Abstract
The genus status of Urocricetus was defined recently based on morphological and molecular data. Even though the amount of evidence for a separate phylogenetic position of this genus among Cricetinae continues to increase, there is still no consensus on its relationship to other groups. Here we give the first comprehensive description of the U. kamensis karyotype (2n = 30, NFa = 50) including results of comparative cytogenetic analysis and detailed examination of its phylogenetic position by means of numerous molecular markers. The molecular data strongly indicated that Urocricetus is a distant sister group to Phodopus. Comparative cytogenetic data showed significant reorganization of the U. kamensis karyotype compared to karyotypes of all other hamsters investigated earlier. The totality of findings undoubtedly means that Urocricetus belongs to a separate divergent lineage of Cricetinae.
Similar content being viewed by others
Introduction
The Kam hamster Urocricetus kamensis (Satunin, 1903 [imprint 1902]) is a member of the lineage commonly known as Tibetan hamsters, which is one of the few rodent taxa endemic to the Qinghai-Tibet plateau where it is found at altitudes up to 5200 m above the sea level1. Over the past century, the classification of Tibetan hamsters based on morphological characteristics has been controversial2, 3. Superficial similarity with another group of small hamsters affiliated with the genus Cricetulus Milne-Edwards, 1867 (e.g. C. barabensis (Pallas, 1773); C. longicaudatus (Milne-Edwards, 1867)) has led to the situation where Tibetan hamsters have been traditionally included within the latter genus, often as the subgenus Urocricetus (Satunin, 1903 [imprint 1902]).
The first available mitochondrial data suggested that the Tibetan-hamster lineage is unrelated to all other Cricetulus members4, 5. Subsequently, this conclusion has been corroborated by phylogenetic reconstructions using a set of five nuclear exons and two mitochondrial genes2. Furthermore, molecular evidence has indicated that Urocricetus is phylogenetically related to dwarf hamsters of the genus Phodopus (Miller, 1910)2, 6. It was suggested that both Phodopus and Urocricetus originated in East Central Asia (east of the Altai–Tian Shan boundary) having separated from each other in the early Late Miocene (~ 10.4 Mya), which is comparable to the time point of the split between chromosomally, morphologically, and genetically divergent Mesocricetus and Cricetus lineages. Accordingly, it has been postulated that Urocricetus definitely deserves the rank of a separate genus2.
Species structure of this genus is unclear. At different times, one to four species of Tibetan hamsters were believed to exit by different authors (e.g., ref.1, 7; for more details see ref.3). Nonetheless, in most checklists, only U. kamensis and U. alticola (Thomas, 1917) have been regarded as valid species (e.g., ref.8), as Cricetulus members. Recent nuclear and mitochondrial data are consistent with the view that U. kamensis and U. alticola are distinct, albeit closely related species2; however, this treatment is opposed by Ding and Liao3, who advocate the monotypy of the genus.
Available molecular data on Urocricetus are limited to sequences of mitochondrial DNA and several exons of nuclear genes. There is absolutely no information on karyotypic organization of genomes of its representatives. In this work, for the first time, we give a complete description of U. kamensis karyotype structure. To precisely determine the phylogenetic position of Urocricetus, we undertook a sequence analysis of fragments of 25 nuclear genes.
Materials and methods
Compliance with ethical standards
The study was carried out in compliance with the ARRIVE guidelines. All experiments were approved by the Ethics Committee on Animal and Human Research at the IMCB SB RAS, Russia (protocol No. 01/20 of 11 February 2020), following all relevant guidelines and regulations. This article does not contain any experiments on human subjects performed by any of the coauthors.
