Abstract
Sorghum damping-off, caused by Fusarium solani (Mart.) Sacc., is a serious disease which causes economic loss in sorghum production. In this study, antagonistic activity of lavender essential oil (EO) at 0.5, 0.75, 1.0, 1.25, 1.5, and 1.6% against F. solani was studied in vitro. Their effects on regulation of three SbWRKY transcription factors, the response factor JERF3 and eight defense-related genes, which mediate different signaling pathways, in sorghum were investigated. Effects of application under greenhouse conditions were also evaluated. The results showed that lavender EO possesses potent antifungal activity against F. solani. A complete inhibition in the fungal growth was recorded for lavender EO at 1.6%. Gas chromatography–mass spectrometric analysis revealed that EO antifungal activity is most likely attributed to linalyl anthranilate, α-terpineol, eucalyptol, α-Pinene, and limonene. Observations using transmission electron microscopy revealed many abnormalities in the ultrastructures of the fungal mycelium as a response to treating with lavender EO, indicating that multi-mechanisms contributed to their antagonistic behavior. Results obtained from Real-time PCR investigations demonstrated that the genes studied were overexpressed, to varying extents in response to lavender EO. However, SbWRKY1 was the highest differentially expressed gene followed by JERF3, which suggest they play primary role(s) in synchronously organizing the transcription-regulatory-networks enhancing the plant resistance. Under greenhouse conditions, treating of sorghum grains with lavender EO at 1.5% prior to infection significantly reduced disease severity. Moreover, the growth parameters evaluated, the activities of antioxidant enzymes, and total phenolic and flavonoid contents were all enhanced. In contrast, lipid peroxidation was highly reduced. Results obtained from this study support the possibility of using lavender EO for control of sorghum damping-off. However, field evaluation is highly needed prior to any usage recommendation.
Similar content being viewed by others
Introduction
Sorghum (Sorghum bicolor (L.) Moench) is among the most important cereal crops worldwide as a source of food/feed, and ethanol production. It is ranked the fifth main grain crop with a total global production of more than 59 million tons1.
Sorghum damping-off, caused by Fusarium solani (Mart.) Sacc., is a serious disease which causes seeds and seedling decay resulting in a significant economic loss in the crop yield2. In addition, F. solani is a toxigenic fungus which produces dangerous mycotoxins such as trichothecenes and fusaric acid affecting human and animal health3,4. The symptoms include seed decay in the soil, discoloration and rotting of the radicles which prevent germination and emergence, and formation of red lesions on the roots of seedlings that do emerge, which, especially at low temperatures, halts their development5. Various chemical fungicides are available for control of damping-off disease such as Mancozeb, Rizolex, and Benomyl6, but the use of chemical fungicides is unfavorable owing to potential deleterious health effects and environmental risks7.
Essential oils (EOs) and plant extracts have been extensively studied by many researchers as alternatives to chemical fungicides due to their ecological safety, and potent antifungal activities against several phytopathogenic fungi8,9,10. In this regard, Ghoneem et al.11 reported full suppression in the fungal growth of Sclerotinia sclerotiorum by clove essential oil at 2%. The rich content of different bioactive components in EOs such as phenols, coumarins, quinines, flavonoids, tannins, and fatty acids provides multifunctional and synergistic antifungal potentialities against plant pathogenic fungi. In addition, these multifunctional bioactive compounds make development of microbial-resistance difficult based on diverse antagonistic modes of action9,12. Moreover, EOs of some medicinal plants may act as elicitors, triggering the plant defense-responses against attacking pathogens8,13.
WRKY proteins represent a pivotal plant family of transcription factors (TF) which work via interconnected signaling networks to synchronously regulate a diverse set of defense-responses against biotic and abiotic stresses, as well as metabolic responses14. Many investigations have implicated WRKY TFs in regulation of defense-responses against different fungal diseases15,16. Overexpression of OsWRKY45 in rice provides resistance against the blast fungus (Magnaporthe oryzae) via triggering salicylic acid (SA)-signaling pathway genes17. Also, overexpression of VvWRKY1 in grapevine elicits expression of jasmonic acid (JA)-signaling pathway genes against the downy mildew fungus (Plasmopara viticola)18. Recently, ninety four SbWRKY TFs were identified in sorghum and classified into three groups according to their binding domains and type of zinc-finger motifs19.
Lavandula angustifolia Mill., frequently known as English lavender, is a flowering shrub which belongs to family Lamiaceae. It has many uses, such as flavoring agent in foods, pharmaceutical uses such as soap, perfumes and cosmetics manufactures, as well as many therapeutic applications owing to its antimicrobial, antioxidant, anxiolytic, antispasmodic, and aphrodisiac properties20. Antifungal activity of lavender EO has been studied against different pathogenic fungi. Xiong et al.21 reported a complete fungal inhibition for Lavender EO at 800 μL/L against Monilinia fructicola. Their study indicated that the utilized antagonistic mechanisms included destruction of the cell membrane, cytoplasmic leakage, and induction of cell apoptosis. In addition, the antifungal activity of lavender EO was reported also against Candida albicans22. The present study aimed to (1) investigate the antagonistic activity of lavender EO against F. solani in vitro, especially effects on ultrastructures, (2) study their effect(s) on regulation of three SbWRKY TFs (1, 19, and 45), Jasmonate and ethylene-response factor 3 (JERF3) and eight defense-related genes, which mediate SA, JA and ethylene (ET)-signaling pathways, in sorghum against Fusarium damping-off, (3) evaluate their biocontrol activity under greenhouse conditions, as well as their effects on the growth and biochemical plant parameters.
