Abstract
The paper deals with the evaluation of the performance of an existing and previously validated CT based radiomic signature, developed in oropharyngeal cancer to predict human papillomavirus (HPV) status, in the context of anal cancer. For the validation in anal cancer, a dataset of 59 patients coming from two different centers was collected. The primary endpoint was HPV status according to p16 immunohistochemistry. Predefined statistical tests were performed to evaluate the performance of the model. The AUC obtained here in anal cancer is 0.68 [95% CI (0.32–1.00)] with F1 score of 0.78. This signature is TRIPOD level 4 (57%) with an RQS of 61%. This study provides proof of concept that this radiomic signature has the potential to identify a clinically relevant molecular phenotype (i.e., the HPV-ness) across multiple cancers and demonstrates potential for this radiomic signature as a CT imaging biomarker of p16 status.
Similar content being viewed by others
Introduction
Human papillomavirus (HPV) is a common virus that can lead to certain types of cancer later in life. Several studies have shown that HPV status of cancer patients plays an important role in treatment outcomes1,2. In particular, HPV + patients with oropharyngeal cancer present a markedly better prognosis and improved response to conventional radiotherapy3. Also, patients with anal cancer show a better prognosis and response to therapy when HPV + 4. Widely accepted methods for detection of HPV infection are in situ hybridization for viral DNA, HPV DNA or RNA PCR, and immunohistochemical investigation of the level of p16 expression, which strongly correlates with HPV infection1,5. However, these methods are time consuming and require biological patient samples. Image data analysis represents a novel non-invasive method of delivering reliable and quality results from standard-of-care medical images. The extraction of clinically relevant information from medical imaging is called radiomics. Radiomics is a rapidly emerging field, introduced in 2012, which concerns with the high-throughput mining of large amounts of quantitative features, derived from medical image such as CT/MRI/PET scans6,7. Radiomics is promising within decision support systems for precision medicine and its potential to predict p16 status in head and neck cancer has been recognized8. Previous studies have reported radiologic differences between p16 positive and negative cases in terms of qualitative radiologist’s readout of perfusion CT9. Furthermore, exploratory radiomic studies have shown that heterogeneity of image-based density is potentially associated with p16 status in oropharyngeal squamous cell carcinoma (OPSCC)10. Most of the studies investigating imaging phenotypes of tumors are based on single center data, which introduce bias to a model and limits its applicability. In particular, factors such as CT scanner, tube voltage, tube current, reconstruction kernel and contrast agent influence the results of quantitative analyses11. In this independent external validation study, we further investigate if a quantitative CT-based radiomic approach can objectively identify the p16 status of anal cancer in addition to OPSCC12, by validating a radiomic signature on patient data from two different institutions. With this study we aim to provide a proof of concept for radiomics to derive molecular information from standard medical images. Moreover, we demonstrate that the already developed and validated radiomics signature for head and neck cancer can also be useful in identification of HPV status of anal cancer patients.
Materials and methods
Patient cohort
Two independent cohorts, for a total of 59 anal cancer patients treated with curative intent by surgery and/or radiation therapy with/without concurrent chemotherapy, were collected from University Hospital of Liège (CHU) (n = 18) and St James's Hospital (TCD). (n = 41) The patients cohort is composed of 12% of stage I, 40% of stage II, 45 of stage III and 3 of stage IV anal cancer. The HPV status of the patients was inferred from p16 immunohistochemistry status (54 + and 5−). All patients underwent pre-treatment CT imaging, according to the standard of care of the treating institution. The gross primary tumor volume (GTV) was manually segmented for each patient by experienced radiologists. Institutional review board approval was obtained from the Ethics committees of the University Hospital of Liege, Trinity College, Cork University Hospital and St. Luke Hospital. The need for informed consent was waived from the same ethics committees since the data were anonymized and retrospectively collected. The present work as been conducted in accordance with the Declaration of Helsinki.