Specimens sampled
The Kam hamsters used in the study were collected in July 2018 in Beizha area, Nangqen county, Qinghai province, China (N31°52′56′′, E96°33′16.2′′ and N31°53′37′′, E96°35′55′′). This site is close to the terra typica of U. kamensis (Satunin, 1903 [imprint 1902]), which was designated originally as “River Moktschjun, district of Mekong, North-Eastern Tibet.” Each collected specimen received its personal field number (see Supplementary Information). One male (Q18-81) and one female (Q18-96) were karyotyped, and Q18-96 was subjected to a molecular cytogenetic analysis. Mitochondrial cytochrome b gene was sequenced in two specimens (Q18-82 and Q18-90), and for specimen Q18-90, the fragments of 25 nuclear DNA loci were sequenced.
Chromosome preparation and staining
Mitotic chromosome suspensions of a U. kamensis male were obtained by the standard mitotic technique from short-term culture of bone marrow after colchicine treatment in vivo following the general protocol9. Only a U. kamensis female was used for the molecular cytogenetic analysis; metaphase chromosome spreads were prepared from primary fibroblast cultures as described previously10, 11. The fibroblastic cell lines were derived from biopsies of lung, rib, and tail tissues. All the cell lines were deposited in the IMCB cell bank (The General Collection of Cell Cultures, No. 0310–2016-0002). Cell lines of Phodopus roborovskii, Phodopus sungorus, and Mesocricetus auratus (the golden hamster) were retrieved from the same cell bank. All cell cultures and chromosome suspensions from them were obtained in the Laboratory of Animal Cytogenetics, IMCB SB RAS, Novosibirsk, Russia.
G-banding was performed by the standard trypsin/Giemsa procedure12. C-banding was carried out following the classic method or a previously published technique13 with some modifications14. CDAG-banding was conducted as described before15.
Microdissection, probe amplification, and labeling
Glass needle–based microdissection was performed on G-banded chromosomes as described earlier16. One copy of each chromosome was collected. Chromosome-specific libraries were created with whole-genome amplification kits (Sigma). After the amplification, DNA was purified by means of nucleic acid purification kits for DNA (BioSilica). DNA libraries were labeled by whole-genome amplification kits (Sigma) according to the manufacturer's protocol. Chromosome-specific probes were created for both homologous chromosomes 13 and homologous chromosomes 14 of the female U. kamensis (Q18-96) and for chromosomes and chromosome regions of M. auratus (MAUR): 6q distal, 6q proximal, 9, 11, 13, 14, 15, and 18 distal.
Fluorescence in situ hybridization (FISH)
The set of chromosome-specific probes and some microdissected painting probes of the golden hamster M. auratus (2n = 44) were described earlier17. The telomeric DNA probe was generated by PCR with oligonucleotides (TTAGGG)5 and (CCCTAA)518. Clones of human ribosomal DNA (rDNA) containing a partial 18S ribosomal gene, the full 5.8S gene, a part of the 28S gene, and two internal transcribed spacers were obtained as described elsewhere19. FISH was performed in accordance with previously published protocols. Images were captured using the VideoTest-FISH software (Imicrotec) with a JenOptic charge-coupled device (CCD) camera mounted on an Olympus BX53 microscope. Hybridization signals were assigned to specific chromosome regions identified by means of G-banding patterns photographed by the CCD camera. All images were processed in Corel Paint Shop Pro X3 (Jasc Software).
Molecular data
Molecular procedures employed for the sequencing of 25 nuclear genes and the mitochondrial Cytb gene are described in detail in Supplementary Information. The nuclear dataset included GenBank sequences of three other Old World hamster species (Cricetulus [barabensis] griseus, Mesocricetus auratus, and Phodopus ex gr. sungorus), which, along with Urocricetus, represent the main four lineages of Cricetinae according to ref.2. The composite outgroup included members of other cricetid subfamilies: Peromyscus sp. (Neotominae), Sigmodon sp. (Sigmodontinae), Ondatra zibethica, and Ellobius talpinus (Arvicolinae). The sequences were aligned by eye in BioEdit version 7.0.9.020 and then trimmed to exclude poorly alignable intronic regions. The length of the final nuclear alignment was 13,125 bp.
