Abstract
Insects entombed in copal, the sub-fossilized resin precursor of amber, represent a potential source of genetic data for extinct and extant, but endangered or elusive, species. Despite several studies demonstrated that it is not possible to recover endogenous DNA from insect inclusions, the preservation of biomolecules in fossilized resins samples is still under debate. In this study, we tested the possibility of obtaining endogenous ancient DNA (aDNA) molecules from insects preserved in copal, applying experimental protocols specifically designed for aDNA recovery. We were able to extract endogenous DNA molecules from one of the two samples analyzed, and to identify the taxonomic status of the specimen. Even if the sample was found well protected from external contaminants, the recovered DNA was low concentrated and extremely degraded, compared to the sample age. We conclude that it is possible to obtain genomic data from resin-entombed organisms, although we discourage aDNA analysis because of the destructive method of extraction protocols and the non-reproducibility of the results.
Similar content being viewed by others
Introduction
Since the first ancient DNA (aDNA) was successfully recovered from an extinct species in 19841, palaeogenetic analyses have revolutionized many fields of research. In particular, bones and teeth have been the most used substrates over the last thirty years. However, current developments in Next Generation Sequencing (NGS) technologies coupled with optimized DNA extraction protocols2,3,4,5,6,7, have recently allowed to perform genomic analysis also using alternative biomaterials8,9,10,11,12,13,14. Nowadays, palaeogenetic researches involve historical and prehistoric findings hailing from archaeological excavations, libraries and museum collections. One of the alternative substrates from which scientists attempted to extract DNA is amber and copal inclusions that can embed organisms older than 30 millions of years (MY)15,16.
Fossilized tree resins represent an important research source, offering numerous opportunities to study biogeography, behavior, extinction and evolutionary changes over multiple generations. Arthropods, and especially insects, are the most abundant and well-preserved life forms in amber and copal, even if plants and rare soft-bodied vertebrates can also be maintained within fossilized resins17,18,19,20. Because amber and copal conserve three-dimensional form, color patterns and microscopic details of the engulfed organisms, such fossils can be easily compared with the relative extant specimens, representing a unique recapitulation of arthropods evolution starting from the Cretaceous era. Moreover, covering more than 200 MY of earth history and being spread throughout several climate zones, resin fossils provide an exceptional source of information to interpret past terrestrial environments and ecosystems21,22,23. Amber and copal are vegetal resins that have been fossilized, through a diagenetic process called amberization that is due to the terpenoid resins transformation24. The essential difference between amber and copal is their states of polymerization and their established ages25. Copal is usually considered an intermediate phase between fresh resin and amber, a completely fossilized resin that takes millions of years to transform from copal26. Resin, secreted from parenchymal plants cells in response to biotic or abiotic host trauma, can entrap living and dead organisms. Despite chemical composition of fossil resins exhibit extensive variations27, the complete and rapid engulfment of the organisms embedded seems to preserve specimen morphology and tissues structure during time. On the other hand, over millions of years of diagenetic events, required to transforms resin into copal before and amber subsequently, minimize the likelihood of aDNA preservation especially in amber specimens. Notably, when DNA is extracted from fossil inclusions, it can be used as a powerful tool to test new evolutionary hypotheses. In entomology, for example, ancient DNA can reveal new phylogenetic relationships between taxa, which are undetectable using other methodologies. However, the preservation of biomolecules in fossilized resins samples is still under debate when considering very old sample inclusions. Stankiewicz et al.28 detected chitin-protein complexes in insect cuticle from sub-fossil resins, while a recent study found amino acids from fossil feathers in amber from almost 100 million years BP29. Additionally, Bada et al.30 uncovered low levels of amino acid racemization rate in amber insect inclusions, implying that peptides could be preserved in these. Although there is no direct correlation between amino acid preservation and DNA survival31, theoretically, within this protective environment, DNA might be better conserved from external degradative processes compared to other substrates. On the other hand, because DNA degrades over time due to the spontaneous chemical breakdown32,33, it could be