Radiomics analysis
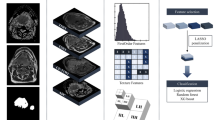
Radiomics is based on the hypothesis that quantitative analysis of medical image data via automatic or semi-automatic software can provide more and better information than that of a physician13. The schematic representation in Fig. 1 depicts the radiomics workflow applied in this study.
The radiomics workflow is divided in four mains steps. First the imaging data are collected, and eventually preprocessed, dividing them in different set (training validation and testing): then the region of interest (ROI) is segmented and annotated manually or (semi) automatically. From this region of interest handcrafted radiomics features are extracted, and divided into Size, Shape, Texture and Intensity features. The radiomics features are then used to train the AI model and the performances are validated in the test set and additionally in an external validation set.
In the external validation study presented here, prior to analysis, all images were resampled to isotropic voxels of 2 mm, using linear interpolation. A total of 37 radiomics features were calculated from five groups: tumor intensity, shape, texture, Wavelet and Laplacian of Gaussian. All features were extracted using the RadiomiX® software (OncoRadiomics SA, Liege, Belgium). Feature descriptions and mathematical definitions can be found in the literature14. Details for the development of the radiomics signature for HPV status in OPSCC are reported elsewhere12. Briefly, the signature was built using regularized logistic regression and showed an AUC of 0.763 [95% CI (0.687–0.839)] in development. The signature class predictions were made with a probability cut-off of 0.5.
Statistical analysis
The signature performances have been evaluated in terms of Precision (1), Sensitivity (2), F1 score (3), Specificity and AUC.
Assuming a 85% prevalence of HPV positive anal cancer patients15, we calculated also Negative predictive value (NPV) and Positive predictive value (PPV). The signature was also evaluated according to the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD)16 and the Radiomics Quality Score (RQS)14. Statistical analysis was performed in R (R Foundation for Statistical Computing; v. 3.3.3).
Results
The model performance on the validation data set of anal cancer patients (n = 59) presents an AUC of 0.681 [95% CI (0.328–1.000)]. The ROC plot and the confusion matrix for the validation set are shown in Fig. 2. Classification performance plot, assuming a disease prevalence of 85%, is reported in Fig. 3.
The signature showed discriminative power also for anal cancer, predicting the probability of p16 + (HPV +), or better the “HPV-ness” of the tumor. Table 1 reports the performance parameters of the external validation in the anal cancer cohort compared to the original validation in OPSSC.
The RQS of the original signature, developed in OPSSC, is 50%. The additional validation with an external dataset, brings the RQS of the signature to 61%. Additionally, the original signature for OPSSC was TRIPOD level 2a (56%) while the new proposed model is a TRIPOD level 4 (57%) signature (See Supplementary Information).
Discussion
In this study we validated a CT based radiomic signature to predict the p16 status of anal cancer patients. The study provides a proof of concept that molecular information can be inferred from standard medical images by means of radiomics. Previous exploratory radiomic studies that indicated a correlation between HPV infection and heterogeneity of imaging-based tumor density have focused on head and neck cancer9,17 and were performed without validation, or only using data from a single institution, for both model development and validation. This is a major problem for the reliability of prediction models based on radiomics signature7. Previously published studies report that HPV positive tumors are more homogenous in CT density18,19. The homogeneity in turn can represent one of the reasons why HPV + tumors have a better therapeutic outcome and prognosis20. We show that the signature which was previously developed and validated in head and neck cancer, also shows discrimination power for anal cancer patients. This suggests that it would be possible to cautiously generalize these findings on tumor radiomics features beyond OPSCC. Furthermore, this study provides an additional insight into p16 (HPV) imaging phenotype. We observed that the p 16 positive tumors are characterized by lower contrast uptake, lower minimum density, and higher changes in the intensity of adjacent voxels. It is worth noting that is not possible to distinguish between p16 + and p16- anal tumors by visual inspection of the CT scan alone, and it has been proven a difficult task also for oropharyngeal cancer21. The difference in prevalence of anal cancer is important to consider in order to assess the performance of the signature. The prevalence of HPV positive anal cancers is 85%15 and needs to be taken into account in the implementation of the signature as decision support tool for clinicians. The main aim of such signature applied to anal cancer would be the identification of HPV-negative tumors, which can be assessed using the specificity and the NPV (Fig. 2).