Phylogenetic analysis
Phylogenetic trees were reconstructed based on nuclear concatenation under maximum likelihood (ML) and Bayesian criteria. The ML reconstructions were conducted in IQTree version 1.621. The ModelFinder routine22 was utilized to determine the optimal partitioning scheme and best-fit substitution models for each subset under the Bayesian information criterion. At the initial stage, all gene × codon position combinations were treated as independent subsets. We tested both edge-linked proportional and edge-unlinked partition models. Clade support was assessed using Ultrafast Bootstrap23 with 10,000 replicates and by aBayes and SH-aLRT tests24; the latter procedure was carried out with 10,000 bootstrap replicates.
To identify a confidence set of trees, we calculated probabilities of all 15 possible topologies for Cricetinae by the approximately-unbiased (AU) test25. The branching pattern of outgroups was fixed to that in the ML tree. The test was performed in IQTree with 100,000 bootstrap replicates.
For single-gene analyses, ML trees were inferred as described above for each of the 25 genes. Gene tree concordance was evaluated by the AU tests contrasting the ML gene-specific topology with the ML tree for concatenated data.
Bayesian tree reconstruction and node age estimation were conducted in BEAST ver. 1.10.426 via concatenated alignment. The partitioning scheme and substitution models were those inferred by the ML procedure under the edge-unlinked partition model. Separate clock models were used for each data subset. Taking into account that the pattern of rate variation is unknown, we applied two relaxed-clock models: the uncorrelated log-normal clock and random local clock. Diffuse uniform priors were used for the mean rate and clock rate parameters. The calibrated Yule model was employed as the tree shape prior. The tree was subjected to secondary calibration for the root of Cricetidae (log-normal distribution with a mean of 18.5 My and standard deviation of 0.85 My) as estimated in ref.27. Chain length was 100 million generations, and parameters were logged to a file at every 50,000th step. Convergence diagnostics were performed via Tracer 1.728.
Results
Description of the U. kamensis karyotype
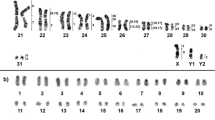
The diploid chromosome number in karyotypes of the male and female was found to be 2n = 30, NFa = 50 (Fig. 1). The autosomal set consists of five pairs of metacentrics (No. 1, 2, 4, 11, and 13), six pairs of submetacentrics (No. 5, 7, 8, 10, and 12), and three pairs of acrocentrics (No. 3, 6, and 9); pair No. 14 turned out to be heteromorphic and is composed of a small meta- and submetacentric. The X chromosome is represented by a medium-size acrocentric. The Y chromosome proved to be the smallest acrocentric in the male karyotype (Fig. 1).
C-banding revealed heterochromatic blocks in pericentromeric regions of all autosomes and X chromosomes (Fig. 1b). The submetacentric homolog of pair No. 14 carries a clearly visible C-positive block in the distal part of the q-arm. The X chromosome contains slightly visible interstitial C-blocks. The Y chromosome was found to be completely C-positive.
CMA3-positive interstitial blocks were seen in chromosomes 2 and X (Fig. 2a). Chromosomes 3 and 6 carry large bright CMA3-positive blocks. The smaller homolog of pair No. 14 has a distal CMA3-positive region, whereas the bigger one contains a 4′,6-diamidino-2-phenylindole (DAPI)-positive block in the q-arm (Fig. 2a). The hybridization indicated that the smaller homolog carries a cluster of rDNA (Fig. 2b). Two clusters of rDNA were also detected in the p-arm and a distal part of the q-arm of chromosome 6. A telomeric probe did not detect any interstitial localization (Fig. 2b).
(a) CDAG-banding of U. kamensis chromosomes; the blue arrow points to a block of AT-rich heterochromatin on one of the homologs of the pair of chromosomes 14, and the green arrow indicates a block of GC-rich heterochromatin on the other homolog of this pair. (b) Localization of telomeric (green) and rDNA (red) probes on U. kamensis chromosomes; red arrows mark rDNA clusters. G-banded metaphases are shown on the left.
