardiac adverse consequences, we screened in the current study the cardiac function of irradiated mice after treatment with either sorafenib or compound 4d. First, CK enzyme activity was determined as it is one of the earliest diagnostic markers for cardiac dysfunction. Significant increments in CK enzymatic activity by 48.35% and 35.88% in IRR group and IRR + sorafenib group, respectively, had been recorded (Fig. 3). Even though compound 4d did not significantly lower CK level compared to the IRR non-treated group, it was intriguing to see that CK activity of compound 4d group was still not significantly different from normal values (Fig. 3).
Effect of daily treatment (3 days) of sorafenib or compound 4d [50 mg/ kg, p.o] on heart creatine kinase activity (CK, ng/mg) in 8 Gy irradiated mice. All values are plotted as mean ± S.E. * represented significance from Normal at P ≤ 0.05, # represented significance from IRR at P ≤ 0.05, s represented significance from IRR + sorafenib at P ≤ 0.05.
In this investigation, serum levels of LDH and catalase were tested to examine the state of general oxidative stress and tissue damage. LDH is found in most organ tissues and has a functional role in gluconeogenesis and DNA metabolism. Its elevation is indicative of various disorders, including cardiac illnesses, and is a helpful diagnostic marker for tissue damage, as well as energy homeostasis22,57. One of the essential antioxidant enzymes is catalase, as it breaks down cellular hydrogen peroxide to create water and oxygen, reducing oxidative stress to a significant degree, and its deficiency may contribute to the development of a variety of disorders58.
Both IRR and IRR + sorafenib groups experienced substantial increases in serum LDH by 86.66% and 106.06%, respectively, while IRR + compound 4d group exhibited a non-significant deviation from normal LDH values. Furthermore, a significant decrease in serum LDH of compound 4d treated group by 40.83%, compared to sorafenib treated group, revealed better protection from tissue damage following radiation exposure. In the same line, IRR and IRR + sorafenib groups had shown substantial decreases in serum catalase activities by 8.22% and 6.19%, respectively while the IRR + comp 4d group had normal catalase activity (Fig. 4). However, this might be explained by the antioxidant properties of sulfur contains compounds, which could reduce the production of endogenous ROS in biological tissue59,60.
Effect of daily treatment (3 days) of sorafenib or compound 4d [50 mg/ kg, p.o] on serum lactate dehydrogenase (LDH, IU/ml) and Catalase activity (IU/ml) in 8 Gy irradiated mice. All values are plotted as mean ± S.E. * represented significance from Normal at P ≤ 0.05, # represented significance from IRR at P ≤ 0.05, s represented significance from IRR + sorafenib at P ≤ 0.05.
In the introduced work, γ-irradiation induced a significant increase in cardiac TNF-α by 137.69%, compared to normal values (Fig. 5). Myocardial inflammation can develop shortly after exposure to γ- radiation and can often last for years. However, a number of variables, including adhesion molecules (I-CAM & V-CAM), interleukins, leukotrienes, and prostaglandins, contribute to the pathogenicity of these inflammatory situations 29.
Effect of daily treatment (3 days) of sorafenib or compound 4d [50 mg/ kg, p.o] on heart levels of caspase-9 (ng/mg Tp) and TNF-α (pg/ mg Tp) in 8 Gy irradiated mice. All values are plotted as mean ± S.E. * represented significance from Normal at P ≤ 0.05, # represented significance from IRR at P ≤ 0.05, s represented significance from IRR + sorafenib at P ≤ 0.05.
It was worth noting that irradiated mice given sorafenib or compound 4d demonstrated considerable reduction in cardiac TNF-α by 31.87% and 35.75%, respectively, in comparison to the control irradiated group (Fig. 5). Another study suggested that sorafenib, as a tyrosine kinase inhibitor, has a role in NF-κB inactivation via inhibiting p-38 phosphorylation, which ultimately results in the suppression of inflammatory cytokines. This attribute could explain the above findings 61.
Caspase-9 had been tested in this work as a pro-apoptotic marker. Results showed that administration of compound 4d to irradiated mice significantly decreased Caspase-9 by 20.63%, compared to control irradiated animals (Fig. 5). However, cardiac caspase-9 levels were significantly increased in the IRR and IRR + sorafenib groups, by 44.2% and 29.83%, respectively, over normal values (Fig. 5). ROS generated from radiation induced activation of Bax which disrupts the permeability of mitochondrial membrane and release cytochrome c into the cytosol, followed by activation of pro-apoptotic caspase-9, initiating caspase-3 cascade leading finally to apoptosis 29,62,63. Moreover, the apoptotic effect of sorafenib on cardiomyocytes could be attributed to the affinity of sorafenib to different tyrosine kinase receptors subtypes 63. We speculated that the sulfapyridine group incorporated in compound 4d, has a favorable effect on oxidative state as demonstrated by catalase activity, this could account for the considerable reduction of cardiomyocytes' caspase-9 and, consequently, reduced cardiac apoptotic state. This comes in accordance with Couto and co-workers 64 who reported antioxidant effect of sulfapyridine.
The suppression activity of the chosen, promising sulfapyridine derivative 4d and sorafenib on hepatic VEGF expression in irradiated mice was further estimated and compared. However, it was difficult for antiangiogenic compounds to demonstrate their full effectiveness in presence of γ-radiation, as the later encourages tissue vascularization and perfusion in order to improve oxygenation for further production of ROS needed for DNA damage in tumor cells 65. In the current study, the expression of VEGF in liver tissue was evaluated after daily administration with either compound 4d or sorafenib for 3 days following γ- irradiation (8 Gy). Results obtained showed a strong positive expression of VEGF in irradiated animals, which was promisingly suppressed upon treatment with sorafenib as well as compound 4d (Fig. 6).
Molecular modelling
In silico evaluation using molecular docking was performed in order to confirm and interpret the preliminary VEGFR-2 inhibition results for the most potent newly synthesized compound 4d. MOE software was used with human VEGFR-2 (PDB: 1ASD). Docking study was performed for the most active compound ; N-(pyridin-2-yl)-4-(thiazolo[4,5-b]quinoxalin-2(3H)-ylidene-amino) benzenesulfonamide (4d), within the active binding site of VEGFR-2 in order to have more perception about the binding mode, taking into consideration main pharmacophoric features required for binding in order to downregulate VEGFR-2 activity as reported42.
It could be noticed that structure of compound 4d occupies mainly the catalytic site of VEGFR-2 receptor, which potentially could be reason for its selectivity. It displayed better binding energy at -7.933 compared to -6.953 of pazopanib (supplementary data). It fits through the following binding interactions (Fig. 7): Quinoxaline ring occupies hinge region. Pyrimidine of the quinoxaline occupied the gatekeeper region as central aromatic linker. At the same time, it forms arene binding interaction with leu 840 in DFG domain (HBD-HBA pharmacophore). Sulfur of thiazole ring forms additional main H- bond with Glu 917 in DFG domain, Sulfonamide moiety forms additional 2- H–bonds with Glu 885 and Phe 1047. Phenyl ring of pazopanib forms only one arene interaction with Leu 840, while quinoxaline ring of 4d forms two arene interactions with Leu 840. These additional interactions are missing in pazopanib binding interactions. which could be responsible for the more stability of 4d-VEGFR-complex. Terminal pyridine ring forms the essentiale arene interaction with Asp 1046 and occupies the allosteric lipophilic pocket.
