Abstract
The soil-borne pathogen Rhizoctonia solani infects a broad range of plants worldwide and is responsible for significant crop losses. Rhizoctonia solani AG3-PT attacks germinating potato sprouts underground while molecular responses during interaction are unknown. To gain insights into processes induced in the fungus especially at early stage of interaction, transcriptional activity was compared between growth of mycelium in liquid culture and the growing fungus in interaction with potato sprouts using RNA-sequencing. Genes coding for enzymes with diverse hydrolase activities were strongly differentially expressed, however with remarkably dissimilar time response. While at 3 dpi, expression of genes coding for peptidases was predominantly induced, strongest induction was found for genes encoding hydrolases acting on cell wall components at 8 dpi. Several genes with unknown function were also differentially expressed, thus assuming putative roles as effectors to support host colonization. In summary, the presented analysis characterizes the necrotrophic lifestyle of R. solani AG3-PT during early interaction with its host.
Similar content being viewed by others
Introduction
The soil-borne fungus Rhizoctonia solani (J. G. Kühn) [teleomorph Thanatephorus cucumeris (A. B. Frank) Donk] belongs to the phylum Basidiomycota. R. solani is a soil-borne plant pathogenic fungus with worldwide distribution and infects many economically important crops like rice, soybean, potato, maize, sugar beet, cabbage, tomato, and lettuce1. R. solani is a species complex of various groups called anastomosis groups (AG), some of which are subdivided into additional subgroups2,3. Hyphae of isolates belonging to the same AG are able to anastomose. In total, 13 different AGs of R. solani have been described with their subgroups differing in morphological and genetic characteristics4,5. Members of the different AGs and the various subgroups show a distinct degree in host specificity6,7.
Diseases on potato caused by R. solani occur in all areas, where potatoes are grown and affect the qualitative and quantitative yield of potato tubers. About 30% of tuber yield loss has been reported8. Rhizoctonia solani AG3 was found to be the predominant AG associated with potato9,10,11,12,13, and the so-called R. solani AG3-potato type (PT) was developed14. Infection of potato by R. solani AG3-PT may result in the characteristic disease symptoms of stem canker and black scurf15. Shortly after planting, necroses on germinating sprouts are the typical symptoms of stem canker, and results in late emergence of potato plants in the field, lower numbers of stems, shorter stolons and deformation of progeny tubers16. It is postulated that necrotic lesions on the stolons disrupt the delivery of photosynthates leading to this characteristic tuber phenotype17. Black scurf symptoms appear later in the season, when sclerotia start to cover ripening potato tubers8. These tuber-borne R. solani inocula can be the main source of primary infection causing stem canker symptoms of below-ground plant tissue of the next crop. Increasing incidence of this plant disease and a current lack of effective control measures requires an improved understanding of the biology behind the processes.
Based on their feeding strategies, plant pathogenic fungi are divided into biotrophs, which feed on living host tissue and necrotrophs, which kill the host tissue and feed on the remainders18. So-called hemi-biotrophs use both strategies initially infecting the host biotrophically and then shifting to the necrotrophic stage19. Necrotrophic pathogens show aggressive and wide-ranging virulence strategies that result in host cell death20. To this end necrotrophs secrete proteinaceous, non-ribosomal peptides, metabolite toxins, and cell wall-degrading enzymes to induce host cell necrosis and leakage of nutrients21. These virulence mechanisms targeting diverse host cellular processes are countered by a complex host response involving cellular, histological, biochemical, and molecular answers. In early stages of necrotrophic interaction, plant host cell death is associated with production of various secondary metabolites, antimicrobial peptides, and hormones, accumulation of reactive oxygen species, callose, and cell wall modifications21. The soil-borne pathogen R. solani is considered to be a necrotrophic plant pathogen with broad host-range. However, recent studies on R. solani AG-1 IA causing sheath blight on rice and Brachipodium distachyon suggest a hemi-biotrophic nature of the R. solani isolate, since protective effect of the plant can be induced by treatment with salicylic acid22.
At present, there is only limiting information about the molecular responses of R. solani AG3 during the pathogenic interaction with its host plant. Histological observations of potato in response to R. solani infection were described23, and there is some information of phytotoxins (3-methylthiopropionic acid, 3-methylthioacrylic acid) produced by R. solani AG3 with relation to disease symptoms24. The expression of selected pathogenesis-related genes of the potato plants´ response upon infection has been analyzed25 and there is information of selected R. solani AG3 genes whose expression is correlated with the interaction process26,27. However, RNA sequencing technologies can give a deeper insight into the gene regulatory networks mediating disease outcomes. Dual sequencing of host–pathogen interactions provides a snapshot of the underlying transcriptional programs from both the host and the pathogen. For instance, such analysis showed that the important pathogen Septoria tritici on wheat alters gene activity to suppress plant defense response during the biotrophic phase before changing to a necrotrophic life style28. So far, RNAseq experiments are often limited to in vitro infection systems that do not fully reflect the conditions of the in vivo environment. For instance, a comprehensive transcriptome analysis of the soil-borne pathogen R. solani has been performed with the R. solani AG1-IB isolate 7/3/14 grown on root exudates29, during interaction with its host plant lettuce in an in vitro leaf model system30,31 and with R. solani AG1-IA during infection of soybean using a detached leaf assay32. A global transcriptome profiling with R. solani AG3-PT isolate Rhs1AP also carried out in vitro during interaction with plant associated bacteria (Serratia plymuthica AS13 and S. proteamaculans S4) revealed major shifts in gene expression with a simultaneous alteration of primary metabolic processes and activation of defense and attack mechanisms33.