Comparative cytogenetics
The set of chromosome-specific probes of the golden hamster contains several mixed probes17. Microdissection was performed to produce individual probes of those chromosomes that hit mixed peaks during flow-sorting. Probes of the following chromosomes of M. auratus or their regions were constructed: 6q distal, 6q proximal, 9, 11, 13, 14, 15, and 18 distal.
Most probes of M. auratus (MAUR), when localized on chromosomes of U. kamensis(UKAM), yielded an obvious signal (Fig. 3). No unambiguous signal was detectable on the pair of chromosomes 14 of U. kamensis. To determine the region of homology, microdissection of chromosomes 13 and 14 of U. kamensis from the same metaphase plate was carried out. Four probes (U1–U4) were obtained. The localization of the resultant probes on chromosomes of the original species and golden hamster was performed next (Fig. 3a,b). It was established that probes U1 and U3 are homologous to chromosome UKAM13 and that U2 and U4 are homologous to chromosome UKAM14 (Fig. 3a). It was demonstrated that the pair of chromosomes 14 of U. kamensis shares a homology region with the interstitial region of the q-arm of chromosome 4 of M. auratus. A part of chromosome 13 of U. kamensis is homologous to the interstitial region of the q-arm of chromosome 3 of M. auratus (Figs. 3b). Overall, 38 autosomal conserved segments were uncovered in the U. kamensis karyotype owing to the observed localization of the set of chromosome-specific probes of M. auratus (Fig. 1a).
Examples of FISH data. Localization of U. kamensis microdissection-derived probes: (a), U1 (green) and U2 (red) on chromosomes of U. kamensis; (b) U3 (green) and U4 (red) on chromosomes of M. auratus. Localization of M. auratus chromosome–specific probes on chromosomes of U. kamensis: (c) 14 (green) and 16 (red); (d) 9 (green) and 16 (red); (e) 4 (green) and 10 (red); (f) 3 (green) and 11 (red); (g) 9 (green) and 5 (red); and (h), 18 (green) and 7 (red). (i) Localization of microdissection-derived probes of M. auratus 16 (red) and M11 (green) on chromosomes of P. roborovskii. G-banded metaphases are presented on the left.
The new microdissection-derived probes of M. auratus and their subsequent localization on chromosomes of two Phodopus species (P. roborovskii and P. sungorus) allowed us to correct the previously published chromosome map for the genus29. It was revealed that earlier, in a comparison of banding patterns, the correspondence of the regions homologous to chromosomes MAUR11-14, and 16 was determined incorrectly. We demonstrated that PROB5p is homologous to MAUR16/14 instead of MAUR15, PROB10 is homologous to MAUR6/14 but not MAUR6/11, and PROB16 is homologous to MAUR14 rather than MAUR11 (Fig. 4). Besides, an additional tiny fragment of MAUR5 was uncovered in the distal part of the p-arm of chromosome PROB7. All the detected changes are also characteristic of homologous regions of P. sungorus chromosomes (PSUN) and, we believe, P. campbelli chromosomes. Microdissection-derived probe UKAM13 hybridized with PSUN5p, PSUN10, PROB11, and PROB15. Probe UKAM14 hybridized with interstitial parts of PSUN1q and PROB4 (data not shown).
The male karyotype of P. roborovskii with corrected localization of M. auratus probes (black lines) on G-banded chromosomes. Centromere positions are marked by black points. Black arrows indicate the regions in which the localization of probes changed relative to those published previously29.
Multilocus phylogenetic trees and molecular dating
The PartititonFinder routine subdivided the alignment of 25 nuclear genes into ten and five subsets for the edge-linked proportional and edge-unlinked models, respectively. The partitioning schemes and optimal substitution models are listed in Supplementary Information. The Bayesian information criterion score was lower for the edge-linked model (76,247.2 versus 76,812.4), consequently indicating a lack of pronounced variation in branch length proportions among the subsets.