preserved in resins and young copals but almost certainly not in amber. Few studies have been performed in order to explore the possibility to extract uncontaminated DNA from copal and amber, yielding contrasting results15,34,35,36 (and references therein). Nevertheless, in 2013, Penney and colleagues37 were unable to obtain any convincing evidence for the preservation of endogenous aDNA in two copals with different age, dated to “post-Bomb” and 10,612 ± 62 cal. BP respectively, thus concluding that DNA is not preserved in this type of material, neither in the recent ones. On the contrary, molecules of endogenous DNA seem to be present in insects embedded in modern resins, albeit in small quantities. Recently, Peris et al.38 were able to amplify DNA from ambrosia beetles recovered in 6 and 2 year old resins, using a PCR-based method. Despite these positive results, access to ancient DNA molecules in older resins is still a challenge. In this work, we explored the possibility to obtain endogenous aDNA molecules from insects preserved in copal applying NGS technologies and experimental protocols specifically designed for recovering and analyzing highly degraded ancient DNA. This approach is more suitable than PCR-based method, allowing to distinguish ancient endogenous molecules from modern contaminants, as well as to evaluate DNA preservation status by fragmentation and deamination patterns analysis. The same procedures were successfully adopted to genotype very ancient specimens, such as Neanderthals, Denisova and Homo heidelbergensis39,40,41,42,43. Because the possibility of extracting insect aDNA from amber samples, dated millions of years ago, is still unknown37, we performed our analysis on two Colombian copal insect inclusions dated back 60 years. Moreover, aDNA isolation from insect inclusions requires a destructive sampling; for this reason, the relatively recent material we analyzed represent a suitable opportunity, in order to preserve rare and very old species stored in museum collections.
Materials and methods
We analyzed two insects embedded in two Colombian sub-fossil resins, labeled C101 and C67 respectively (Fig. 1). Only one complete insect was present in each copal and no other equivalent specimens are available for further analyses. Copal were acquired by C.Z. for commercial use and the provided information regarding the regional location of the samples refer to Vélez—Santander (Colombia). To our knowledge, copals were untreated with high pressure heat to increase their durability and commercial appeal, which may have compromised the DNA preservation. C.Z. authorized sampling to conduct the scientific research. Direct radiocarbon dating was performed on each copal at the Beta Analytic testing Laboratory (London). Before sampling for molecular analysis, inclusions were morphologically characterized in order to identify their taxonomical attribution (Table 1).
Genetic analysis was performed in a dedicated aDNA facility, at the Laboratory of Molecular Anthropology and Paleogenetics (University of Florence, Italy), where no experiments have been previously performed on entomological samples. Preventive measures were taken to avoid contamination during all the experimental procedures44,45, and negative controls were processed during each step.
To remove environmental contaminants, copal was washed with 30% bleach and UV-irradiated (254 nm) for 30 min for each side. Sampling was carried out as described in Penney et al.37, with some modifications: each specimen was trimmed to a small cube using a micro-drill with disposable disk saw, taking care not to expose the insects. Inclusions were extracted dissolving, through rotation, the remaining resin in 5 mL chloroform for 24 h at RT conditions46, to minimize the DNA degradation risk. Each fully intact insect was drawn out from the dissolved copal and washed 3 times with 500 µL of Molecular Biology Grade water (ddH2O). Samples were finely crushed with a sterile micropestel and were subsequently incubated overnight at 55 °C in 1 mL of Digestion Buffer, which is specific for DNA extraction coming from chitinous tissues (10 mM Tris buffer pH 8.0; 10 mM NaCl; 5 mM CaCl2; 2 mM EDTA pH 8.0; 2% SDS; 25 mM DTT and 0.25 mg/ml Proteinase K)7. After pelleting, supernatant was extracted using a protocol properly designed for short fragments recovery2. DNA was quantified using QubitTM 4 Fluorometer (dsDNA High Sensitivity Kit). Twenty microliters of the extracts were used to prepare the libraries, following a custom double-indexing protocol optimized for ancient samples47,48. No uracil DNA glycosylase (UDG) treatment was performed in order to retain misincorporation profiles. Libraries concentration was determined with the Agilent 2100 Bioanalyzer (DNA 1000 chip). Afterwards libraries were pooled in equimolar amount with other samples and finally sequenced with an Illumina NovaSeq 6000 (SP flowcell) with a single-end 1 × 100 + 8 + 8 cycles at the Laboratorio di Genomica Avanzata, Univerity of Florence.