Including data from different institutions introduces variability in image acquisition and reconstruction, which affects radiomic features22. Besides variability in CT imaging, demographic differences also have to be considered. Developing a model on a single cohort is unlikely to capture the diversity that exists across data from different centers, resulting in a model with poor generalizability, unsuited for routine clinical use. Since the original radiomics signature was developed on a heterogeneous dataset, coming from 5 different institutions, the robustness and the widespread application was greatly improved. The patient cohort for the anal cancer validation was also acquired from two different centers with different scanners and image acquisition parameters (Table S1). Even considering the small number of data available, the model has retained enough discriminating power to correctly classify 70% of the anal cancer patients.
Another open question is related to the rate of false positive in the immunohistochemical test for 16p. Part of the OPSCC patients that test positive for p16 immunohistochemistry are in fact HPV DNA negative23. This is also true for anal cancer patients24. It is worth noting that HPV + status does not imply per definition p16 + status and vice versa. Prognosis of HPV + /p16 + is therefore not the same as HPV − /p16 + , most likely because in the latter case tumors are not HPV induced. Furthermore, model class predictions (i.e. predicting either HPV positive or HPV negative), were made with a probability cut-off of 0.5, meaning that the costs for false-positives and false-negatives were considered equal. In clinical practice, false positive have a much higher cost in term of patient management and healthcare quality and should be minimized. To achieve a clinically acceptable level of accuracy, further development and validation would be needed, including HPV DNA testing. The radiomics HPV prediction model, while reliable, should not supersede the traditional clinical decision-making approach, based on universally accepted methodology. However, radiomics has the potential to serve as a time-efficient, complementary method for HPV screening, also, for non-oropharyngeal SCCs25,26. Radiomics approaches can be used to perform retrospective biomarker studies on HPV status where tissue samples are not available or in countries where HPV testing is not routinely performed. Furthermore, additional improvement in inferring tumor HPV status may be achieved when combining radiomics with clinical features27.
Conclusion
The discriminating power of the radiomics signature for p16 status determination, developed for OPSCC, was also validated for anal cancer patients. These preliminary but encouraging results may pave the road for further generalization of CT image features of HPV related tumors. The use of a larger cohort with p16 and HPV DNA test data, as well as the inclusion of other possible cancer types which shown a correlation with HPV status would be instrumental in this regard.
Data availability
The datasets used and/or analyzed during the current study is available from the corresponding author on reasonable request.
References
Hasegawa, Y. et al. Human papilloma virus in non-small cell lung cancer in never smokers: A systematic review of the literature. Lung. Cancer 83, 8–13. https://doi.org/10.1016/j.lungcan.2013.10.002 (2014).
Clifford, G. 5—Pooled analysis of HPV infection in paired anal and cervical samples, by HIV status. Papillomavirus Res. 5, s2-3. https://doi.org/10.1016/j.pvr.2018.07.006 (2018).
Yan, F. et al. The evolution of care of cancers of the head and neck region: State of the science in 2020. Cancers (Basel) https://doi.org/10.3390/cancers12061543 (2020).
Mai, S. et al. Prognostic relevance of hpv infection and p16 overexpression in squamous cell anal cancer. Int. J. Radiat. Oncol. 93, 819–27. https://doi.org/10.1016/j.ijrobp.2015.08.004 (2015).
Kiyuna, A. et al. High-risk type human papillomavirus infection and p16 expression in laryngeal cancer. Infect. Agent. Cancer 14, 8. https://doi.org/10.1186/s13027-019-0224-y (2019).
Aerts, H. J. W. L. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006. https://doi.org/10.1038/ncomms5006 (2014).
Aerts, H. J. W. L. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2, 1636–1642. https://doi.org/10.1001/jamaoncol.2016.2631 (2016).
Bagher-Ebadian, H. et al. Application of radiomics for the prediction of HPV status for patients with head and neck cancers. Med. Phys. 47, 563–575. https://doi.org/10.1002/mp.13977 (2020).