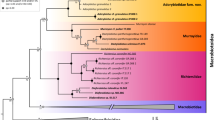
Both the ML tree and Bayesian tree (Fig. 5) contained Phodopus + Urocricetus and Cricetulus + Mesocricetus clades. This branching pattern is robustly supported by fast bootstrapping, Bayesian posterior probabilities, and P values from the aBayes and SH-aLRT tests. The AU test indicated that the 99.9% credible set contains a single tree.
ML phylogeny of Cricetinae as deduced from concatenated alignment of 25 nuclear genes. Clade support is shown at the nodes [top numbers: ML fast bootstrap via edge-linked model (%)/ML fast bootstrap via edge-unlinked model (%); bottom numbers: SH-aLRT support (%)/aBayes support]. Representatives of subfamilies Arvicolinae, Sigmodontinae, and Neotominae serve as outgroups.
The gene trees for the 25 markers had various topologies with only 10 genes reproducing the same branching pattern as that in the concatenated tree. The Phodopus + Urocricetus clade was present in 17 gene trees. Nevertheless, according to the AU tests, none of the gene-specific topologies was significantly better than that of the concatenated tree at P < 0.05. Therefore, all variations among the examined genes can be attributed to insufficient information content in certain alignments rather than to a significant conflict among loci.
According to the node age reconstruction in BEAST (Figure S1), the crown age of Cricetinae dates back to the Middle/Late Miocene boundary (11.2–12.0 Mya). The inferred time of divergence between Phodopus and Urocricetus (9.2–10.1 Mya) supports an early Late Miocene origin of these lineages.
The obtained two Cytb sequences were identical and were found to share the highest similarity with GenBank sequences having accession numbers MK047375, MK047376, and MH142301, differing from them only by seven substitutions. All these sequences belong to clade A as determined in ref.3, which we believe to correspond to true U. kamensis occurring in eastern Tibet and Nanshan.
Discussion
Urocricetus is one of the few genera of small mammals endemic to Tibet3. Numerous studies show that Tibetan fauna species have undoubtedly gone through habitat fragmentation and population fluctuations during glacial cold phases [see reviews in refs.3, 30]. It can be assumed that unstable environmental and climatic conditions along with the presence of high-altitude terrains acting as barriers have contributed to rapid chromosomal and molecular evolution.
Comparative cytogenetics
Karyotype variation within Urocricetus remains unstudied. The only previously published piece of information on the chromosome number is the indication that U. alticola is characterized by 2n = 22 as reported previously1. Nonetheless, the latter publication contains no reference to the original cytogenetic study, and therefore this report needs verification. Here we revealed that U. kamensis has 2n = 30, which is higher than 2n in hamsters of the Cricetulus group, where 2n = 20–24 is characteristic for all studied species.
The results of comparative chromosome painting convincingly showed that the U. kamensis karyotype is very different from that of the rest of cricetine hamsters in terms of the number of detected syntenic blocks and the type of identified M. auratus chromosome associations (Fig. 1a). The 38 autosomal conserved segments revealed in the U. kamensis karyotype is not only greater than this number in representatives of genera Cricetulus and Phodopus but also greater than that in all representatives of the subfamily17, 29.
In the U. kamensis karyotype, we detected some chromosome associations previously identified in karyotypes of other Cricetinae. MAUR17/19 is characteristic for all cricetine hamsters except Mesocricetus29. On the other hand, in karyotypes of Cricetulus and Allocricetulus, researchers have registered a full fragment of chromosome MAUR1917, 29, 31, 32, whereas in the U. kamensis karyotype, the association is formed by a part of MAUR19 (Fig. 1a). Chromosome MAUR19 is represented by three fragments in the genome of this species. Fission of this chromosome is also characteristic of Phodopus and Tscherskia, where MAUR19 was detected in two fragments [Fig. 4 and Ref.29].