According to sample specific index sequences, raw reads were demultiplexed. EAGER pipeline49 was used for the initial reads quality control, the adapter trimming and for mapping the reads to the human reference genome in order to monitor and remove human contamination. FastQC50 was used to perform the quality control of the reads. Adapter sequences were trimmed with Clip&Merge v. 1.7.449 and, fragments less than 30 base pairs (bp), were discarded. BWA v. 0.7.1051, set with aDNA specific parameters (-l 1000, -n 0.01, -o 2)52, was applied to map filtered reads to the human reference genome (Hg19, NCBI Build 38, December 2017). PCR duplicates were removed using DeDup49, and reads presenting a mapping quality lower than 30 (filtered with SAMtools-1.753) were also omitted from the analysis. In order to identify the DNA sequences contained in our copal sample, we downloaded the entire NCBI database (version 4) (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov) and built the related BLAST54 database files (version 2.9 database). Then we aligned our copal sequences previously processed with the de-novo assembler ABySS 2.055 (parameters: -t 64 -pe openmpi 32) against the NCBI BLAST database. The xml BLAST results were parsed using a script written in Python 3 (https://github.com/tacclab/NCBIXML_parser) that used the module Bio.blast and the NCBIXML class in order to obtain a table including the sequence length, the taxonomic group, the corresponding database entry, the organism description, the length of the matching, BLAST Bit-score, E-value, the query start, the query end, the subject start and the subject end (See Supplementary Table S1). To validate our previous results we preprocessed the set of DNA sequences contained in our xml BLAST file with MEGAN656. MEGAN computed and analyzed the taxonomical content of our dataset, employing the NCBI taxonomy to summarize and order the output.
The actual degradation of the embedded insects DNA was assessed by deamination and length patterns analysis. It has been shown that, over time, cytosines principally located at the ends of the fragments are deaminated to uracils57, which are sequenced as thymine residues. The C to T misincorporation patterns at the 5′-end of the molecule can be used to test the authenticity of aDNA sequences58,59. Since the complete genome of our identified insect is not available in the NCBI database, reads were aligned against rRNAs and mitochondrial genes related to identified the insect species Ogcodes basalis (See Supplementary Tables S1 and S2). Ancient DNA damage patterns were examined using MapDamage260, setting –l 99 and –forward. As 100 bp single-end sequencing was carried out, mapDamage was performed on fragments up to 99 bp long in order to be sure about the full fragment sequence. Finally, cytochrome oxidase I (COI) sequence was called using mpileup and vcfutils.pl of the SAMtools-1.7 package53 and phylogenetically analyzed, to confirm the species-level identity of the copal insect inclusion. The software MUSCLE61 was used to align our COI sequence to that of six different Ogcodes species available in NCBI database (Supplementary Table S3). Nemestrinus sp. and Stratiomyidae sp. were used as out-groups. A phylogenetic tree was built with the Maximum Parsimony method (Subtree-Pruning-Regrafting algorithm) using MEGAX62. The tree was constructed with 99% partial deletion and phylogeny was tested with the bootstrap method with 1000 replications. Tree was then visualized by FigTree 1.4.4 (available from http://tree.bio.ed.ac.uk/software/figtree/).