Conclusion
Based on quinoxaline scaffold and pharmacophore features needed for VEGFR-2 inhibition, herein 10 novel thiazoloquinoxaline derivatives coupled with different sulfonamide moieties 4(a-j) were designed and synthesized. Compounds 4d and 4f. demonstrated strong VEGFR-2 inhibition, and both of them had antiproliferative activity against the HepG2 cell line, indicating in-vitro potential efficacy against HCC. Moreover, compound 4d showed more in-vitro myocardium cytoprotective effect than the commonly used anti-HCC, sorafenib. The therapeutic outcomes of compound 4d and sorafenib were extended for studying in irradiated mice, where both treatments inhibited hepatic VEGF expression. The main advantage of using compound 4d in the current work is the low radiation-cardiotoxic potential, as it diminished the elevated pro-apoptotic, pro-inflammatory and oxidative mediators in myocardium of irradiated mice. Collectively, the current study revealed the efficacy of the newly synthesized N-(pyridin-2-yl)-4-(thiazolo[4,5-b]quinoxalin-2(3H)-ylideneamino) benzene-sulfonamide (4d) as down regulator to VEGFR-2 activity and a cytotoxic agent against HCC. Also, compound 4d has a significant influence on lowering radiation-induced deleterious effects on cardiac tissue, indicating its potential as a cardio-safe VEGFR-2 inhibitor.
Experimental
Chemistry
General
Uncorrected melting points were recorded using a Stuart melting point device (Stuart Scientific, Redhill, UK) and were transported in open capillary tubes. The infrared (IR) spectra of the substances were captured using an FTIR Shimadzu spectrometer (Shimadzu, Tokyo, Japan). TMS was utilised as an internal Standard and DMSO-d6 as the solvent for recording 1H NMR and 13C NMR spectra with a Bruker (400 MHz for 1H NMR and 100 MHz for 13C NMR) spectrometer. On the HP Model MS-5988, mass spectra were conducted (Hewlett Packard, Palo, Alto, Calofornia, USA). For acquiring the results of the microanalyses, a Carlo Erba 1108 Elemental Analyzer (Heraeus, Hanau, Garmany) was employed. In order to check the completion of the reaction, pre-coated SiO2 gel (HF254, 200 mesh) aluminium plates (Merk, Daemstadt, Germany) were employed as the TLC. CAS number for reagents used are mentioned between brackets following each reagent.
General procedure for the synthesis of N-(substituted)-4-thioureidobenzenesulfonamide derivatives 2 (a–j)
A mixture of benzenesulfonamide derivatives 1 (63–74-1, 144–80-9, 57–67-0, 144–83-2, 68–35-9, 723–46-6, 72–14-0, 127–79-7, 57–68-1, 122–11-2) (a-j), respectively (0.01 mol) in diluted HCl 10% (20 ml) was refluxed for 1 h. Ammoniumthiocyanate (1762–95-4) (0.01 mol, 0.76 gm) was then added and the mixture was refluxed for 5 h. The reaction mixture was cooled, poured onto ice water, the precipitated solid was filtered, washed with water, dried and crystallized from ethanol (64–17-5) to give compounds 2(a-j), respectively.
4-Thioureidobenzenesulfonamide (2a)49
N-((4-Thioureidophenyl)sulfonyl) acetamide (2b)
Yield % 75; m.p.; 181–183 °C; IR (KBr,cm-1): 3420, 3323, 3275 (NH2, 2NH), 3062 (CH arom.), 2915, 2835 (CH aliph.), 1795 (C=O), 1351 (C=S), 1210, 1115 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.06 (s, 3H, CH3), 6.46 (d, 2H, Ar–H, J = 10.88 Hz), 7.20 (d, 2H, Ar–H, J = 8.08 Hz), 8.13 (s, 2H, 2NH, D2O exch.), 8.46 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 21.8 (CH3), 123.0 (2), 129.8 (2), 136.8, 140.8, 171.7 (C=O), 181.8 (C=S). MS (m/z): 273(M+). Analysis calculated for: C9H11N3O3S2 (273): C, 39.55; H, 4.06; N, 15.37; found: C, 39.23; H, 4.33; N, 15.58.
N-Carbamimidoyl -4-thioureidobenzenesulfonamide (2c)
Yield % 86; m.p.; 220–222 °C; IR (KBr,cm-1): 3395–3145 (3NH, 2NH2), 3070 (CH arom.), 1345 (C=S), 1232, 1129 (SO2); 1H NMR (DMSO-d6, δ, ppm): 6.08 (d, 2H, Ar–H, J = 17.28 Hz), 7.49 (d, 2H, Ar–H, J = 7.48 Hz), 8.40 (s, 2H, 2NH, D2O exch.), 8.48 (s, H, NH, D2O exch.), 8.48 (s, 4H, 2NH2, D2O exch.) . 13C NMR (DMSO-d6, δ, ppm): 121.2 (2), 129.6 (2), 136.4, 139.9, 159.0 (C=NH), 181.3 (C=S). MS (m/z): 273(M+). Analysis calculated for: C8H11N5O2S2 (273): C, 35.15; H, 4.06; N, 25.62; found: C, 34.86; H, 3.87; N, 25.87.
N-(Pyridin-2-yl) -4-thioureidobenzenesulfonamide (2d)
Yield % 90; m.p.; 180–182 °C; IR (KBr,cm-1): 3320, 3284, 3160 (NH2, 2NH), 3077 (CH arom.), 1350 (C=S), 1244, 1110 (SO2); 1H NMR (DMSO-d6, δ, ppm): 6.35–6.42 (m, 3H, Ar–H), 6.49 (d, H, CH-pyridine, J = 3.16 Hz), 7.55–7.68 (m, 3H, Ar–H), 7.86 (d, H, CH-pyridine, J = 6.96 Hz), 8.64 (s, 2H, 2NH, D2O exch.), 8.66 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 109.5, 119.6, 123.3 (2), 129.9 (2), 136.4, 139.3, 143.0, 148.4 (CH-pyridine), 152.7 (NH-C-pyridine), 181.8 (C=S). MS (m/z): 308(M+). Analysis calculated for: C12H12N4O2S2 (308): C, 46.74; H, 3.92; N, 18.17; found: C, 46.46; H, 4.25; N, 17.96.
N-(Pyrimidin-2-yl) -4-thioureidobenzenesulfonamide (2e)
Yield % 82; m.p.; > 260 °C; IR (KBr,cm-1): 3330, 3270, 3120 (NH2, 2NH), 3050 (CH arom.), 1343 (C=S), 1238, 1125 (SO2); 1H NMR (DMSO-d6, δ, ppm): 6.35 (d, 2H, Ar–H, J = 8.76 Hz), 6,39 (t, H, CH-pyrimidine, J = 6.16 Hz), 7.59 (d, 2H, Ar–H, J = 5.28 Hz), 8.17 (d, 2H, 2 CH-pyrimidine, J = 10.48 Hz), 8.75 (s, 2H, 2NH, D2O exch.), 8.89 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 115.8, 121.7 (2), 129.9 (2), 136.2, 142.1, 156.2 (2), 169.3 (NH–C-pyrimidine), 181.0 (C=S). MS (m/z): 309(M+). Analysis calculated for: C11H11N5O2S2 (309): C, 42.71; H, 3.58; N, 22.64; found: C, 42.39; H, 3.79; N, 22.83.