In karyotypes of all hamsters except Mesocricetus, association MAUR3/13 containing a complete fragment of chromosome 3 has been reported17, 29, 31, 32. By contrast, in U. kamensis and Phodopus karyotypes, the association includes only one of the three fragments of chromosome 3 identified in the genome of the species under study (Fig. 1a)29. A similar situation was observed here with the MAUR3/11 association found in karyotypes of absolutely all cricetine hamsters. It is important to note that in the karyotype of U. kamensis, the association manifested itself as MAUR11/3/11, whereas in the P. roborovskii karyotype it looks like MAUR3/11/3 (Figs. 1a and 4). The question whether these associations have a common evolutionary origin remains open.
Association MAUR4/10, represented by complete fragments of chromosomes 4 and 10, has been identified in the Allocricetulus curtatus karyotype31. Of note, it is present in two more cricetine species. In Phodopus karyotypes, it contains only a fragment of chromosome 1029, but in the karyotype of U. kamensis, it features a fragment of chromosome 4 (Figs. 1a and 3e).
All representatives of Cricetulus and Allocricetulus have association MAUR5/9/14/16/15 in their karyotypes17, 29, 31, 32. Some components of this association have been documented in karyotypes of other hamsters, e.g., association MAUR5/9/14 in Phodopus29. Nevertheless, it has been shown that this association contains other parts of M. auratus chromosomes. It is likely that associations MAUR5/9, 9/14/16/9, and 15/16 found in the U. kamensis karyotype have independent evolutionary origins. Updating previously published data29 allowed us to determine precisely that in the karyotypes of U. kamensis and Phodopus species, MAUR14 is represented by three parts, whereas the number of MAUR16 fragments differs. Thus, in U. kamensis, we identified two fragments homologous to MAUR16, and both Phodopus species possess three (Figs. 1a and 4).
The discovery of an additional fragment of MAUR5 in Phodopus species led to the identification of a new association in their karyotypes (MAUR5/20). A similar association is typical for the U. kamensis karyotype, where it was found in the p-arm of chromosome 10 in our study. Previously, association MAUR5q/20 has also been registered in the karyotype of Tscherskia triton29, however, it is still unknown whether these associations contain identical fragments of chromosome MAUR5.
Our FISH data show that although some associations are shared by Urocricetus with other hamsters, and could, in fact, belong to the karyotype of their common ancestor, there is no unambiguous chromosomal synapomorphy supporting an association of Urocricetus with any other cricetine lineage.
Molecular phylogeny of Urocricetus
Molecular data convincingly support the hypothesis that Urocricetus is a member of a highly divergent lineage that is a sister group to Phodopus thereby corroborating previous results based on a less representative sampling (five nuclear genes2). According to our molecular-clock analysis, the four major lineages of Cricetinae (Phodopus, Urocricetus, Mesocricetus, and Cricetus:Tscherskia: Cricetulus clade) diverged in quick succession in the earliest Late Miocene, which apparently matches transition to a colder and more arid climate (middle Miocene transition33). Relatively short time intervals corresponding to basal internodes in the cricetine tree (1.3–2.0 My) may account for the lack of chromosomal synapomorphies for the Phodopus + Urocricetus clade.
Considering the intrageneric relationships, it should be emphasized that—contrary to the interpretation by Ding and Liao3 who regard Urocricetus as monotypic—the available genetic data (both nuclear and mitochondrial) are consistent with recognition of two species within the genus based on the genetic species concept34. Thus, mitochondrial phylogeographic data support the existence of two major clades that can be assigned to traditionally accepted U. kamensis and U. alticola. These clades are separated by the Cytb K2P distance of 6.7%, which falls within the divergence range between indisputable sister species of hamsters: i.e., Phodopus sungorus/Ph. campbelli (4.5%35), Mesocricetus auratus/M. raddei (6.0%35), and Cricetulus barabensis sensu lato/C. sokolovi (8.5%36). According to nuclear-gene analysis, the split between U. kamensis and U. alticola dates back to the end of the Early Pleistocene (940 Kya2),which is also suggestive of the species level divergence. Moreover, mitochondrial data from ref.3 reveal significant structuring within the U. alticola mitochondrial clade that consists of three main subclades supposedly corresponding to three subspecies: U. a. alticola, U. a. lama (Bonhote, 1905), and U. a. tibetanus (Thomas, 1922). At present, no morphological, chromosomal, or nuclear multilocus data are available that can elucidate the interrelationships among these taxa.