Results
We performed two direct radiocarbon analyses for our samples which both date to post-Bomb (Table 1). The initial qualitative and quantitative analysis showed differences in the DNA preservation degree between the two inclusions. DNA from C101 sample was 0.065 ng/µL, while DNA was not quantifiable for C67. Samples were converted into genomic libraries. After a first attempt starting from 20 µL of the extract, library was repeated for C67 starting from 30 µL of the same extracts. In both case, it was not possible to obtain a good quality library. According to these results, C67 was excluded from further analysis, thus C101 was successively sequenced. 33,271,931 reads were generated by shotgun sequencing (Table 2). Reads mapped to the human genome were 8220 (0.025%)–7430 (0.022%) after removing duplicates (Supplementary Table S4)—and they represent modern contamination, as shown by deamination profiles (Supplementary Fig. S1).
Using ABySS 2.0, we assembled the non-human reads from the Fastq files, obtaining 18 assembled sequences with a length greater than 200. Seven sequences have also an E-value and a Bit-score less than 0.01 and greater than 394 respectively. Among the top seven sequences, the Ogcodes basalis species is the mostly identified organism by the BLAST algorithm (Supplementary Table S1), which was compared against all the genomes included in the NCBI repository. The vast majority of the hits associated to this species gave significant BLAST Bit-score higher than 190. Contigs showing lower scores have not been considered because they represent sequences phylogenetically associated to Ogcodes genus, such as the Lysiphlebus testaceipes wasp, the Culicoides sonorensis midge or the Bactrocera cucurbitae fly (see Supplementary Table S1). Same results were obtained using MEGAN6 (Fig. 2), where the genus Ogcodes belong to the class with the highest assigned value. We also found one hit matching with Wolbachia sp, with high Bit-score (298) and E-value less than 0.01 that showed typical features of aDNA (Supplementary Fig. S2).
Because O. basalis is the best represented taxon relative to other species of the genus in NCBI database, we performed a phylogenetic analysis among Ogcodes exemplars (Supplementary Table S3) based on COI sequence data, to confirm the attribution obtained with BLAST algorithm. We reconstructed the 64.98% of the COI with an average depth of coverage of 11.01. The phylogenetic position of the sample was assessed using Maximum Parsimony tree (Supplementary Fig. S3). C101 shows strong affinity with the O. basalis, confirming previous results.
Afterwards, raw reads were mapped against the genomic sequences, retrieved from NCBI database, of the O. basalis species (Supplementary Table S1). Despite incompleteness of the reference sequence, 277,876 mapped reads were obtained. However, after the duplicates removal, only 9845 reads were retained, showing a high value (27.1) of cluster factor (ratio of reads before and after PCR duplicate removal) (Table 2 and Supplementary Table S2). Since this measure is likely related to the library complexity level, high value of cluster factor indicates a low quality/quantity of the starting genomic material. The aforementioned value is significantly higher in the mapping statistic obtained for O. basalis than in the one gained for Homo sapiens (Supplementary Table S4). However, this result can be considered reasonable when looking at the different length of the analyzed genomes. Considering that the partial genome of the O. basalis used as reference has a length of 16,243 bp, the filtered reads obtained after duplicate removal are consistent to achieve a deep coverage (53.92%) of the entire sequence annotated. On the contrary, the number of reads mapping on the human reference genome are extremely low if compared to its extent. For this reason, experimental amplification is unlikely to have led to an over-representation of the initial human molecules, but it could have determined the high cluster factor for O. basalis. Comparable values of duplication rate were reached in Weyrich et al.10 where they compared the mapping statistics obtained for bacterial and human mitochondrial genomes which are characterized by a similar length.
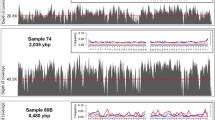
Mapped reads displayed typical features of aDNA: short fragments length (average value of 88.96 bp), and high rate of C to T deamination (21.93%) at the 5′ termini of the molecules (Table 2 and Fig. 3), confirming the authenticity of data. Extraction and library negative controls yielded a total of 2,250,680 of reads, whereas only 56 of them mapped to the human genome. No reads associated to O. basalis or other fly insects were retrieved.