Results
Screening for antifungal activity of lavender EO in vitro
Antifungal activity of lavender EO was assessed in vitro against F. solani at 0, 0.5, 0.75, 1, 1.25, 1.5 and 1.6% (Fig. 1). The mean reductions in the fungal growth are presented in Table 1. All tested concentrations exhibited inhibitory effects in varying extents compared with the control treatment. Growth inhibition was elevated with the increment in the concentration of lavender EO. The highest growth inhibition (97.6%) was obtained at 1.5% recording 2 mm radial growth compared with the control and the lowest growth inhibition was recorded at the concentration of 0.5%. The minimum inhibitory concentration (MIC) was obtained at 1.6%, while 50% inhibitory concentration (IC50) was recorded at 0.65%.
Scanning electron microcopy observations (SEM)
The antifungal effects of lavender EO on morphology of F. solani were examined using SEM to investigate its antagonistic mechanisms. SEM observations of the untreated fungus showed normal, intact, thick, and regular mycelia with smooth surface (Fig. 2a). In addition, typical, long, unbranched, and smooth-surfaced aerial conidiophores bearing false heads of ellipsoidal, non-septated microconidia were also observed (Fig. 2b,c). In contrast, mycelia of F. solani treated with lavender EO showed dramatic alterations in their morphology including severe collapse, distortion, shrinking, squashing, deformation, and rough surface. Furthermore, no conidia were detected in the treated fungal mycelia (Fig. 2d–f).
Scanning electron micrographs showing effects of lavender essential oil on morphology of Fusarium solani. The untreated fungus shows normal, intact, thick, and regular mycelia with smooth surface (a), typical, long, unbranched, and smooth-surfaced aerial conidiophores bearing false heads of ellipsoidal, non-septated microconidia (b and c). Mycelia of F. solani treated with lavender EO show severe collapse, distortion, shrinking, squashing, deformation, and rough surface with no observed conidia (d–f).
Transmission electron microcopy observations (TEM)
Observation of untreated (control) hyphae of F. solani using TEM exhibited normal ultrastructure. Thin cell walls and plasmalemma embracing the cytoplasm with electron-lucent lipid globules, nucleus, and vacuoles were noted (Fig. 3a,b). In contrast, TEM observations of F. solani hyphae treated with lavender EO showed considerable ultrastructural alterations. Thick cell walls and plasmalemma enclosing an electron-dense cytoplasm were observed. Large vacuoles containing electron-dense materials and absence of the lipid globules were also noted (Fig. 3c,d).
Transmission electron micrographs of a cross section in hyphae of Fusarium solani. Where, (a) and (b) show untreated hyphae (control). Note cell wall (W), plasmalemma (P), cytoplasm (C), lipid globules (L), nucleus (N) and vacuoles (V), while, (c) and (d) show hyphae treated with lavender essential oil at 1.25%. Note a thick plasmalemma (TP), big vacuoles (BV), electron-dense cytoplasm (short arrows), thick wall (TW) and vacuoles contain electron-dense materials (long arrow).
Gas chromatography–mass spectrometry (GC–MS)
The chemical composition of lavender EO was analyzed via GC–MS (Fig. 4). Twenty-eight compounds in varying proportions were identified (Table 2). The major components in lavender EO included; linalool (31.1%), linalyl anthranilate (16.8%), benzyl acetate (12.9%), and 1,8-cineole (10.1%). Other components were identified in intermediate proportions including α-Terpineol acetate (4.92%), ɣ-Terpineol (3.89%), α-Terpineol (3.15%), α-Pinene (2.85%), Dihydrocarveol (2.34%), Dihydromyrcenol (1.81%), and Limonene (1.04%), while, the rest components were present in minor ratios.
Transcript levels of three SbWRKY TFs and nine defense-related genes
Transcriptional expression profiles of three SbWRKY TFs, JERF3 and eight defense-related genes in sorghum shoot were studied 3 and 6 days post emergence (dpe) (Fig. 5). In every case, EO in the presence of the pathogen increased the level of expressions as measured by mRNA levels and uninoculated controls had the least mRNA. Inoculated plants in the absence of EO were also higher than controls and in most but not all cases expression induced by EO alone was also above the control level. Of all studied genes, SbWRKY1 was the highest expressed gene followed by JERF3. For SbWRKY1 expression, infection of sorghum plants with F. solani or treating with lavender EO induced their transcript level, but the transcriptional expression in the infected plants was much higher (21-fold at 3 dpe) than in the EO treated-plants when compared with the untreated control plants. However, the highest expression level was recorded for the infected plants also treated with lavender EO (43-fold at 3 dpe). For all treatments, the expression level of SbWRKY1 at 6 dpe was lower than that at 3 dpe. The expression level of JERF3 came in second after SbWRKY1 and was triggered by infection with F. solani and/or treating with lavender EO, compared with the untreated control plants, but the expression level of the dual treatment was higher than the single treatments recording 29- and 28-fold at 3 and 6 dpe, respectively. Concerning PR1, PR2, PR3, PR5, PR12, SbWRKY19 and SbWRKY45, infection with F. solani and/or treating with lavender EO induced the gene expression level at 3 and 6 dpe in varying degrees. Regarding PAL1, AFPRT, and GST1, the untreated-infected sorghum plants or infected plants which were treated with EO showed considerable up-regulation in the transcript level of the three genes, but the dual treatment was more highly induced than the infection-alone treatment. In contrast, sorghum plants treated with lavender EO did not exhibit any significant difference in the expression level of these three genes, when compared with the untreated control plants. In all expression profiles, the transcript level of the studied genes reduced from 3 to 6 dpe.