Conclusion
Considering the high level of genetic variation within Urocricetus and taking into account that Palearctic hamsters are characterized by a high rate of chromosomal evolution—so that in other cricetine genera, chromosomal data have played a major role in the discovery of cryptic species—one may expect that future studies on karyotypic variation within Tibetan hamsters may provide important insights into the taxonomy and evolutionary history of this genus.
References
Smith, A. T. & Xie, Y. A Guide to the Mammals of China. (Princeton University Press, Princet, 2008).https://doi.org/10.1515/9781400834112.1.
Lebedev, V. S. et al. Molecular phylogenetics and taxonomy of dwarf hamsters Cricetulus Milne-Edwards, 1867 (Cricetidae, Rodentia): Description of a new genus and reinstatement of another. Zootaxa 4387, 331–339 (2018).
Ding, L. & Liao, J. Phylogeography of the Tibetan hamster Cricetulus kamensis in response to uplift and environmental change in the Qinghai-Tibet Plateau. Ecol. Evol. 9, 7291–7306 (2019).
Kang, C. et al. The complete mitochondrial genome of Cricetulus kamensis (Rodentia: Cricetidae). Mitochondrial DNA 27, 976–977 (2016).
Lebedev, V. S., Ivanova, N. V., Pavlova, N. K. & Poltoraus, A. B. Molecular phylogeny of the Palearctic hamsters. in Systematics, phylogeny and paleontology of small mammals. Proceedings, International Conference, devoted to the 90-th anniversary of Prof. I. M. Gromov (eds. Averianov, A. & Abramson, N.) 114–118 (Zoological Institute RAS, 2003).
Ding, L., Zhou, Q., Sun, Y., Feoktistova, N. Y. & Liao, J. Two novel cricetine mitogenomes: Insight into the mitogenomic characteristics and phylogeny in Cricetinae (Rodentia: Cricetidae). Genomics 112, 1716–1725 (2020).
Wang, S. & Cheng, C.-L. Notes on Chinese hamsters (Cricetinae). Acta Zool. Sin. 19, 61–68 (1973).
Musser, G. G. & Carleton, M. D. Superfamily Muroidea. in Mammal Species of the World: A Taxonomie and Geographic Reference (eds. Wilson, D. E. & Reeder, D. M.) 894–1532 (John Hopkins Univ. Press, 2005).
Ford, C. E. & Hamerton, J. L. A colchicine, hypotonic citrate, squash sequence for mammalian chromosomes. Biotech. Histochem. 31, 247–251 (1956).
Stanyon, R. & Galleni, L. A rapid fibroblast culture technique for high resolution karyotypes. Bolletino di Zool. 58, 81–83 (1991).
Romanenko, S. A. et al. Segmental paleotetraploidy revealed in sterlet (Acipenser ruthenus) genome by chromosome painting. Mol. Cytogenet. 8, 90 (2015).
Seabright, M. A rapid banding technique for human chromosomes. Lancet 2, 971–972 (1971).
Sumner, A. T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75, 304–306 (1972).
Gladkikh, O. L. et al. Rapid karyotype evolution in Lasiopodomys involved at least two autosome - Sex chromosome translocations. PLoS ONE 11, 0167653 (2016).
Lemskaya, N. A. et al. A combined banding method that allows the reliable identification of chromosomes as well as differentiation of AT- and GC-rich heterochromatin. Chromosom. Res. 26, 307–315 (2018).
Yang, F., Trifonov, V., Ng, B. L., Kosyakova, N. & Carter, N. P. Generation of paint probes from flow-sorted and microdissected chromosomes. in Fluorescence In Situ Hybridization (FISH) (ed. Liehr, T.) 63–79 (2017). doi:https://doi.org/10.1007/978-3-662-52959-1_6.