Deamination, base frequency and fragmentation patterns observed on Ogcodes basalis. Misincorporation plot (a, bottom plot) shows the percent frequency of C to T mismatches at 5′ read ends (in red print) and complementary G to A mismatches at the 3′ ends (in blue print); base composition plots (a, upper plots), describe the base frequency outside and in the read. Length distribution plot (b) show the frequency of read lengths.
Discussion
In this work, we applied the most innovative molecular and bioinformatics techniques in order to analyze two resin-entombed insects and their DNA preservation in Colombian copal. DNA was successfully extracted at very low concentration from sample C101 (0.065 ng/µL), while no genetic material was retrieved from the second one (C67). Despite the few amount of the starting genetic material, C101 sample yielded enough quantity of DNA to assess its preservation degree in the sub-fossil resin. One of the main issues in aDNA studies is contamination; DNA molecules from people who handle the samples can easily exceed the small amounts of endogenous aDNA63,64,65,66. In our case, the number of reads which mapped to the human genome, was very low, 8202 (0.025%) and 7430 (0.022%) before and after duplicates removal (Supplementary Table S4), suggesting that resin prevents and protects the inner environment and the endogenous DNA from external contaminants. BLAST alignment of our non-human assembled contigs, revealed significant percentages of similarity with O. basalis species (Supplementary Table S1). Similar results were obtained with MEGAN656 where Ogcodes is the class with the highest assigned value (Fig. 2). Taxonomic attribution was also confirmed by phylogenetic analysis of the COI sequence. Maximum Parsimony tree shows clearly that C101 has a significantly higher affinity to O. basalis when compared to other Ogcodes exemplars (Supplementary Fig. S3). Ogcodes is the largest and widely distributed genus of the spider-parasite Acroceridae family, likely evolved during the late Triassic and greatly diversified during the Cretaceous67. Species belong to the genus show typical globose body, moderately small head and reduced wing venation. This group is relatively well represented in the fossil record, with some species being quite common in Baltic and Dominican amber deposits68. On the contrary, no Ogcodes exemplars are described in Colombian copal assemblages until now. Concordantly, same result was obtained with the observation of the phenotypic characteristics of the insect body embedded in the copal. This analysis identified our sample as belonging to the order Diptera and sub-order Brachycera that are the taxa to whom the genus Ogcodes belongs (Table 1). Interestingly, we also found one hit matching with Wolbachia sp, a gram-negative bacteria genus, that usually infects flying insects, especially species belonging to the Diptera order69. Because reads showed clear signatures of damage (Supplementary Fig. S2), it is reasonable to assume that this sequences arise from pathogens already present in the analyzed sample. Considering that typical microbial DNA molecules coming from environmental matrixes (e.g. soil) were not identified, our data can confirm that resins protect inclusions from external contamination. Although these are promising results, high proportion of low significance scores hits (see Supplementary Table S1) were detected. Probably, they are sequences derived from extremely degraded DNA. Indeed, when we performed a detailed analysis of the reads mapping Ogcodes genus, we found high degradation levels of the endogenous DNA (Table 2 and Fig. 3). First, the high value of cluster factor (27.1) indicates a low complexity levels in the sequenced library. Since this parameter is correlated with the quality and the quantity of the starting DNA70, these results lead us to question about the preservation of the endogenous genetic material. Another possible explanation for the high value of cluster factor is the suboptimal procedure applied to release the embedded insect. Indeed, Peris et al.38 found that chloroform used for dissolving resin surrounding the specimen could compromise DNA concentration. Secondly, damage pattern observed at the ends of the fragments (21.93% at 5′ end) highlights a high level of degradation (Fig. 3). Sawyer et al.59 reported a strong positive correlation between C to T substitutions and the age of the samples for bone remains; for this reason, nucleotide misincorporation patterns can be used as a valuable criteria to distinguish recent DNA sources from ancient ones. However, given the young age of the copal, molecules from O. basalis showed C to T frequencies higher than other samples with the same age range59. Indeed, despite their age, while C67 sample did not provide any genetic material, endogenous DNA extracted from C101 was low concentrated and extremely degraded.