Histograms showing relative transcriptional expression levels of three SbWRKY transcription factors and some defense-related genes in sorghum plants infected with Fusarium solani and/or treated with lavender essential oil at 1.5% after 3 and 6 days post-emergence (dpe). C: untreated control, P: infected with F. solani, EO: treated with lavender essential oil at 1.5%, and P + EO: infected with F. solani and treated with lavender essential oil at 1.5%. In each time for each studied gene, columns superscripted with the same letter are not significantly different according to Duncan’s multiple range test (P ≤ 0.05). Each value represents the mean of three biological replicates; each sample was analyzed in triplicate. Error bars represent standard errors.
Hierarchical clustering analysis
Hierarchical clustering heat map of transcriptional expression of the investigated genes in sorghum shoot is illustrated in Fig. 6. As seen from the heat map, all tested treatments are grouped into two main clusters, the first represents the untreated control plants, and the lavender-EO-treated plants at 3 and 6 dpe, while the other represents the infected plants whether treated with lavender EO or not at 3 and 6 dpe. In the first cluster, the untreated control plants at both investigated times (3 and 6 dpe) are grouped together in a separate subcluster, while, the lavender-EO-treated plants at the same times are grouped together in the other subcluster. In the second main cluster, the infected plants at the investigated times are grouped together in a separate subcluster, while, the infected plants which treated with lavender EO at 3 and 6 dpe are grouped together in another separate subcluster. Concerning the gene clustering, all genes are grouped into two main clusters, the transcription factor SbWRKY45 is grouped in a separate out-cluster revealing its unique behavior, while, the other main cluster included all the other investigated genes. Moreover, the hierarchical clustering heat map shows 5 two-genes-clusters between GST1-PAL1, PR5-PR2, AFPRT-PR12, PR3-PR1, and SbWRKY19-JERF3. In general, the hierarchical clustering expression exhibited high up-regulation of the investigated genes in case of the infection treatments, whether treated with lavender EO or not. The maximum transcription levels were observed for the infected plants treated with lavender EO at 3 dpe.
Hierarchical clustering heat map of transcriptional expression of three SbWRKY transcription factors and some defense-related genes in sorghum plant infected with Fusarium solani and/or treated with lavender essential oil at 1.5% after 3 and 6 days post-emergence (dpe). Where, C: untreated control, P: infected with F. solani, EO: treated with lavender essential oil at 1.5%, and P + EO: infected with F. solani and treated with lavender essential oil at 1.5%.
Disease assessment
Disease assessment data of the infected sorghum seedlings in response to treatment with lavender EO at different concentrations are presented in Table 3. The data indicated that the infection with F. solani caused damping-off of sorghum leading to up to 92% mortality, when compared with the untreated control treatment. Typical symptoms of Fusarium damping-off were recorded, including seed rotting, and pre- and post-emergence damping-off. In contrast, treating of sorghum grains with lavender EO prior to infection with F. solani led to a reduction in the disease severity, which increased with increased EO concentration. In this regard, the best result was recorded for the sorghum grains treated with lavender EO at 1.5% prior to the infection (17.7% mortality), which was essentially identical to treatment with the chemical fungicide.
Effect on plant growth
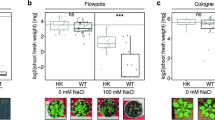
Results of the growth parameter evaluations obtained from the greenhouse experiment in response to treating with lavender EO at different concentrations and infection with F. solani are presented in Table 4. Infection of sorghum plants with F. solani led to a considerable reduction in growth, shoot weight and root weight at 30 and 45 dap, when compared with the untreated control plants. In contrast, treating of sorghum grains with lavender EO significantly enhanced the growth of sorghum plants compared with the untreated control plants. The growth promoting effect increased with the increment in EO concentration. The highest growth parameters were recorded for the sorghum plants treated with lavender EO at 1.5% harvested at both 30 and 45 dap. Compared to the treatment with the chemical fungicide, sorghum plants treated with lavender EO prior to infection with F. solani showed higher growth for plant height, shoot and root dry weights, than the untreated-infected sorghum plants. In this regard, the growth enhancing effect is directly proportional to the EO concentration at 30 and 45 dap.
Effects on activities of antioxidant enzymes
Effects of lavender EO on activities of different antioxidant enzymes of sorghum plants infected with F. solani are shown in Table 5. Data obtained indicated that infection of sorghum plants with F. solani led to an induction in the activities of all studied enzymes when compared with the untreated control plants at 30 and 45 dap. In general, activity of catalase (CAT) and superoxide dismutase (SOD) enzymes in sorghum plants at 30 dap was higher than that at 45 dap, while, activity of ascorbate peroxidase (APX) and polyphenol oxidase (PPO) enzymes increased from 30 to 45 dap. Treating of sorghum plants with lavender EO at different concentrations significantly triggered activity of all tested enzymes compared with the untreated control. However, the inducing effect resulting from infection was more than that of the lavender EO treatments at both studied times. For all studied enzymes, the highest enzyme activity was recorded for the infected sorghum plants also treated with lavender EO at 1.5%, when compared with the treatment of the chemical fungicide. In this regard, the inducing effect on enzymes activity is directly proportional to the EO concentration at 30 and 45 dap.