Romanenko, S. A. et al. Reciprocal chromosome painting between three laboratory rodent species. Mamm. Genome 17, 1183–1192 (2006).
Ijdo, J. W., Wells, R. A., Baldini, A. & Reeders, S. T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 19, 4780 (1991).
Maden, B. E. H. et al. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem. J. 246, 519–527 (1987).
Hall, T. A. BioEdit: a User-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT.Nucleic Acids Symp. Ser .41, 95-98
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Minh, B. Q., Nguyen, M. A. T. & von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195 (2013).
Anisimova, M., Gil, M., Dufayard, J.-F., Dessimoz, C. & Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699 (2011).
Shimodaira, H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Lebedev, V. et al. Cryptic variation in mole voles Ellobius (Arvicolinae, Rodentia) of Mongolia. Zool. Scr. 49, 535-548 (2020).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901-904 (2018).
Romanenko, S. A. et al. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroidea, Rodentia) inferred from chromosomal painting and banding comparison. Chromosom. Res. 15, 283–297 (2007).
Yang, S., Dong, H. & Lei, F. Phylogeography of regional fauna on the Tibetan Plateau: A review. Prog. Nat. Sci. 19,789–799 (2009).
Romanenko, S. A. et al. Comparative cytogenetics of hamsters of the genus Allocricetulus Argyropulo 1932 (Cricetidae, Rodentia). Cytogenet. Genome Res. 139, 2582–2666 (2013).
Poplavskaya, N. S. et al. Karyotype evolution and phylogenetic relationships of cricetulus sokolovi orlov et Malygin 1988 (Cricetidae, Rodentia) inferred from chromosomal painting and molecular data. Cytogenet. Genome Res. 152, 65–72 (2017).
Holbourn, A., Kuhnt, W., Schulz, M. & Erlenkeuser, H. Impacts of orbital forcing and atmospheric carbon dioxide on Miocene ice-sheet expansion. Nature 438, 483–487 (2005).
Bradley, R. D. & Baker, R. J. A test of the genetic species concept: cytochrome-b sequences and mammals. J. Mammal. 82, 960–973 (2001).
Neumann, K. et al. Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene. Mol. Phylogenet. Evol. 39, 135–148 (2006).
Poplavskaya, N. et al. Phylogeographic structure in the chromosomally polymorphic rodent Cricetulus barabensis sensu lato (Mammalia, Cricetidae). J. Zool. Syst. Evol. Res. 57, 679–694 (2019).
Acknowledgements
The authors gratefully acknowledge the resources provided by the “Molecular and Cellular Biology” core facility of the IMCB SB RAS. The English language was corrected by shevchuk-editing.com.
Funding
This research was funded by the Russian Science Foundation (grant No. 19–14-00034 to A.S.G.) and the Russian Foundation for Basic Research (grant No. 20–54-53003 to A.V.S.).
Author information
Authors and Affiliations
Contributions
S.A.R. established the cell lines; prepared the chromosome suspensions; labeled and prepared the probes for FISH; carried out the FISH experiments, microdissection, banding, probe amplification, and microscopy analysis; and wrote the manuscript. V.S.L. performed the phylogenetic analysis. A.A.B. did the gene sequencing. S.V.P. obtained the chromosome suspensions from bone marrow, carried out the banding, and contributed to the drafting of the manuscript. N.A.S. extracted Cot DNA. N.Y.F., Q.J., S.Y., and A.V.S. collected the materials. A.V.S. and A.S.G. conceived and supervised the project. A.S.G. discussed the results with S.A.R. and revised the manuscript. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Romanenko, S.A., Lebedev, V.S., Bannikova, A.A. et al. Karyotypic and molecular evidence supports the endemic Tibetan hamsters as a separate divergent lineage of Cricetinae. Sci Rep 11, 10557 (2021). https://doi.org/10.1038/s41598-021-89890-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89890-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.