N-(5-Methylisoxazol-3-yl) -4-thioureidobenzenesulfonamide (2f.)
Yield % 79; m.p.; > 260 °C; IR (KBr,cm-1): 3355, 3290, 3263 (NH2, 2NH), 3063 (CH arom.), 2978, 2812 (CH aliph.), 1348 (C=S), 1225, 1145 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.41 (s, 3H, CH3), 6.11 (s, H, CH-isoxazole), 6.54 (d, 2H, Ar–H, J = 4.44 Hz), 7.61 (d, 2H, Ar–H, J = 7.28 Hz), 8.30 (s, 2H, 2NH, D2O exch.), 8.87 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 12.8 (CH3), 95.3 (CH-isoxazole), 121.0 (2), 130.4 (2), 136.2, 141.4, 150.3 (NH–C-ixoxazole), 169.8 (C–CH3), 181.5 (C=S). MS (m/z): 312(M+). Analysis calculated for: C11H12N4O3S2 (312): C, 42.30; H, 3.87; N, 17.94; found: C, 42.11; H, 4.13; N, 18.26.
N-(Thiazol-2-yl)- 4-thioureidobenzenesulfonamide (2g) 50
N-(4-Methylpyrimidin-2-yl) -4-thioureidobenzenesulfonamide (2h)
Yield % 77; m.p.; > 260 °C; IR (KBr,cm-1): 3349, 3277, 3125 (NH2, 2NH), 3077 (CH arom.), 2923, 2845 (CH aliph.), 1342 (C=S), 1222, 1115 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.46 (s, 3H, CH3), 6.34 (d, 2H, Ar–H, J = 5.76 Hz), 6.92 (d, H, CH-pyrimidine, J = 8.32 Hz), 7.58 (d, 2H, Ar–H, J = 4.76 Hz), 8.41 (d, H, N–CH-pyrimidine, J = 9.56 Hz), 8.61 (s, 2H, 2NH, D2O exch.), 8.89 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 23.8 (CH3), 110.5, 123.5 (2), 129.8 (2), 134.4, 141.0 (CH-pyrimidine), 157.1 (CH-pyrimidine), 166.4 (C–CH3), 168.2 (NH–C-pyrimidine), 181.1 (C=S). MS (m/z): 323(M+). Analysis calculated for: C12H13N5O2S2 (323): C, 44.57; H, 4.05; N, 21.66; found: C, 44.24; H, 3.78; N, 21.94.
N-(4,6-Dimethylpyrimidin-2-yl) -4-thioureidobenzenesulfonamide (2i)
Yield % 83; m.p.; 220–222 °C; IR (KBr,cm-1): 3388, 3213, 3165 (NH2, 2NH), 3059 (CH arom.), 2916, 2855 (CH aliph.), 1349 (C=S), 1208, 1130 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.31 (s, 6H, 2 (CH3)), 6.39 (d, 2H, Ar–H, J = 6.68 Hz), 6.92 (s, H, CH-pyrimidine), 7.56 (d, 2H, Ar–H, J = 9.60 Hz), 8.37 (s, 2H, 2NH, D2O exch.), 8.74 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 24.1 (2) (2CH3), 110.4 (CH-pyrimidine), 123.7 (2), 129.9 (2), 135.9, 141.6, 166.5 (2) (2 (C–CH3)), 168.9 (NH–C-pyrimidine), 181.3 (C=S). MS (m/z): 337(M+). Analysis calculated for: C13H15N5O2S2 (337): C, 46.27; H, 4.48; N, 20.76; found: C, 45.92; H, 4.74; N, 21.13.
N-(2,6-Dimethoxypyrimidin-4-yl) -4-thioureidobenzenesulfonamide (2j)
Yield % 74; m.p.; 198–200 °C; IR (KBr,cm-1): 3340, 3282, 3163 (NH2, 2NH), 3086 (CH arom.), 2895, 2810 (CH aliph.), 1346 (C=S), 1210, 1123 (SO2); 1H NMR (DMSO-d6, δ, ppm): 3.87 (s, 3H, OCH3), 3.89 (s, 3H, OCH3), 5.94 (s, H, CH-pyrimidine), 6.54 (d, 2H, Ar–H, J = 9.00 Hz), 7.57 (d, 2H, Ar–H, J = 6.52 Hz), 8.23 (s, 2H, 2NH, D2O exch.), 8.68 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 54.3 (OCH3), 54.9 (OCH3), 84.6 (CH-pyrimidine), 122.2 (2), 129.9 (2), 135.4, 141.5, 162.4 (NH–C-pyrimidine), 164.0 (C–OCH3), 171.0 (C–OCH3), 181.1 (C=S). MS (m/z): 369(M+). Analysis calculated for: C13H15N5O4S2 (369): C, 42.27; H, 4.09; N, 18.96; found: C, 41.95; H, 4.24; N, 19.18.
General procedure for N-(substituted) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino)benzenesulfonamide derivatives 4 (a–j)
The N-(substituted)-4-thioureidobenzenesulfonamide derivatives 2 (a–j) (0.01 mol) were refluxed with dichloroquinoxaline 3 (1.97 gm, 0.01 mol), in ethanol (20 ml) for 8 h. The reaction mixture was cooled, then the precipitated solid was filtered, washed with ethanol, dried and crystallized from ethanol to afford compounds 4(a-j), respectively.
4-(Thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino)benzenesulfonamide (4a)
Yield % 80; m.p.; 165–167 °C; IR (KBr,cm-1): 3289, 3272 (2NH), 3080 (CH arom.), 1262, 1130 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.43 (s, 2H, NH2, D2O exch.), 7.46 (d, 2H,2CH–Ar–H, J = 5.00 Hz), 7.48–7.68 (m, 6H, Ar–H), 7.88 (s, H, NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 120.4 (2), 123.3, 123.5, 126.2, 126.4 (2), 129.9, 136.2 (N=C–S), 136.3, 136.6, 143.7 (C–SO2), 150.3, 161.4 (C=N), 163.6 (N=C–NH). MS (m/z): 357 (M+). Analysis calculated for: C15H11N5O2S2 (357): C, 50.41; H, 3.10; N, 19.59; found: C, 50.57; H, 3.43; N, 19.41.