Conclusion
Resin fossils represent a unique palaeobiological resource. Given their peculiar fossilization process and the exceptional morphological and biochemical preservation, they were thought to be the best means to obtain genetic material from ancient specimens. Although previous studies claim the impossibility to recover endogenous DNA from copal and amber, our positive results demonstrate that also sub-fossil resin inclusions, even though with cautions, may be useful in aDNA researches. Indeed, we were able to identify the taxonomic status of the specimen embedded in a Colombian copal following the most innovative molecular techniques developed to analyze highly degraded DNA. At the same time, we ascertained and described a high level of DNA degradation, despite the young age of the specimen. For this reason, because of the destructive method of DNA extraction protocols and the non-reproducibility of the results, we strongly discourage aDNA analysis from amber-preserved fossils, even if our study was carried out on copal inclusion.
Data availability
Raw data are available at the NCBI Sequence Read Archive under Accession Number PRJNA633577.
References
Higuchi, R. et al. DNA sequences from the quagga, an extinct member of the horse family. Nature 312, 282–284 (1984).
Dabney, J. et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U. S. A. 110, 15758–15763 (2013).
Hansen, H. B. et al. Comparing ancient DNA preservation in petrous bone and tooth cementum. PLoS ONE 12, e0170940 (2017).
Hagan, R. W. et al. Comparison of extraction methods for recovering ancient microbial DNA from paleofeces. Am. J. Phys. Anthropol. 171, 275–284 (2020).
Epp, L. S., Zimmermann, H. H. & Stoof-Leichsenring, K. R. Sampling and extraction of ancient DNA from sediments. In Ancient DNA. Methods in Molecular Biology (eds Shapiro, B. et al.) 31–44 (Humana Press, 2019).
Modi, A. et al. Combined methodologies for gaining much information from ancient dental calculus: testing experimental strategies for simultaneously analysing DNA and food residues. Archaeol. Anthropol. Sci. 12, 10 (2020).
Campos, P. F. & Gilbert, M. T. P. DNA extraction from keratin and chitin. In Ancient DNA. Methods in Molecular Biology (eds Shapiro, B. et al.) 57–63 (Humana Press, 2019).
Adler, C. J. et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the neolithic and industrial revolutions. Nat. Genet. 45, 450-455e1 (2013).
Warinner, C. et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 46, 336–344 (2014).
Weyrich, L. S. et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 544, 357–361 (2017).
Slon, V. et al. Neandertal and Denisovan DNA from Pleistocene sediments. Science 356, 605–608 (2017).
Teasdale, M. D. et al. The York Gospels: a 1000-year biological palimpsest. R. Soc. Open. Sci. 4, 170988 (2017).
Boast, A. et al. Coprolites reveal ecological interactions lost with the extinction of New Zealand birds. Proc. Natl. Acad. Sci. U. S. A. 115, 1546–1551 (2018).
Zarrillo, S. et al. The use and domestication of Theobroma cacao during the mid-Holocene in the upper Amazon. Nat. Ecol. Evol. 2, 1879–1888 (2018).
Cano, R. J. et al. Amplification and sequencing of DNA from a 120–135 million-year-old weevil. Nature 363, 536–538 (1993).
DeSalle, R. et al. DNA sequences from a fossil termite in Oligo-Miocene amber and their phylogenetic implications. Science 257, 1933–1936 (1992).
Sherratt, E. et al. Amber fossils demonstrate deep-time stability of Caribbean lizard communities. Proc. Natl. Acad. Sci. U. S. A. 112, 9961–9966 (2015).