Effects on lipid peroxidation, total phenolic and flavonoid contents
Effects of lavender EO on lipid peroxidation, total phenolic and flavonoid contents of sorghum plants infected with F. solani are presented in Table 6. Results of biochemical analyses of sorghum plants showed that infection with F. solani led to significant elevations in lipid peroxidation, total phenolics and flavonoids at 30 and 45 dap, when compared with the untreated control plants. In contrast, treating with lavender EO at different concentrations did not affect lipid peroxidation of sorghum plants. Treating of the infected sorghum plants with lavender EO significantly reduced the lipid peroxidation, compared with the treatment of the chemical fungicide. This effect is directly proportional to the EO concentration at 30 and 45 dap. The lipid peroxidation in sorghum plants at 30 dap was higher than that at 45 dap. Regarding total phenolic and flavonoid contents, the data showed that treating of sorghum plants with lavender EO at different concentrations significantly increased both categories of compounds in direct proportional relationship at 30 and 45 dap. The highest levels of phenolics and flavonoids were recorded for the infected sorghum plants treated with lavender EO at 1.5%, compared with the chemical fungicide treatment at 30 and 45 dap. In general, levels of both increased from 30 to 45 dap in all sorghum plants treatments.
Discussion
The present work aimed to evaluate the effect of lavender EO in regard to their antifungal activity against F. solani in vitro, and their resistance-inducing activity against Fusarium damping-off in sorghum, especially on SbWRKY TFs. In vitro, the results indicated that lavender EO possesses concentration dependent antifungal activity against F. solani. This result is in agreement with findings reported by Bahmani and Schmidt23, and Behmanesh et al.22. Antifungal activity of EOs and extracts from different medicinal plants, including lavender EO, has been reported by many researchers8,9. The chemical composition of medicinal plants comprises various bioactive phytochemicals such as coumarins, flavonoids, terpenes, anthocyanins, and tannins, which may contribute to the fungitoxic activity. Different mechanisms have been described in this concern including interfering with permeability and integrity of fungal cell walls and plasma membranes, suppression of metabolic enzymes, and DNA damage24. GC–MS analysis of lavender EO showed existence of some bioactive constituents with a known antifungal background including linalool as the main bioactive component, in addition to linalyl anthranilate, α-terpineol, 1,8-cineole (eucalyptol), α-Pinene, and limonene. Most of the antifungal activity of lavender EO is attributed to linalool, simply because it is the most abundant bioactive component. Recent researchers have reported a potent antifungal activity for linalool25. It’s fungitoxic effect can be explained in the light of interference with cell wall biosynthesis and disrupting permeability of plasmalemma26. In addition, α-terpineol has been reported also as a potent antifungal agent and its antimycotic effect has been suggested to be due to its activity on cytoplasmic degeneration and hyphal distortions27. These antifungal effects were observed in our TEM observations. In this regard, the TEM observations revealed many abnormalities in cellular ultrastructure of F. solani treated with lavender EO such as thickening of cell wall and plasmalemma, indicating that multi-mechanisms contributed to the observed antagonistic behavior, that are compatible with a result loss of permeability. Thickening of the cell wall and plasmalemma leads to restriction of the cellular exchange of ions and molecules with the surrounding medium, which finally results in cell death8. In addition, another antifungal mechanism was observed by TEM, which is cytoplasmic coagulation. This effect is correlated with the impairment of the plasmalemma, followed by condensation and coagulation of the cytoplasm and finally cell death28. Absence of lipid globules in the treated F. solani hyphae was also observed with TEM. Lipid droplets play important roles in the fungal cell as energy reserves, preventing lipotoxicity, and regulating some physiological processes29. Absence of lipid globules reveals that the fungal cell is suffering stress conditions.
At the molecular level, twelve genes including three SbWRKY TFs, JERF3 and eight defense-related genes, representing SA-, JA- and ET-signaling pathways, were selected in this study as pathway reporter genes. Transcriptional expression levels of these genes were investigated in sorghum shoot in response to application of lavender EO and/or infection with F. solani at 3 and 6 dpe. The obtained results demonstrated that SbWRKY1 was the highest expressed gene followed by JERF3, which suggest probable primary role(s) in the plant resistance in response to these treatments. Plants are subjected to multiple environmental stresses, including pathogenic fungi, and energetically respond to these challenges to survive. In order to overcome the stresses encountered, plants initiate transcriptional cascades through cellular signaling pathways. These pathways interact in coordination with each other via signaling molecules leading to stimulation of the defensive-gene-regulatory networks30. Transcription factors, such as WRKY proteins, play important roles in synchronously organizing the transcription-regulatory-networks enhancing the plant responses against biotic and abiotic stresses16. WRKY TFs bind to W-boxes found in the stress-inducible promoters of many defense-related genes in plants. The W-boxes exist in clusters, suggesting coordinated interactions of several WRKY TFs31. In this regard, WRKY1 TF has been reported as a key element mediating induced resistance against infection with Alternaria solani in wild tomato (Solanum arcanum)32. WRKY1 regulates SA-signaling pathway via interaction with NPR1 gene (Natriuretic Peptide Receptor 1), which functions as a master regulator in the orchestration of the plant-defense-responses, controlling expression of more than 2000 defense-related genes33,34. JERF3, which functions as a key element of ET/JA-signaling pathways, activates multiple defense responses via binding to the GCC box located in the promoters of some defense-related genes35. In this regard, Zhang et al.36 reported the involvement of ERF3 in triggering an array of defense responses against Blumeria graminis in wheat at early stages via the SA-signaling pathway, and against F. graminearum or Rhizoctonia