N-((4-(Thiazolo[4,5-b]quinoxalin-2(3H)ylideneamino) phenyl) sulfonyl)-acetamide (4b)
Yield % 76; m.p.; 245–247; IR (KBr,cm-1): 3316, 3218 (2NH), 3055 (CH arom.), 2910, 2840 (CH aliph.), 1770 (C=O), 1230, 1108 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.10 (s, 3H, CH3), 7.47 (d, 2H, Ar–H, J = 7.76 Hz), 7.59–7.88 (m, 6H, Ar–H), 8.20 (s, H, NH-SO2, D2O exch.), 8.64 (s, H, NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 21.2 (CH3), 121.3 (2), 124.2, 124.4, 124.6, 129.8 (2), 130.0, 135.5 (N=C–S), 135.8, 136.2, 136.6 (C–SO2), 152.1, 161.8 (C=N), 162.1 (N=C–NH), 171.3 (C=O). MS (m/z): 399 (M+). Analysis calculated for: C17H13N5O3S2 (399): C, 51.12; H, 3.28; N, 17.53; found: C, 51.31; H, 3.54; N, 17.19.
N-Carbamimidoyl -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino) benzene-esulfonamide (4c)
Yield % 69; m.p.; > 260 °C; IR (KBr,cm-1): 3380–3150 (3NH, NH2), 3063 (CH arom.), 1220, 1111 (SO2); 1H NMR (DMSO-d6, δ, ppm): 7.44 (d, 2H, Ar–H, J = 4.40 Hz), 7.57–7.89 (m, 6H, Ar–H), 8.18 (s, 3H, 3NH, D2O exch.), 8.34 (s, 2H, NH2, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 123.4 (2), 124.5, 126.0, 127.9, 129.5 (2), 129.8, 136.2 (N=C–S), 137.6, 138.8, 141.1 (C–SO2), 151.8, 158.4 (NH=C–NH2), 162.2 (C=N), 163.1 (N=C–NH). MS (m/z): 399 (M+). Analysis calculated for: C16H13N7O2S2 (399): C, 48.11; H, 3.28; N, 24.55; found: C, 47.83; H, 3.59; N, 24.79.
N-(pyridin-2-yl) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino) benzene-sulfonamide (4d)
Yield % 72; m.p.; 196–198 °C; IR (KBr,cm-1): 3312, 3282 (2NH), 3078 (CH arom.), 1255, 1115 (SO2); 1H NMR (DMSO-d6, δ, ppm): 6.71 (t, H, CH-pyridine, J = 7.96 Hz), 6.79 (d, H, CH-pyridine, J = 4.08 Hz), 7.40–7.63 (m, 9H, Ar–H), 7.85 (d, H, CH-pyridine, J = 8.40 Hz), 8.58 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 110.1, 117.5, 121.7 (2), 124.2, 126.3, 126.4, 129.9 (2), 130.0, 135.5 (N=C–S), 136.2, 136.3 (2), 136.6 (C–SO2), 148.6, 151.7, 153.2 (C-pyridine), 161.4 (C = N), 164.1 (N=C–N). MS (m/z): 434 (M+). Analysis calculated for: C20H14N6O2S2 (434): C, 55.29; H, 3.25; N, 19.34; found: C, 55.18; H, 3.46; N, 19.09.
N-(Pyrimidin-2-yl) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino) benzene-sulfonamide (4e)
Yield % 67; m.p.; 223–225 °C; IR (KBr,cm-1): 3295, 3276 (2NH), 3055 (CH arom.), 1228, 1108 (SO2); 1H NMR (DMSO-d6, δ, ppm): 6.84 (t, H, CH-pyrimidine, J = 7.32 Hz), 7.04–7.25 (m, 8H, Ar–H), 8.64 (d, 2H, 2 CH-pyrimidine, J = 12.00 Hz), 8.83 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 117.5, 120.4 (2), 123.2, 126.2, 127.1, 129.9 (2), 129.9, 136.5 (N=C–S), 139.4, 140.5 (C-SO2), 140.8, 150.1 , 157.1 (2), 161.7 (C=N), 163.6 (N=C–NH), 168.5 (NH–C-pyrimidine). MS (m/z): 435(M+). Analysis calculated for: C19H13N7O2S2 (435): C, 52.40; H, 3.01; N, 22.51; found: C, 52.14; H, 3.32; N, 22.76.
N-(5-Methylisoxazol-3-yl) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino) benzene-sulfonamide (4f.)
Yield % 81; m.p.; 186–188 °C; IR (KBr,cm-1): 3293, 3285 (2NH), 3094 (CH arom.), 2945, 2835 (CH aliph.), 1315, 1105 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.27 (s, 3H, CH3), 6.12 (s, H, CH-isoxazole), 7.13 (d, 2H, Ar–H, J = 1.36 Hz), 7.42–7.66 (m, 6H, Ar–H), 8.17 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 12.4 (CH3), 95.4 (CH- isoxazole), 121.7 (2), 124.4, 126.0, 127.9, 129.6 (2), 129.8, 136.1 (N=C–S), 137.3, 137.6 (C–SO2), 139.1, 150.4 (C- isoxazole-NH), 150.8, 161.6 (C=N), 162.3 (N=C–NH), 169.4 (CH-CH3). MS (m/z): 438 (M+). Analysis calculated for: C19H14N6O3S2 (438): C, 52.04; H, 3.22; N, 19.17; found: C, 51.73; H, 3.45; N, 19.36.
N-(Thiazol-2-yl)- 4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino) benzene-sulfonamide (4g)
Yield % 86; m.p.; 238 -240 °C; IR (KBr,cm-1): 3295, 3279 (2NH), 3083 (CH arom.), 1285, 1110 (SO2). 1H NMR (DMSO-d6, δ, ppm): 6.28 (d, H, S-CH-thiazole, J = 7.92 Hz), 6.86 (d, H, N–CH-thiazole, J = 7.64 Hz), 7.06 (d, 2H, Ar–H, J = 7.28 Hz), 7.16–7.53 (m, 6H, Ar–H), 8.07 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 112.1 (S-CH-thiazole), 121.1 (2), 123.5, 123.6, 126.3, 129.8 (2), 129.9, 136.1 (N=C–S), 136.3 (N–CH-thiazole), 136.5, 138.4 (2), 150.3, 161.9 (C=N), 162.4 (N=C–NH), 169.8 (NH-C-thiazole). MS (m/z): 440 (M+). Analysis calculated for: C18H12N6O2S3 (441): C, 49.08; H, 2.75; N, 19.08; found: C, 49.39; H, 2.48; N, 18.91.
N-(4-Methylpyrimidin-2-yl) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino)-benzenesulfonamide (4h)
Yield % 77; m.p.; 199–201 °C; IR (KBr,cm-1): 3305, 3268 (2NH), 3089 (CH arom.), 2910, 2812 (CH aliph.), 1259, 1125 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.43 (s, 3H, CH3), 6.83 (d, H, CH-pyrimidine, J = 2.00 Hz), 7.46–7.67 (m, 8H, Ar–H), 7.97 (d, H, N–CH-pyrimidine, J = 3.28 Hz), 8.19 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 23.2 (CH3), 110.5, 121.4 (2), 123.7, 126.3, 126.4, 129.9 (2), 130.0, 135.5 (N=C–S), 137.3, 137.6 (C–SO2), 137.8, 153.20, 156.31 (CH-pyrimidine), 161.17 (C=N), 163.21 (N=C–N), 169.24 (C–CH3), 169.92 (N–CH-pyrimidine). MS (m/z): 449(M+). Analysis calculated for: C20H15N7O2S2 (450): C, 53.44; H, 3.36; N, 21.81; found: C, 53.31; H, 3.74; N, 22.02.