Bogowicz, M. et al. Computed tomography radiomics predicts hpv status and local tumor control after definitive radiochemotherapy in head and neck squamous cell carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 99, 921–928. https://doi.org/10.1016/j.ijrobp.2017.06.002 (2017).
Yu, K. et al. Radiomic analysis in prediction of human papilloma virus status. Clin. Transl. Radiat. Oncol. 7, 49–54. https://doi.org/10.1016/j.ctro.2017.10.001 (2017).
Shafiq-ul-Hassan, M. et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 44, 1050–1062. https://doi.org/10.1002/mp.12123 (2017).
Leijenaar, R. T. H. et al. Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: A multicenter study. Br. J. Radiol. 91, 1–8. https://doi.org/10.1259/bjr.20170498 (2018).
Guiot, J. et al. A review in radiomics: Making personalized medicine a reality via routine imaging. Med. Res. Rev. https://doi.org/10.1002/med.21846 (2021).
Lambin, P. et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 14, 749–762. https://doi.org/10.1038/nrclinonc.2017.141 (2017).
Hartwig, S. et al. Estimation of the epidemiological burden of HPV-related anogenital cancers, precancerous lesions, and genital warts in women and men in Europe: Potential additional benefit of a nine-valent second generation HPV vaccine compared to first generation HPV va. Papillomavirus Res. 1, 90–100. https://doi.org/10.1016/j.pvr.2015.06.003 (2015).
Collins, G. S., Reitsma, J. B., Altman, D. G. & Moons, K. G. M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. BMC Med. 13, 1. https://doi.org/10.1186/s12916-014-0241-z (2015).
Mungai, F. et al. CT assessment of tumor heterogeneity and the potential for the prediction of human papillomavirus status in oropharyngeal squamous cell carcinoma. Radiol. Med. 124, 804–811. https://doi.org/10.1007/s11547-019-01028-6 (2019).
Buch, K. et al. Using Texture analysis to determine human papillomavirus status of oropharyngeal squamous cell carcinomas on CT. Am. J. Neuroradiol. https://doi.org/10.3174/ajnr.A4285 (2015).
Forghani, R. et al. Head and neck squamous cell carcinoma: Prediction of cervical lymph node metastasis by dual-energy CT texture analysis with machine learning. Eur. Radiol. 29, 6172–6181. https://doi.org/10.1007/s00330-019-06159-y (2019).
Zhang, J., Zhang, Y. & Zhang, Z. Prevalence of human papillomavirus and its prognostic value in vulvar cancer: A systematic review and meta-analysis. PLoS ONE 13, e0204162 (2018).
Cantrell, S. C. et al. Differences in imaging characteristics of HPV-positive and HPV-negative oropharyngeal cancers: A blinded matched-pair analysis. Am. J. Neuroradiol. https://doi.org/10.3174/ajnr.A3524 (2009).
Zhao, B. et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci. Rep. 6, 23428. https://doi.org/10.1038/srep23428 (2016).
Rietbergen, M. M. et al. Molecular characterization of p16-immunopositive but HPV DNA-negative oropharyngeal carcinomas. Int. J. Cancer 134, 2366–2372. https://doi.org/10.1002/ijc.28580 (2014).
Sun, G. et al. The prognostic value of HPV combined p16 status in patients with anal squamous cell carcinoma: A meta-analysis. Oncotarget; 9(8), 8081 (2017).
Brown, P. J. et al. Prediction of outcome in anal squamous cell carcinoma using radiomic feature analysis of pre-treatment FDG PET-CT. Eur. J. Nucl. Med. Mol. Imaging 46, 2790–2799. https://doi.org/10.1007/s00259-019-04495-1 (2019).
Saeed, H. et al. MRI-based radiomic fingerprint in cervical cancer: A new predictor for progression-free survival. Brachytherapy 18, S13. https://doi.org/10.1016/j.brachy.2019.04.033 (2019).