cerealis at late stages via ET/JA-signaling pathways. In this study, one of the most interesting results obtained by the hierarchical clustering analysis is the single clustering of SbWRKY45 away from the other studied genes revealing its unique behavior. This result is in agreement with that obtained by Shimono et al.37 who reported the vital role of OsWRKY45 in triggering plant resistance against the blast fungus (Magnaporthe grisea) on rice. The same concept was reported by Qiu and Yu38 against Pyricularia oryzae and Xanthomonas oryzae on rice, and in Arabidopsis. The WRKY45-induced resistance included overexpression of some PR genes, particularly, PR1 and PR2 (markers of systemic acquired resistance). In addition, he confirmed the mediation of OsWRKY45 to SA-signaling pathway. Likewise, WRKY19 has been also reported to be involved in induction of plant resistance against powdery mildew of barley39. It is known that induction of SA-signaling pathway leads to overexpression of the pathogenesis-related (PR) genes PR1, PR2, and PR5, while, triggering JA-signaling pathway induces PR3, PR4, and PR12 genes40. In this regard, data obtained in this study revealed overexpression of PR1 (antifungal), PR2 (β-1,3-glucanase), and PR5 (Thaumatin-like protein) which are SA-responsive defense genes. This result is in accordance with the reported overexpression of WRKY genes. PR1 proteins are highly abundant in plants during biotic- and abiotic-stress responses and have been widely used as a defense marker. Unlike PR2 and PR5 proteins, which have known antifungal enzymatic activities, the antifungal mechanism of PR1 proteins remains unclear. However, recent studies have suggested multiple roles of PR1 proteins including sterol-binding activity, hypersensitivity response (cell death), and harboring an embedded defense-signaling peptide (CAP-Derived Peptide 1)41. PR2 encodes the lytic enzyme β-1,3-glucanase, which hydrolyz β‐1,3‐glycosidic bond in the 1,3-glucan molecules, degrading the cell walls of attacking phytopathogenic fungi42. PR5 encodes antifungal protein which exhibits fungal-cell-wall-lytic activity (glucan binding and glucanase activities), xylanase inhibitor activity, and pathogen recognition (binding with the pathogen cell surface)43. The two-genes-clustering (PR2 and PR5) with similar patterns obtained in this study can be explained in the light of their shared glucanase activities and the same SA-signaling pathway. Overexpression of these PR genes implicate a role for SA-signaling in sorghum resistance against F. solani. Data from the quantitative Real-Time PCR (qRT-PCR) obtained in this study revealed overexpression of PR3 (chitinase 15), and PR12 (Plant defensin 1), which are JA-responsive defense genes. PR3 encodes the antifungal enzyme chitinase, which catalyze hydrolysis of β-1,4 bonds between N-acetylglucosamine subunits of chitin molecules, the main constituent of the fungal cell wall. PR1 and PR3 proteins synergistically inhibit the fungal growth as a plant defense response42. Clustering of PR1 and PR3 observed in this study is in accordance with the synergism reported between the two proteins in the literature. PR12 encodes antifungal and cytotoxic proteins, which have significant roles in plant resistance against wide range of phytopathogenic fungi44. Overexpression of these PR genes revealed the participation of the JA-signaling pathway in sorghum resistance against F. solani. PAL1 is the key gene in the phenylpropanoid pathway regulating biosynthesis of an array of antifungal polyphenolic compounds in plant including flavonoids, lignins, and chlorogenic acid45. GST1 encodes the antioxidant-defense enzyme, which involved in the detoxification function against xenobiotics through binding with glutathione46. In addition to PR genes, PAL1 and GST1 are also defense genes, which regulated by WRKY transcription factors. The overexpression of all studied genes was supported with the elevated activities of the estimated antioxidant enzymes and total phenol content explaining the synergistic effect of lavender EO and infection with F. solani in triggering the sorghum resistance.
Materials and methods
Fungal inoculum, sorghum cultivar, and lavender shrubs
A virulent isolate of the fungus F. solani (GenBank accession no.: KJ831188), isolated from sorghum seedling showing damping-off symptoms, was used in this study. The fungal isolate was maintained on potato dextrose agar (PDA) slants and kept at 4 °C until use. For inoculum preparation, fungal spores from 7-days-old PDA cultures of F. solani were harvested using sterile water and the spore suspension was adjusted at 1 × 106 spore mL−1. Sorghum grains cv. Giza 15, obtained from the Central Administration for Seed Certification, Egypt, were used in the greenhouse experiment. The used lavender shrubs were originally cultivated in the Experimental Nursery of the Medicinal and Aromatic Plants Research Department, Horticultural Research Institute, Agriculture Research Center, Giza, Egypt. The shrubs were harvested in July 2020, identified by Prof. Ibrahim A. Mashaly, and deposited at the Herbarium of Faculty of Science, Mansoura University, Mansoura, Egypt under the deposition number (Mans 0303075019).
Essential oil extraction
Lavender EO was extracted from 200 g of air-dried lavender flowers via hydro-distillation for 60 min using Clevenger apparatus as described by Zheljazkov et al.47. The EO was then filtered and stored in dark bottle at 4 °C until use.
Screening for antifungal activity of lavender EO in vitro
Antifungal activity of lavender EO was assessed against mycelial growth of F. solani in vitro using agar plate technique. PDA plates supplemented with lavender EO at final concentrations of 0.5, 0.75, 1.0, 1.25, and 1.5% were used. The tested concentrations were prepared by adding suitable volumes of EO to 100 mL Erlenmeyer flasks containing melted PDA medium before solidification and 0.5% Tween-80. PDA plates supplemented with a synthetic fungicide (nystatin) at 50 µg/mL were used as positive control. Untreated PDA plates were used as negative control. The PDA plates were inoculated in the centers with 7-mm-diameter discs taken from active margins of 7-days-old culture of F. solani. For each treatment, three plates were used. All plates were incubated in dark at 25 ± 2 °C until full fungal growth was obtained in the control plate. Diameter of the fungal colony in each plate was measured and the reduction percentage in mycelial growth was calculated as follows:
where C = colony diameter of the control plate, and T = colony diameter of the treated plate.