N-(4,6-Dimethylpyrimidin-2-yl) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylideneamino) benzenesulfonamide (4i)
Yield % 75; m.p.; 246–248 °C; IR (KBr,cm-1): 3375, 3266 (2NH), 3077 (CH arom.), 2896, 2813 (CH aliph.), 1232, 1105 (SO2); 1H NMR (DMSO-d6, δ, ppm): 2.33 (s, 6H, 2 (CH3)), 6.97 (s, H, CH-pyrimidine), 7.46–7.68 (m, 8H, Ar–H), 8.14 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 24.0 (2) (2CH3), 110.3 (CH-pyrimidine), 119.8 (2), 123.4, 123.7, 126.3, 129.8 (2), 129.9, 136.8 (N=C–S), 137.1, 137.3 (C–SO2), 137.8, 150.7, 160.1 (C–N), 163.4 (N = C–NH), 167.3 (2) (2 (C–CH3), 169.8 (C-pyrimidine). MS (m/z): 463(M+). Analysis calculated for: C21H17N7O2S2 (464): C, 54.41; H, 3.70; N, 21.15; found: C, 54.08; H, 3.91; N, 21.44.
N-(2,6-Dimethoxypyrimidin-4-yl) -4-(thiazolo[4,5-b]quinoxalin- 2(3H) -ylidene-amino)-benzenesulfonamide (4j)
Yield % 80; m.p.; 249–251 °C; IR (KBr,cm-1): 3296, 3230 (2NH), 3086 (CH arom.), 2916, 2839 (CH aliph.), 1222, 1117 (SO2); 1H NMR (DMSO-d6, δ, ppm): 3.75 (s, 3H, OCH3), 3.87 (s, 3H, OCH3), 5.92 (s, H, CH-pyrimidine), 7.41–7.75 (m, 8H, Ar–H), 8.24 (s, 2H, 2NH, D2O exch.). 13C NMR (DMSO-d6, δ, ppm): 54.4 (OCH3), 54.7 (OCH3), 82.41(CH-pyrimidine), 121.7 (2), 126.2, 126.3, 126.4, 129.9 (2), 130.0, 137.4 (N=C–S), 137.8, 138.1 (C–SO2), 138.4, 151.4, 160.8 (C=N), 162.2 (N=C–NH), 162.4 (NH–C-pyrimidine), 165.7 (CH-C–OCH3), 174.7 (N–C–OCH3). MS (m/z): 495 (M+). Analysis calculated for: C21H17N7O4S2 (496): C, 50.90; H, 3.46; N, 19.79; found: C, 50.73; H, 3.74; N, 20.12.
Biological evaluation
In vitro assay
In-vitro VEGFR-2 kinase assay
The VEGFR-2 Kinase Assay Kit [Catalog no., 40325, BPS Bioscience®, USA] is designed to measure VEGFR-2 kinase activity for screening and profiling applications using Kinase-Glo® MAX as a detection reagent. The VEGFR-2 Kinase Assay Kit comes in a convenient 96-well format, with enough purified recombinant VEGFR2 enzyme and VEGFR2 substrate. The assay protocol was carried out according to manufacturer instructions.
HepG2 & H9C2 cells culture and viability assay
Hepatocellular carcinoma (HepG2) and rat heart/myocardium (H9C2) were obtained from Nawah Scientific Inc., (Mokattam, Cairo, Egypt). Cells were maintained in DMEM media supplemented with 100 mg/mL of streptomycin (3810–74-0), 100 units/mL of penicillin (113–98-4) and 10% of heat-inactivated fetal bovine serum in humidified, 5% (v/v) CO2 atmosphere at 37 °C.
Cell viability was assessed by SRB assay 66,67. Aliquots of 100 μL cell suspension (5 × 10^3 cells) were in 96-well plates and incubated in complete media for 24 h. Cells were treated with another aliquot of 100 μL media containing compound 4d and Sorafenib (475,207–59-1) (Cipla Co.®, India) at various concentrations. After 72 h of drug exposure, cells were fixed by replacing media with 150 μL of 10% TCA (76–03-9) and incubated at 4 °C for 1 h. The TCA solution was removed, and the cells were washed 5 times with distilled water. Aliquots of 70 μL SRB (3520–42-1) solution (0.4% w/v) were added and incubated in a dark place at room temperature for 10 min. Plates were washed 3 times with 1% acetic acid (64–19-7) and allowed to air-dry overnight. Then, 150 μL of TRIS (77–86-1) (10 mM) was added to dissolve protein-bound SRB stain; the absorbance was measured at 540 nm using a BMG LABTECH®- FLUOstar Omegammicroplate reader (Ortenberg, Germany).
In vivo assay
Animals
Male Swiss albino mice (25–35 g) were obtained from the animal breeding unit of the National Center of Radiation Research and technology (NCRRT), Cairo, Egypt.
Ethical approval
This study was conducted in accordance with the regulations approved by the ethics committee standards and guidelines issued by the U.S. National Health Institutes in the National Research Center (NIH publication No. 4A/22) and the usage of experimental animals licensed by NCRRT animal care committee. The current study also adheres to the Arrive guidelines for reporting in-vivo experiments.
Acute toxicity test
For the study of the compound 4d's oral toxicity, 30 male Swiss albino mice were used. They were randomly allocated into 5 groups; the first group of mice (n = 6) received a vehicle and was used as control, while the other four groups, each received a different acute dose of compound 4d dissolved in 0.05% Tween 80 (9005–65-6) (200, 500, 800, and 1000 mg/kg p.o.). Up to 14 days after treatment, clinical toxicity symptoms (death, more than 20% weight loss, diarrhea, lethargy, tremors, and convulsions) were noted 68.
Irradiation
Whole body γ-irradiation of mice was performed at the NCRRT using the Gamma Cell-40 biological irradiator with a Cesium-137 irradiator unit, produced by the atomic energy of Canada limited (Sheridan science and technology park, Mississauga, Ontario, Canada). Mice were exposed to an acute dose of 8 Gy according to Yoo et al. 69.
Experimental design
A total 32 mice were randomly allocated into 4 groups as follows: 1- control normal [Control] mice received vehicle (10% DMSO). 2- Irradiated mice (8 Gy) received vehicle [IRR]. 3- Irradiated mice treated with Sorafenib (50 mg/ kg body wt./ p.o)70 for 3 successive days [IRR + sorafenib]. 4- Irradiated mice treated with Compound 4d (50 mg/ kg body wt./ p.o) for 3 successive days, [IRR + cmp 4d]. After 3 days, all animals were anesthetized with ketamine (80 mg/ kg), blood samples were collected via retro-orbital plexus, and then serum samples were separated after centrifugation. Heart and liver samples were dissected and rapidly washed with cool saline then heart samples were stored at -80 °C till assay time. One lobe of liver was embedded in 10% formalin for immunohistochemistry assay.