Sadowski, E. M. et al. Carnivorous leaves from Baltic amber. Proc. Natl. Acad. Sci. U. S. A. 112, 190–195 (2015).
Xing, L. et al. A feathered dinosaur tail with primitive plumage trapped in mid-Cretaceous amber. Curr. Biol. 26, 3352–3360 (2016).
Rikkinen, J., Grimaldi, D. A. & Schmidt, A. R. Morphological stasis in the first myxomycete from the Mesozoic, and the likely role of cryptobiosis. Sci. Rep. 9, 19730 (2019).
Peñalver, E. et al. Thrips pollination of Mesozoic gymnosperms. Proc. Natl. Acad. Sci. U. S. A. 109, 8623–8628 (2012).
Cai, C. et al. Beetle pollination of cycads in the mesozoic. Curr. Biol. 28, 2806-2812.e1 (2018).
Bao, T., Wang, B., Li, J. & Dilcher, D. Pollination of Cretaceous flowers. Proc. Natl. Acad. Sci. U. S. A. 116, 24707–24711 (2019).
Labandeira, C. Amber. Paleont. Soc. Pap. 20, 163–215 (2014).
Solórzano-Kraemer, M. M. et al. A revised definition for copal and its significance for palaeontological and Anthropocene biodiversity-loss studies. Sci. Rep. 10, 19904 (2020).
Clifford, D. J. & Hatcher, P. G. Structural transformations of polylabdanoid resinites during maturation. Org. Geochem. 23, 407–418 (1995).
Lambert, J. B., Santiago-Blay, J. A., Wu, Y. & Levy, A. J. Examination of amber and 490 related materials by NMR spectroscopy. Magn. Reson. Chem. 53, 2–8 (2015).
Stankiewicz, B. A. et al. Chemical preservation of plants and insects in natural resins. Proc. Biol. Sci. 265, 641–647 (1998).
McCoy, V. E. et al. Ancient amino acids from fossil feathers in amber. Sci. Rep. 9, 6420 (2019).
Bada, J. L. et al. Amino acid racemization in amber-entombed insects: implications for DNA preservation. Geochim. Cosmochim. Acta. 58, 3131–3135 (1994).
Collins, M. J. et al. Is amino acid racemization a useful tool for screening for ancient DNA in bone?. Proc. R. Soc. B 276, 2971–2977 (2009).
Allentoft, M. E. et al. The half-life of DNA in bone: measuring decay kinetics in 158 dated fossils. Proc. R. Soc. B 279, 4724–4733 (2012).
Kistler, L. et al. A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Res. 45, 6310–6320 (2017).
DeSalle, R., Barcia, M. & Wray, C. PCR jumping in clones of 30-million-year-old DNA fragments from amber preserved termites (Mastotermes electrodominicus). Experientia 49, 906–909 (1993).
Poinar, G. O., Poinar, H. N. & Cano, R. J. DNA from amber inclusions. In Ancient DNA (eds Herrmann, B. & Hummel, S.) 92–103 (Springer, 1994).
Austin, J. J. et al. Problems of reproducibility: does geologically ancient DNA survive in amber-preserved insects?. Proc. Roy. Soc. Lond. Ser. B 264, 467–474 (1997).
Penney, D. et al. Absence of ancient DNA in sub-fossil insect inclusions preserved in ‘Anthropocene’ Colombian copal. PLoS ONE 8, e73150 (2013).
Peris, D. et al. DNA from resin-embedded organisms: past, present and future. PLoS ONE 15, e0239521 (2020).
Reich, D. et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010).
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012).
Prüfer, K. et al. The complete genome sequence of a Neandertal from the Altai Mountains. Nature 505, 43–49 (2014).
Prüfer, K. et al. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017).
Meyer, M. et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507 (2016).
Gilbert, M. T., Bandelt, H. J., Hofreiter, M. & Barnes, I. Assessing ancient DNA studies. Trends Ecol. Evol. 20, 541–544 (2005).