SEM
To investigate effects of lavender EO on morphology of F. solani using SEM, one block from F. solani culture on PDA plate and another one block from F. solani culture treated with lavender EO were separately processed using tissue processor (Leica Biosystems, Inc.). The two blocks were fixed using osmium oxide, dehydrated using ethanol, and dried using a critical point drier (EMS 850), and then coated with gold using a sputter coater (EMS 550). Morphology of the fungal structures was observed using SEM (JEOL JSM-6510LV).
TEM
To investigate effects of lavender EO on ultrastructure of F. solani using TEM, samples of the treated plates were fixed using 3% glutaraldehyde in phosphate buffer at pH 6.8, followed with 1% osmium tetroxide, then dehydrated in a gradual ethanol series as described by Alberto et al.48. The dehydrated specimens were embedded in plastic epoxy resin and cut to ultra-thin sections using Reichert ultramicrotome, and stained with uranyl acetate followed by lead citrate. The sections were examined using JEOL-TEM (model JEM-1230).
GC–MS
Chemical constituents of lavender EO were identified using a GCMS-QP2010 system (Shimadzu, Japan). The EO sample was injected at flow rate of 1.5 mL min−1 via a DB-5 column (60 m × 0.25 mm, 0.25 μm thick) using helium as a carrier at 300 °C. The ion source temperature was 210 °C, while the interface temperature was 300 °C, at an ionization voltage of 70 eV. Retention time and mass spectra were used to identify the EO composition using the NIST11library (Gaithersburg, USA).
Greenhouse experiment
Plastic pots (15 cm diameter) filled with sterile sandy-clay soil (1:2 v/v) were used. For soil infestation, the spore suspension of F. solani (1 × 106 spore mL−1) was mixed with upper layer soil of the pots at the rate of 2% (v/v) ten days before planting. Sorghum grains were soaked separately in lavender EO at different concentrations (1, 1.25, and 1.5%) for 2 h, then air-dried before planting. For positive control, sorghum grains were treated with the chemical fungicide Rhizolex-T as seed dressing at the recommended dose of 3 g kg−1 grains. For each treatment, ten sorghum grains were sown in each pot. Pots planted with surface-sterilized sorghum grains were used as a control treatment. Four replicates were used for each treatment. All pots were regularly irrigated as required, arranged in a complete randomized design, and kept under greenhouse conditions (27/21 °C day/night temperature, 65% humidity). The applied treatments were designated as follows: C: untreated control, EO1.0, EO1.25, EO1.5: treated with lavender EO at 1.0, 1.25, and 1.5% respectively, P: infected with F. solani, P + EO1.0, P + EO1.25, and P + EO1.5: infected with F. solani and treated with lavender EO at 1.0, 1.25, and 1.5% respectively, and P + F: infected with F. solani and treated with chemical fungicide. All pots were evaluated for damping-off incidence 45-days after planting (dap). Percentages of seed rot, pre- and post-emergence damping-off were recorded.
Expression analysis of the defense-related genes
Total RNA extraction and cDNA synthesis
Total RNA was extracted from fresh sorghum shoot at 3 and 6 days post seedling emergence (dpe) using RNeasy Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. The extracted RNA was incubated with DNase for 1 h at 37 °C and quantified using a NanoDrop 1000 spectrophotometer (Thermo Scientific, USA).
For cDNA synthesis, RT-PCR kit (Qiagen, Germany) was used according to the manufacturer’s instructions. The reaction mixture (20µL) contained 2.5 µL dNTPs (2.5 mM), 5 µL 5×-buffer with MgCl2, 4 µL oligo (dT) primer (20 pmol µL−1), 0.2 µL reverse transcriptase enzyme (Omniscript RT, Qiagen, Germany) and 2 µL RNA. PCR amplification was performed using a thermal cycler (Promega, Germany), at 42 °C for 2 h and 65 °C for 20 min, the cDNA was then stored at − 20 °C until used.
qRT-PCR
The reaction mixture included 1 µL of template, 12.5 µL of SYBR Green Master Mix (Bioline, Germany), 1 µL of forward primer, 1 µL of reverse primer, and sterile RNase free water for a total volume of 20 µL. A β-actine gene was used as a reference gene. Sequences of used primers are presented in Table 7. The real time PCR program was performed using a Rotor-Gene-6000-system (Qiagene, USA) as follows: one cycle at 95 °C for 10 min, 40 cycles (95 °C for 20 s, 58 °C for 25 s and 72 °C for 30 s). For each sample, three biological and three technical replicates were performed. The comparative CT method (2−ΔΔCT) was used to analyze the relative mRNA expression levels according to Livak and Schmittgen49.
Evaluation of plant growth
For each treatment, three plants were carefully uprooted 30 and 45 dap, washed with tap water to remove soil particles, and evaluated for plant height, shoot and root dry weights. Dry weights were measured after the samples oven-dried at 80 °C for 48 h.
Biochemical analyses
Preparation of crude plant extract
Samples of plant roots (2 g) were ground and homogenized in 5 mL of 100 mM phosphate buffer (pH 7). The homogenate was centrifuged at 15,000 rpm for 20 min, then the supernatant was collected and used as crude extract for next enzyme assays and biochemical analyses. The protein content was estimated for the assayed enzymes according to Bradford50.
Assay of enzymes activities
Activity of CAT was determined according to Aebi51. Activity of SOD was determined according to Misra and Fridovich52. Activity of ascorbate peroxidase enzyme (APX) was determined according to Nakano and Asada53. Activity of PPO was determined according to Duan et al.54.
Lipid peroxidation, and total phenolic and flavonoid contents
Lipid peroxidation was measured as described by Heath and Packer55. Total phenolic content was estimated according to Slinkard and Singleton56. Total flavonoid content was determined as described by Wang et al.57.