ELISA assay of heart creatine kinase (CK), tumor necrosis factor-alpha (TNF-α) and Caspase-9
Heart tissues of mice were weighted and minced then homogenized in cold fresh lysis buffer, the homogenate was centrifuged for 5 min at 4000 rpm, and then the supernatant was used for assay. Cardiac contents of CK, TNF-α and Caspase-9 were measured according to manufacturer instructions using CK ELISA kit [catalog no. EK730963, AFG Bioscience®, USA], TNF-α ELISA kit [catalog no. SEA626Ra, Cloud-Clone Corp®, USA] and Caspase-9 ELISA kit [catalog no. E-EL-R02054, Elabsciense®, USA], respectively.
Assessment of serum LDH and catalase
Serum LDH was estimated kinetically using Spectrum® coloring assay Kit [Egyptian company for biotechnology, SAE®, Egypt]. It was carried according to manufacturer's instructions; the values were expressed as IU/ml.
Serum catalase was assessed colorimetrically in all groups according to manufacturer instructions using coloring endpoint kit [Biodiagnostic co.®, Egypt].
Expression of VEGF by immunohistochemistry
Immunohistochemical study was carried out on paraffin tissue sections prepared for the detection VEGF, liver tissue sections were incubated separately with anti- VEGF (Gene Tex, USA). The expression of each marker was visualized by the chromogen 3,3-diaminobenzidine tetrahydrochloridem (DAB, Sigma Chemical Co.).
Statistical analysis
Data were expressed as mean ± standard error (SE). Statistical analysis was carried out using one-way analysis of variance (ANOVA) test followed by Tukey- Kramer multiple comparison’s test. GraphPad Prism® software package was used to carry out all statistical tests. Figures were drawn using the Microsoft Excel program.
Molecular modelling
The molecular docking study for compound 4d 's structure and pazopanib was carried out using the Molecular Operating Environment (MOE) software, version 2014.090. The energy of the compound was minimised using the HamiltonianForce FieldMMFF94x. The partial charges of the forcefield were computed. The conformational stochastic of compound 4d was analysed using the default settings. The X-ray crystal structure of VEGFR-2 in complex with sorafenib as ligand (PDB ID: 4ASD) was obtained from http://www.rscb.org/pdb. The Protein Data Bank (pdb) protein–ligand complex was prepared for docking. After threedimensional protonation of the enzyme, the system was optimised. Protein repeated chains and co-crystallized water molecules were removed. The binding pocket was determined and isolated, and the backbone was then hidden. The results were validated by redocking the sorafenib ligand. MOEDOCK was used to determine the most stable conformers' flexible docking of the ligandrigid receptor. The alpha triangle placement method and London dG as a function were used for scoring. Using the same scoring function, forcefield refinement was applied to the obtained poses. 50 of the most stable docking models of the ligand with the highest scored conformation were retained.
Data availability
The datasets generated and/or analysed during the current study are available in the supplementary files.
References
Ferlay, J. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359-386. https://doi.org/10.1002/ijc.29210 (2015).
Song, M. J. Hepatic artery infusion chemotherapy for advanced hepatocellular carcinoma. World J. Gastroenterol. 21, 3843–3849. https://doi.org/10.3748/wjg.v21.i13.3843 (2015).
Parikh, N. D., Cuneo, K. & Mendiratta-Lala, M. Radiation therapies for the treatment of hepatocellular carcinoma. Clin. Liver Dis. (Hoboken) 17, 341–346. https://doi.org/10.1002/cld.1060 (2021).
Hourdequin, K. C., Schpero, W. L., McKenna, D. R., Piazik, B. L. & Larson, R. J. Toxic effect of chemotherapy dosing using actual body weight in obese versus normal-weight patients: A systematic review and meta-analysis. Ann. Oncol. 24, 2952–2962. https://doi.org/10.1093/annonc/mdt294 (2013).
Sowa, Y. & Sakai, T. Development of novel epigenetic molecular-targeting agents. Nihon Rinsho 73, 1263–1267 (2015).
Wang, H. T. & Xia, M. A meta-analysis of efficacy and safety of sorafenib versus other targeted agents for metastatic renal cell carcinoma. Medicine (Baltimore) 98, e13779. https://doi.org/10.1097/md.0000000000013779 (2019).
Llovet, J. M. et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390. https://doi.org/10.1056/NEJMoa0708857 (2008).
Zhang, P. et al. Cost-effectiveness of sorafenib as a first-line treatment for advanced hepatocellular carcinoma. Eur. J. Gastroenterol. Hepatol. 27, 853–859. https://doi.org/10.1097/meg.0000000000000373 (2015).
Gauthier, A. & Ho, M. Role of sorafenib in the treatment of advanced hepatocellular carcinoma: An update. Hepatol. Res. 43, 147–154. https://doi.org/10.1111/j.1872-034X.2012.01113.x (2013).
Jensen, B. C. et al. Effects of the kinase inhibitor sorafenib on heart, muscle, liver and plasma metabolism in vivo using non-targeted metabolomics analysis. Br. J. Pharmacol. 174, 4797–4811. https://doi.org/10.1111/bph.14062 (2017).
Qin, S. et al. Recent advances on anti-angiogenesis receptor tyrosine kinase inhibitors in cancer therapy. J. Hematol. Oncol. 12, 27. https://doi.org/10.1186/s13045-019-0718-5 (2019).
Chen, J. et al. Potential molecular, cellular and microenvironmental mechanism of sorafenib resistance in hepatocellular carcinoma. Cancer Lett. 367, 1–11. https://doi.org/10.1016/j.canlet.2015.06.019 (2015).
Wang, X. Q. et al. Bioavailability and pharmacokinetics of sorafenib suspension, nanoparticles and nanomatrix for oral administration to rat. Int. J. Pharm. 419, 339–346. https://doi.org/10.1016/j.ijpharm.2011.08.003 (2011).
Wu, C. & Shemisa, K. Sorafenib-associated heart failure complicated by cardiogenic shock after treatment of advanced stage hepatocellular carcinoma: A clinical case discussion. Case Rep. Cardiol. 2017, 7065759. https://doi.org/10.1155/2017/7065759 (2017).
Adhikari, A. et al. Anticancer drug-induced cardiotoxicity: Insights and pharmacogenetics. Pharmaceuticals (Basel) https://doi.org/10.3390/ph14100970 (2021).
Bouitbir, J., Panajatovic, M. V. & Krähenbühl, S. Mitochondrial toxicity associated with imatinib and sorafenib in isolated rat heart fibers and the cardiomyoblast H9c2 cell line. Int. J. Mol. Sci. https://doi.org/10.3390/ijms23042282 (2022).
Florescu, M., Cinteza, M. & Vinereanu, D. Chemotherapy-induced Cardiotoxicity. Maedica (Bucur) 8, 59–67 (2013).
Yang, B. & Papoian, T. Preclinical approaches to assess potential kinase inhibitor-induced cardiac toxicity: Past, present and future. J. Appl. Toxicol. 38, 790–800. https://doi.org/10.1002/jat.3584 (2018).
Neves, K. B., Montezano, A. C., Lang, N. N. & Touyz, R. M. Vascular toxicity associated with anti-angiogenic drugs. Clin. Sci. (Lond.) 134, 2503–2520. https://doi.org/10.1042/cs20200308 (2020).