Willerslev, E. & Cooper, A. Ancient DNA. Proc. Biol. Sci. 272, 3–16 (2005).
Penney, D., Wadsworth, C. & Green, D. I. Extraction of inclusions from (sub)fossil resins, with description of a new species of stingless bee (Hymenoptera: Apidae: Meliponini), in quaternary Colombian copal. Paleontol. Contrib. 2013, 7:1–6 (2013).
Meyer, M. & Kircher, M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 6, pdb.prot5448 (2010).
Modi, A. et al. Complete mitochondrial sequences from Mesolithic Sardinia. Sci. Rep. 7, 42869 (2017).
Peltzer, A. et al. EAGER: efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016).
Andrews, S. FastQC: a quality control tool for high throughput sequence data (2010). Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Schubert, M. et al. Improving ancient DNA read mapping against modern reference genomes. BMC Genomics 13, 178 (2012).
Li, H. et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Altschul, S. et al. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Jackman, S. D. et al. ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27, 768–777 (2017).
Huson, D. H. et al. MEGAN community edition: interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016).
Hofreiter, M., Jaenicke, V., Serre, D., Haeseler, A. & Pääbo, S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 9, 4793–4799 (2011).
Briggs, A. W. et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U. S. A. 104, 14616–14621 (2007).
Sawyer, S., Krause, J., Guschanski, K., Savolainen, V. & Pääbo, S. Temporal patterns of nucleotide misincorporations and DNA fragmentation in ancient DNA. PLoS ONE 7, e34131 (2012).
Jonsson, H., Ginolhac, A., Schubert, M., Johnson, P. L. & Orlando, L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29, 1682–1684 (2013).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Kumar, S. et al. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018).
Noonan, J. P. et al. Genomic sequencing of pleistocene cave bears. Science 309, 597–599 (2005).
Green, R. E. et al. Analysis of one million base pairs of Neanderthal DNA. Nature 444, 330–336 (2006).
Garcia-Garcera, M. et al. Fragmentation of contaminant and endogenous DNA in ancient samples determined by shotgun sequencing; prospects for human palaeogenomics. PLoS ONE 6, e24161 (2011).
Llamas, B. et al. From the field to the laboratory: Controlling DNA contamination in human ancient DNA research in the high-throughput sequencing era. STAR 3, 1–14 (2017).
Wintertona, S. L., Brian, M. W. & Evert, I. S. Phylogeny and Bayesian divergence time estimations of small-headed Xies (Diptera: Acroceridae) using multiple molecular markers. Mol. Phylogenet. Evol. 43, 808–832 (2007).
Gillung, J. P. & Wintertona, S. L. Evolution of fossil and living spider flies based onmorphological and molecular data (Diptera, Acroceridae). Syst. Entomol. 44, 820–841 (2019).
Klasson, L. et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc. Natl. Acad. Sci. U. S. A. 106, 5725–5730 (2009).
Daley, T. & Smith, A. D. Predicting the molecular complexity of sequencing libraries. Nat. Methods 10, 325–327 (2013).
Acknowledgements
The authors thank Prof. Günter Bechly for morphological analysis. This work was supported by the facilities of Department of Excellence fund to Department of Biology University of Florence (2018–2022), and SID2017 Project code BIRD171214 from MAPS Department at University of Padova.
Author information
Authors and Affiliations
Contributions
C.T., D.C., A.M. and S.V. conceived the project. C.Z. provided the samples. A.M., C.Ve., S.V. and M.L. designed the sequencing experiments. C.T., A.M, G.M.T. and C.V. carried out data analysis. C.T. and A.M. wrote the paper with the input of all coauthors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Modi, A., Vergata, C., Zilli, C. et al. Successful extraction of insect DNA from recent copal inclusions: limits and perspectives. Sci Rep 11, 6851 (2021). https://doi.org/10.1038/s41598-021-86058-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-86058-9
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.