Statistical analyses
Statistical significances were analyzed using the software CoStat (version 6.4). Comparisons between the means were performed using Duncan’s multiple range test58 at P ≤ 0.05. The hierarchical clustering analysis was performed using BioVinci Software (Bioturing, San Diego, CA, USA) (Supplementary Figure 1).
Ethics declaration
Authors confirm that all the methods and experiments were carried out in accordance with relevant guidelines and regulations.
Change history
16 March 2022
The original online version of this Article was revised: In the original version of this Article the Funding section was incomplete. The Funding section now reads: “This research did not receive specific grants from funding agencies in the public, commercial, or not-for-profit sectors. Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).”
References
FAOSTAT Statistics Division (2021). http://www.fao.org/faostat/en/#data/QC (2021).
Lamichhane, J. R. et al. Integrated management of damping-off diseases. A review. Agron. Sustain. Dev. 37, 10 (2017).
Eltariki, F. E. M., Alhoot, M. A. & Tiwari, K. Occurrence and prevalence of mycotoxigenic Fusarium solani in onion samples collected from different regions of Libya. Asian J. Biol. Sci. 11, 192–196 (2018).
Munkvold, G. P. Fusarium species and their associated mycotoxins BT—mycotoxigenic fungi: methods and protocols. In (eds. Moretti, A. & Susca, A.) 51–106 (Springer New York, 2017). https://doi.org/10.1007/978-1-4939-6707-0_4
Frederiksen, R. A. Compendium of Sorghum Diseases. Mycologia Vol. 79, (American Phytopathological Society, 1987).
Hudge, B. V. Management of damping-off disease of soybean caused by Pythium ultimum Trow. Int. J. Curr. Microbiol. Appl. Sci. 4, 799–808 (2015).
Zubrod, J. P. et al. Fungicides: An overlooked pesticide class?. Environ. Sci. Technol. 53, 3347–3365 (2019).
Rashad, Y. M., Aseel, D. G. & Hafez, E. E. Antifungal potential and defense gene induction in maize against Rhizoctonia root rot by seed extract of Ammi visnaga (L.) Lam. Phytopathol. Mediterr. 57, 73–88 (2018).
Ghoneem, K. M., Saber, W. I. A., El-Awady, A. A., Rashad, Y. M. & Al-Askar, A. A. Alternative preservation method against Sclerotium tuber rot of Jerusalem artichoke using natural essential oils. Phytoparasitica 44, 341–352 (2016).
Baka, Z. A. M. & Rashad, Y. M. Alternative control of early blight of tomato using plant extracts from Acacia nilotica, Achillea fragrantissima and Calotropis procera. Phytopathol. Mediterr. 55, 121–129 (2016).
Ghoneem, K. M., Saber, W. I. A., Aml, A. E. A., Rashad, Y. M. & Al-Askar, A. A. Clove essential oil for controlling white mold disease, sprout suppressor and quality maintainer for preservation of Jerusalem artichoke tubers. Egypt. J. Biol. Pest Control 26, 601–608 (2016).
Hyldgaard, M., Mygind, T. & Meyer, R. L. Essential oils in food preservation: Mode of action, synergies, and interactions with food matrix components. Front. Microbiol. 3, 12 (2012).
Moutassem, D., Belabid, L., Bellik, Y., Ziouche, S. & Baali, F. Efficacy of essential oils of various aromatic plants in the biocontrol of Fusarium wilt and inducing systemic resistance in chickpea seedlings. Plant Prot. Sci. 55, 202–217 (2019).
Banerjee, A. & Roychoudhury, A. WRKY proteins: Signaling and regulation of expression during abiotic stress responses. Sci. World J. 2015, 807560 (2015).
Shimono, M. et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol. Plant Pathol. 13, 83–94 (2012).
Phukan, U. J., Jeena, G. S. & Shukla, R. K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 7, 760 (2016).
Inoue, H. et al. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc. Natl. Acad. Sci. USA. 110, 9577–9582 (2013).
Marchive, C. et al. Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 8, e54185 (2013).
Baillo, E. H. et al. Genome-wide identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench). PLoS ONE 15, e0236651 (2020).
Prusinowska, R. & Śmigielski, K. B. Composition, biological properties and therapeutic effects of lavender (Lavandula angustifolia L). A review. Herba Pol. 60, 56–66 (2014).
Xiong, X. et al. Antifungal mechanisms of lavender essential oil in the inhibition of rot disease caused by Monilinia fructicola in postharvest flat peaches. Can. J. Microbiol. 67, 724–736 (2021).
Behmanesh, F. et al. Antifungal effect of lavender essential oil (Lavandula angustifolia) and clotrimazole on Candida albicans: An in vitro study. Scientifica 2015, 1–5 (2015).
Bahmani, M. & Schmidt, O. Plant essential oils for environment-friendly protection of wood objects against fungi. Maderas Cienc. y Tecnol. 20, 325–332 (2018).
Kumar, A., Khan, F. & Saikia, D. Exploration of medicinal plants as sources of novel anticandidal drugs. Curr. Top. Med. Chem. 19, 2579–2592 (2019).
Dias, I. J. et al. Antifungal activity of linalool in cases of Candida spp. isolated from individuals with oral candidiasis. Braz. J. Biol. 78, 368–374 (2018).
Hsu, C. C., Lai, W. L., Chuang, K. C., Lee, M. H. & Tsai, Y. C. The inhibitory activity of linalool against the filamentous growth and biofilm formation in Candida albicans. Med. Mycol. 51, 473–482 (2013).
Kong, Q. et al. Antifungal mechanisms of α-terpineol and terpene-4-alcohol as the critical components of Melaleuca alternifolia oil in the inhibition of rot disease caused by Aspergillus ochraceus in postharvest grapes. J. Appl. Microbiol. 126, 1161–1174 (2019).