Camarda, N., Travers, R., Yang, V. K., London, C. & Jaffe, I. Z. VEGF Receptor inhibitor-induced hypertension: Emerging mechanisms and clinical implications. Curr. Oncol. Rep. 24, 463–474. https://doi.org/10.1007/s11912-022-01224-0 (2022).
Brewster, L. M., Haan, Y. C., Zwinderman, A. H., van den Born, B. J. & van Montfrans, G. A. CK (creatine kinase) is associated with cardiovascular hemodynamics: The HELIUS study. Hypertension 76, 373–380. https://doi.org/10.1161/hypertensionaha.120.14675 (2020).
Zhu, W. et al. Serum level of lactate dehydrogenase is associated with cardiovascular disease risk as determined by the framingham risk score and arterial stiffness in a health-examined population in China. Int. J. Gen. Med. 15, 11–17. https://doi.org/10.2147/ijgm.S337517 (2022).
Rim, C. H., Park, S., Shin, I. S. & Yoon, W. S. Is the concurrent use of sorafenib and external radiotherapy feasible for advanced hepatocellular carcinoma? A meta-analysis. Cancers (Basel) https://doi.org/10.3390/cancers13122912 (2021).
Wild, A. T. et al. Concurrent versus sequential sorafenib therapy in combination with radiation for hepatocellular carcinoma. PLoS ONE 8, e65726. https://doi.org/10.1371/journal.pone.0065726 (2013).
Koutroumpakis, E. et al. Modern radiotherapy and risk of cardiotoxicity. Chemotherapy 65, 65–76. https://doi.org/10.1159/000510573 (2020).
Bergom, C. et al. Past, present, and future of radiation-induced cardiotoxicity: Refinements in targeting, surveillance, and risk stratification. JACC CardioOncol 3, 343–359. https://doi.org/10.1016/j.jaccao.2021.06.007 (2021).
Halle, M., Hall, P. & Tornvall, P. Cardiovascular disease associated with radiotherapy: Activation of nuclear factor kappa-B. J. Intern. Med. 269, 469–477. https://doi.org/10.1111/j.1365-2796.2011.02353.x (2011).
Rousseau, M. et al. RhoA GTPase regulates radiation-induced alterations in endothelial cell adhesion and migration. Biochem. Biophys. Res. Commun. 414, 750–755. https://doi.org/10.1016/j.bbrc.2011.09.150 (2011).
Tapio, S. Pathology and biology of radiation-induced cardiac disease. J. Radiat. Res. 57, 439–448. https://doi.org/10.1093/jrr/rrw064 (2016).
Sharma, A., Deep, A., Marwaha, M. G. & Marwaha, R. K. Quinoxaline: A chemical moiety with spectrum of interesting biological activities. Mini Rev. Med. Chem. 22, 927–948. https://doi.org/10.2174/1389557521666210927123831 (2022).
Khandan, M., Sadeghian-Rizi, S., Khodarahmi, G. & Hassanzadeh, F. Synthesis and cytotoxic evaluation of some novel quinoxalinedione diarylamide sorafenib analogues. Res. Pharm. Sci. 13, 168–176. https://doi.org/10.4103/1735-5362.223802 (2018).
Rakesh, K. P. et al. Recent development of sulfonyl or sulfonamide hybrids as potential anticancer agents: A key review. Anticancer Agents Med. Chem. 18, 488–505. https://doi.org/10.2174/1871520617666171103140749 (2018).
Sayed, A. M. et al. Design, synthesis, molecular docking, in silico ADMET profile and anticancer evaluations of sulfonamide endowed with hydrazone-coupled derivatives as VEGFR-2 inhibitors. Bioorg. Chem. 108, 104669. https://doi.org/10.1016/j.bioorg.2021.104669 (2021).
Saleh, N. M., El-Gaby, M. S. A., El-Adl, K. & Abd El-Sattar, N. E. A. Design, green synthesis, molecular docking and anticancer evaluations of diazepam bearing sulfonamide moieties as VEGFR-2 inhibitors. Bioorg. Chem. 104, 104350. https://doi.org/10.1016/j.bioorg.2020.104350 (2020).
Stotz, M. et al. Molecular targeted therapies in hepatocellular carcinoma: Past, present and future. Anticancer Res. 35, 5737–5744 (2015).
Yau, T. et al. Phase I dose-finding study of pazopanib in hepatocellular carcinoma: Evaluation of early efficacy, pharmacokinetics, and pharmacodynamics. Clin. Cancer Res. 17, 6914–6923. https://doi.org/10.1158/1078-0432.Ccr-11-0793 (2011).
Zhu, X. D. et al. Antiangiogenic effects of pazopanib in xenograft hepatocellular carcinoma models: Evaluation by quantitative contrast-enhanced ultrasonography. BMC Cancer 11, 28. https://doi.org/10.1186/1471-2407-11-28 (2011).
Santoni, M. et al. Risk of gastrointestinal events with sorafenib, sunitinib and pazopanib in patients with solid tumors: A systematic review and meta-analysis of clinical trials. Int. J. Cancer 135, 763–773. https://doi.org/10.1002/ijc.28544 (2014).
Boudou-Rouquette, P. et al. Clinical pharmacology, drug–drug interactions and safety of pazopanib: A review. Expert Opin. Drug Metab. Toxicol. 12, 1433–1444. https://doi.org/10.1080/17425255.2016.1225038 (2016).
Chen, Y.-F. & Fu, L.-W. Mechanisms of acquired resistance to tyrosine kinase inhibitors. Acta Pharmaceutica Sinica B 1, 197–207 (2011).
El Newahie, A. M. S. et al. Design and synthesis of new quinoxaline derivatives as anticancer agents and apoptotic inducers. Molecules https://doi.org/10.3390/molecules24061175 (2019).
Ghorab, M. M. et al. Novel sulfonamide derivatives carrying a biologically active 3,4-dimethoxyphenyl moiety as VEGFR-2 inhibitors. Chem. Pharm. Bull. (Tokyo) 64, 1747–1754. https://doi.org/10.1248/cpb.c16-00614 (2016).
El-Gazzar, M. G., El-Hazek, R. M., Zaher, N. H. & El-Ghazaly, M. A. Design and synthesis of novel pyridazinoquinazoline derivatives as potent VEGFR-2 inhibitors: In vitro and in vivo study. Bioorg. Chem. 92, 103251. https://doi.org/10.1016/j.bioorg.2019.103251 (2019).
Lee, K. et al. Pharmacophore modeling and virtual screening studies for new VEGFR-2 kinase inhibitors. Eur. J. Med. Chem. 45, 5420–5427. https://doi.org/10.1016/j.ejmech.2010.09.002 (2010).
Yu, P. et al. NSK-01105 inhibits proliferation and induces apoptosis of prostate cancer cells by blocking the Raf/MEK/ERK and PI3K/Akt/mTOR signal pathways. Tumour Biol. 36, 2143–2153. https://doi.org/10.1007/s13277-014-2824-x (2015).
Chen, F. et al. Evolution in medicinal chemistry of sorafenib derivatives for hepatocellular carcinoma. Eur. J. Med. Chem. 179, 916–935. https://doi.org/10.1016/j.ejmech.2019.06.070 (2019).