Chan, M. W. et al. Morphologic and topographic radiologic features of human papillomavirus-related and –unrelated oropharyngeal carcinoma. Head Neck 39, 1524–1534. https://doi.org/10.1002/hed.24764 (2017).
Acknowledgements
The authors thank Fabio Bottari, PhD, of Radiomics for providing medical writing support in accordance with Good Publication Practice (GPP3) guidelines (http://www.ismpp.org/gpp3).
Funding
This work was supported by the European Union’s Horizon 2020 research and innovation programme under grant agreement EuCanImage n° 952103.
Author information
Authors and Affiliations
Contributions
R.T.H. L., S.W., A.V. conceptualize the study, planned the methodological approach and performed the analysis. L.A. V.L.S. performed the statistical analysis and the data visualization. M.L., R.J., C.G., F.K., D.D., R.H. contributed to the collection and curation of the imaging and clinical data. M.O., W.V., J.G., P.L., P.L. supervised the project and acquired and managed the funding needed. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
R. T.H. L. has shares in the company Radiomics and is co-inventor of an issued patent with royalties on radiomics (PCT/NL2014/050728) licensed to Radiomics. R. T.H. L. is a former employee of Radiomics. S. W. reports personal fees from Radiomics, outside the submitted work A.V., L. A., V. S. L. are salaried employees of Radiomics. M. O. reports personal fees from Radiomics, outside the submitted work. J. G. reports, outside the submitted work, research agreements from Radiomics. He is in the permanent SAB of Radiomics for the SALMON trial without any specific consultancy fee for this work. He is co-inventor of one issued patent on radiomics licensed to Radiomics. P.L. reports, within and outside the submitted work, the following disclosures: grants/sponsored research agreements from Radiomics SA, Convert Pharmaceuticals and LivingMed Biotech. He received a presenter fee (in cash or in kind) and/or reimbursement of travel costs/consultancy fee (in cash or in kind) from Radiomics SA, BHV. PL has minority shares in the companies Radiomics SA, Convert pharmaceuticals, Comunicare, LivingMed Biotech and Bactam. PL is co-inventor of two issued patents with royalties on radiomics (PCT/NL2014/050248 and PCT/NL2014/050728), licensed to Radiomics SA; one issued patent on mtDNA (PCT/EP2014/059089), licensed to ptTheragnostic/DNAmito; one non-issued patent on LSRT (PCT/ P126537PC00), licensed to Varian; three non-patented inventions (softwares) licensed to ptTheragnostic/DNAmito, Radiomics SA and Health Innovation Ventures and two non-issued, non-licensed patents on Deep Learning-Radiomics (N2024482, N2024889). grants/sponsored research agreements from Radiomics, ptTheragnostic/DNAmito, Health Innovation Ventures. He received an advisor/presenter fee and/or reimbursement of travel costs/consultancy fee and/or in kind manpower contribution from radiomics SA, BHV, Merck, Varian, Elekta, ptTheragnostic, BMS and Convert pharmaceuticals. Dr Lambin has minority shares in the company Radiomics SA, Convert pharmaceuticals, Comunicare Solutions and LivingMed Biotech, he is co-inventor of two issued patents with royalties on radiomics (PCT/NL2014/050248, PCT/NL2014/050728) licensed to Radiomics SA and one issued patent on mtDNA (PCT/EP2014/059089) licensed to ptTheragnostic/DNAmito, one non issued patent on LSRT (PCT/ P126537PC00) licensed to Varian Medical, three non-patented invention (softwares) licensed to ptTheragnostic/DNAmito, Radiomics SA and Health Innovation Ventures and three non-issues, non licensed patents on Deep & handcrafted Radiomics (US P125078US00, PCT/NL/2020/050794, n° N2028271). W. V. have shares in the company Radiomics. The rest of co-authors have no competing interest to disclose.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Leijenaar, R.T.H., Walsh, S., Akshayaa Vaidyanathan et al. External validation of a radiomic signature to predict p16 (HPV) status from standard CT images of anal cancer patients. Sci Rep 13, 7198 (2023). https://doi.org/10.1038/s41598-023-34162-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34162-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.