Glass, N. L. & Dementhon, K. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 9, 553–558 (2006).
Gao, M., Huang, X., Song, B. L. & Yang, H. The biogenesis of lipid droplets: Lipids take center stage. Prog. Lipid Res. 75, 100989 (2019).
Moreira, X., Abdala-Roberts, L. & Castagneyrol, B. Interactions between plant defence signalling pathways: Evidence from bioassays with insect herbivores and plant pathogens. J. Ecol. 106, 2353–2364 (2018).
Bakshi, M. & Oelmüller, R. Wrky transcription factors jack of many trades in plants. Plant Signal. Behav. 9, e27700 (2014).
Shinde, B. A. et al. WRKY1 acts as a key component improving resistance against Alternaria solani in wild tomato, Solanum arcanum Peralta. Plant Biotechnol. J. 16, 1502–1513 (2018).
Shi, Z., Maximova, S. N., Liu, Y., Verica, J. & Guiltinan, M. J. Functional analysis of the Theobroma cacao NPR1 gene in Arabidopsis. BMC Plant Biol. 10, 248 (2010).
Saleh, A. et al. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169–182 (2015).
Pirrello, J. et al. Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 12, 190 (2012).
Zhang, Z. et al. A novel ERF transcription activator in wheat and its induction kinetics after pathogen and hormone treatments. J. Exp. Bot. 58, 2993–3003 (2007).
Shimono, M. et al. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076 (2007).
Qiu, Y. & Yu, D. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47 (2009).
Meng, Y. & Wise, R. P. HvWRKY10, HvWRKY19, and HvWRKY28 regulate Mla-triggered immunity and basal defense to barley powdery mildew. Mol. Plant-Microbe Interact. 25, 1492–1505 (2012).
Ali, S. et al. Identification and comparative analysis of Brassica juncea pathogenesis-related genes in response to hormonal, biotic and abiotic stresses. Acta Physiol. Plant. 39, 268 (2017).
Breen, S., Williams, S. J., Outram, M., Kobe, B. & Solomon, P. S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879 (2017).
Ji, C. & Kuć, J. Antifungal activity of cucumber β-1,3-glucanase and chitinase. Physiol. Mol. Plant Pathol. 49, 257–265 (1996).
Liu, J. J., Sturrock, R. & Ekramoddoullah, A. K. M. The superfamily of thaumatin-like proteins: Its origin, evolution, and expression towards biological function. Plant Cell Rep. 29, 419–436 (2010).
Thomma, B. P. H. J., Cammue, B. P. A. & Thevissen, K. Plant defensins. Planta 216, 193–202 (2002).
Rashad, Y., Aseel, D., Hammad, S. & Elkelish, A. Rhizophagus irregularis and Rhizoctonia solani differentially elicit systemic transcriptional expression of polyphenol biosynthetic pathways genes in sunflower. Biomolecules 10, 1–20 (2020).
Gullner, G., Komives, T., Király, L. & Schröder, P. Glutathione S-transferase enzymes in plant–pathogen interactions. Front. Plant Sci. 9, 1836 (2018).
Zheljazkov, V. D., Cantrell, C. L., Astatkie, T. & Jeliazkova, E. Distillation time effect on lavender essential oil yield and composition. J. Oleo Sci. 62, 195–199 (2013).
Bianchi, A., Zambonelli, A., D’Aulerio, A. Z. & Bellesia, F. Ultrastructural studies of the effects of Allium sativum on phytopathogenic fungi in vitro. Plant Dis. 81, 1241–1246 (1997).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Bradford, M. M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Aebi, H. Catalase in vitro. Methods Enzymol. 105, 121–126 (1984).
Misra, H. P. & Fridovich, I. The role of superoxide anion in the autoxidation of epinephrine and a simple assay for superoxide dismutase. J. Biol. Chem. 247, 3170–3175 (1972).
Nakano, Y. & Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880 (1981).
Duan, X. et al. Effect of nitric oxide on pericarp browning of harvested longan fruit in relation to phenolic metabolism. Food Chem. 104, 571–576 (2007).
Heath, R. L. & Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198 (1968).
Slinkard, K. & Singleton, V. Total phenol analysis: Automation and comparison with manual methods. Am. J. Enol. Vitic. 28, 49–55 (1977).
Wang, X., Cao, J., Wu, Y., Wang, Q. & Xiao, J. Flavonoids, antioxidant potential, and acetylcholinesterase inhibition activity of the extracts from the gametophyte and archegoniophore of Marchantia polymorpha L. Molecules 21, 360 (2016).
Duncan, D. B. Multiple range and multiple F tests. Biometrics 11, 1–42 (1955).
Acknowledgements
The authors are grateful to Prof. Zakaria Baka (Faculty of Sciences, Damietta University, Egypt) for his kind help in the TEM investigations.
Funding
This research did not receive specific grants from funding agencies in the public, commercial, or not-for-profit sectors. Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Y.M.R. conceived the idea and the design of the work, contributed to the microbiological and ultrastructural studies, statistical analyses of the data, implementation of the greenhouse experiment, and manuscript and photo editing. E.S.A. contributed to the molecular and biochemical investigations, and helped in the greenhouse experiment. D.B.D contributed to the GC–MS analysis and helped in the greenhouse experiment. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rashad, Y.M., Abdel Razik, E.S. & Darwish, D.B. Essential oil from Lavandula angustifolia elicits expression of three SbWRKY transcription factors and defense-related genes against sorghum damping-off. Sci Rep 12, 857 (2022). https://doi.org/10.1038/s41598-022-04903-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-04903-x
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.