Erigür, E. C., Altuğ, C., Angeli, A. & Supuran, C. T. Design, synthesis and human carbonic anhydrase I, II, IX and XII inhibitory properties of 1,3-thiazole sulfonamides. Bioorg. Med. Chem. Lett. 59, 128581. https://doi.org/10.1016/j.bmcl.2022.128581 (2022).
Gaballah, S. T., Amer, H., Hofinger-Horvath, A., Al-Moghazy, M. & Hemida, M. Synthesis, antimicrobial, and docking investigations of remarkably modified sulfathiazole derivatives. Egypt. J. Chem. 63, 171–184 (2020).
Fontanella, C. et al. Clinical advances in the development of novel VEGFR2 inhibitors. Ann. Transl. Med. 2, 123. https://doi.org/10.3978/j.issn.2305-5839.2014.08.14 (2014).
El-Mageed, A. Cytotoxicity of sorafenib in human hepatocellular carcinoma cells. Al-Azhar J. Pharm. Sci. 51, 31–40 (2015).
Chen, J. C. et al. Enhanced cytotoxicity of human hepatocellular carcinoma cells following pretreatment with sorafenib combined with trichostatin A. Oncol. Lett. 17, 638–645. https://doi.org/10.3892/ol.2018.9582 (2019).
Ghorab, M. M., Ragab, F. A., Heiba, H. I., El-Gazzar, M. G. & El-Gazzar, M. G. M. Novel thioureido-benzenesulfonamide derivatives with enaminone linker as potent anticancer, radiosensitizers and VEGFR2 inhibitors. Bioorg. Med. Chem. Lett. 28, 1464–1470. https://doi.org/10.1016/j.bmcl.2018.03.089 (2018).
Sahyon, H. A. E., Ramadan, E. N. M., Althobaiti, F. & Mashaly, M. M. A. Anti-proliferative effects of the combination of Sulfamethoxazole and Quercetin via caspase3 and NFkB gene regulation: An in vitro and in vivo study. Naunyn Schmiedebergs Arch Pharmacol. 395, 227–246. https://doi.org/10.1007/s00210-021-02174-3 (2022).
Das, J., Ghosh, J., Manna, P. & Sil, P. C. Taurine suppresses doxorubicin-triggered oxidative stress and cardiac apoptosis in rat via up-regulation of PI3-K/Akt and inhibition of p53, p38-JNK. Biochem. Pharmacol. 81, 891–909. https://doi.org/10.1016/j.bcp.2011.01.008 (2011).
Ramila, K. C. et al. Role of protein phosphorylation in excitation-contraction coupling in taurine deficient hearts. Am. J. Physiol. Heart Circ. Physiol. 308, H232-239. https://doi.org/10.1152/ajpheart.00497.2014 (2015).
Magné, N. et al. Tomorrow’s targeted therapies in breast cancer patients: What is the risk for increased radiation-induced cardiac toxicity?. Crit. Rev. Oncol. Hematol. 76, 186–195. https://doi.org/10.1016/j.critrevonc.2010.01.012 (2010).
Feng, Y. et al. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 7, 6124–6136. https://doi.org/10.1002/cam4.1820 (2018).
Nandi, A., Yan, L. J., Jana, C. K. & Das, N. Role of catalase in oxidative stress- and age-associated degenerative diseases. Oxid. Med. Cell Longev. 2019, 9613090. https://doi.org/10.1155/2019/9613090 (2019).
Göçer, H., Akıncıoğlu, A., Öztaşkın, N., Göksu, S. & Gülçin, İ. Synthesis, antioxidant, and antiacetylcholinesterase activities of sulfonamide derivatives of dopamine-related compounds. Arch Pharm. (Weinheim) 346, 783–792. https://doi.org/10.1002/ardp.201300228 (2013).
Egbujor, M. C., Garrido, J., Borges, F. & Saso, L. Sulfonamide a valid scaffold for antioxidant drug development. Mini-Rev. Org. Chem. 20, 190–209 (2023).
Li, X. et al. Sorafenib inhibits LPS-induced inflammation by regulating Lyn-MAPK-NF-kB/AP-1 pathway and TLR4 expression. Cell Death Discov. 8, 281. https://doi.org/10.1038/s41420-022-01073-7 (2022).
Park, M. S., De Leon, M. & Devarajan, P. Cisplatin induces apoptosis in LLC-PK1 cells via activation of mitochondrial pathways. J. Am. Soc. Nephrol. 13, 858–865. https://doi.org/10.1681/asn.V134858 (2002).
Grabowska, M. E., Chun, B., Moya, R. & Saucerman, J. J. Computational model of cardiomyocyte apoptosis identifies mechanisms of tyrosine kinase inhibitor-induced cardiotoxicity. J. Mol. Cell Cardiol. 155, 66–77. https://doi.org/10.1016/j.yjmcc.2021.02.014 (2021).
Couto, D. et al. Scavenging of reactive oxygen and nitrogen species by the prodrug sulfasalazine and its metabolites 5-aminosalicylic acid and sulfapyridine. Redox. Rep. 15, 259–267. https://doi.org/10.1179/135100010x12826446921707 (2010).
Goedegebuure, R. S. A., de Klerk, L. K., Bass, A. J., Derks, S. & Thijssen, V. Combining radiotherapy with anti-angiogenic therapy and immunotherapy; a therapeutic triad for cancer?. Front. Immunol. 9, 3107. https://doi.org/10.3389/fimmu.2018.03107 (2018).
Skehan, P. et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J. Natl. Cancer Inst. 82, 1107–1112. https://doi.org/10.1093/jnci/82.13.1107 (1990).
Allam, R. M. et al. Fingolimod interrupts the cross talk between estrogen metabolism and sphingolipid metabolism within prostate cancer cells. Toxicol. Lett. 291, 77–85. https://doi.org/10.1016/j.toxlet.2018.04.008 (2018).
In, O. Acute oral toxicity-Acute oral toxic class method. Guideline 423. Eleventh Addendum to the OECD Guidelines for the Testing (2001).
Yoo, G. S. et al. Radiation-induced abscopal effect and its enhancement by programmed cell death 1 blockade in the hepatocellular carcinoma: A murine model study. Clin. Mol. Hepatol. 27, 144–156. https://doi.org/10.3350/cmh.2020.0095 (2021).
Wei, J. et al. Baicalin enhanced oral bioavailability of sorafenib in rats by inducing intestine absorption. Front. Pharmacol. 12, 761763. https://doi.org/10.3389/fphar.2021.761763 (2021).
Acknowledgements
The authors would like to thank the staff members of the γ‐irradiation unit at the National Center for Radiation Research and Technology (NCRRT) for their cooperation in carrying out the irradiation.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
All authors contribute equally in this manuscript
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El-Hazek, R.M.M., Zaher, N.H., El-Gazzar, M.G.M. et al. Novel VEGFR2 inhibitors with thiazoloquinoxaline scaffold targeting hepatocellular carcinoma with lower cardiotoxic impact. Sci Rep 13, 13907 (2023). https://doi.org/10.1038/s41598-023-40832-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40832-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.