In this study, we are focusing on transcriptome changes in R. solani AG3-PT during interaction with a medium resistant potato cultivar. The used isolate in this study, Ben3, originated from sclerotia on mature potato tubers and was characterized as R. solani AG3-PT. The draft genome of isolate Ben3 was analyzed and the size of the diploid genome was estimated to correspond to 116 Mb34. Gene prediction resulted in 12,567 identified genes, which is in the same range as for other completely sequenced AG3 isolates named Rhs1AP35 and RS-2036. With this global expression studies using RNAseq, we want to understand in its entirety the molecular interaction in the pathosystem S. tuberosum and R. solani AG3-PT. Together with the concomitant transcriptome analysis in the potato host this extensive study will finally broaden the basis for functional and comparative analyses of the host pathogen interaction of R. solani.
Results and discussion
Transcriptome analysis
The interspecific interaction between R. solani AG3-PT isolate Ben3 with the medium resistant potato cultivar 'Arkula' was analyzed on transcriptome level using RNA sequencing. In order to find factors important for the establishment of the interaction and further interaction progression, we used three different samplings: pure mycelium of R. solani AG3-PT isolate Ben3 cultured without being attracted by a growing potato plant (Ben3); potato sprouts at 3 dpi of the tubers with R. solani (early) and 8 dpi (late). At both sampling dates of R. solani in interaction with the potato sprout all emerging sprouts were harvested and used in the analysis. Necrotic lesions on sprouts become first visible at 8 dpi. Subsequently, RNA extraction, RNA sequencing (RNAseq) and mapping of the reads to the genome of isolate Ben334 was performed to calculate reads per kilobase per million mapped reads (RPKM) values. In general, for the inoculated samples between 1 and 23% of each dataset was mapped on the Ben3 draft genome. For the control samples, between 70 and 75% of these datasets could be mapped on the Ben3 genome.
In all three samplings a comparable number of genes could be detected as being expressed (Table 1). In pure mycelium 11,206 genes out of the identified 12,567 genes on the R. solani AG3-PT isolate Ben3 genome were expressed, while reads could be mapped on 10,181 and 9,939 genes at 3 dpi and 8 dpi, respectively. In summary, in all transcriptomes within this experiment 11,287 genes out of the identified 12,567 genes on the R. solani AG3-PT isolate Ben3 genome were expressed in any of the samples analyzed.
As obvious from Table 1, the amounts of total mapped reads differed considerably. This is owed to the fact that in the Ben3 sampling the transcriptome of the pure mycelium of R. solani AG3-PT isolate Ben3 has been sequenced, while in the early and late samplings (3 and 8 dpi) a dual RNAseq approach has been used to access the transcriptomes of both interacting organisms simultaneously30. Since library sizes and the amount of sequences produced from each library were comparable, in the Ben3 sampling almost all of the reads could be mapped to the isolate Ben3 genome. In the dual RNAseq approaches of the samplings at 3 and 8 dpi, only a fraction of the produced reads could be mapped to the Ben3 genome, while the others mainly mapped to the potato genome. Due to these different amounts of total mapped reads to the R. solani AG3-PT isolate Ben3 genome per sampling the following has to be considered: (1) genes with low RPKM value only in Ben3 are not necessarily not expressed when the fungus challenges the plant, it might be just owed to the library sizes; (2) genes that are found to be expressed in all three samplings could be assigned as being commonly expressed; (3) genes that are only found to be expressed in the much smaller libraries of the 3 and 8 dpi samplings can be assumed as being interaction specific.
The summarized mapping results (Table 1) already indicated that there must be many genes commonly expressed in all three samplings. Therefore, a Venn diagram was constructed to compare the three samplings and visualize the calculated number of genes they have in common versus the number of genes that distinguishes the individual samplings (Fig. 1). More than 9,000 genes were found to be expressed in all three samplings, while 871 were exclusively transcribed in the mycelium without plant contact. Expression of 29 genes was found to be in common in the 3 and 8 dpi samples, representing genes only expressed in the presence of the living potato plant. In addition, at 3 dpi 27 genes were exclusively expressed, and sequence reads mapping to a small set of 21 genes were exclusively detectable at 8 dpi. Lists of these genes together with their respective RPKM values and descriptions are given in Supplementary Tables 1 – 4.
From the 29 genes that were commonly expressed in the presence of the living potato plant, four genes are coding for proteins involved in plant cell wall degradation. This matches the virulence strategy of a necrotrophic pathogen with secretion of cell wall-degrading enzymes to induce host cell necrosis and leakage of nutrients21. In addition, one gene coding for a protein involved in protein degradation further supports this line of attack. Moreover, two genes coding for proteins with DNA-binding domains and putative function in transcriptional regulation are also in this list and are likely to play a role in transcriptional regulation of the offence (e.g. Ben3g4553, see Table 2). The majority of the 27 genes exclusively expressed at 3 dpi are coding for hypothetical proteins where no putative function can be assigned. From the 21 genes that were exclusively expressed in later stages of the interaction (8 dpi), eight genes coding for proteins involved in plant cell wall degradation and two are coding for proteases. This again demonstrates the typical strategy of a necrotrophic pathogen.
In summary, a first examination of the transcriptome data of the three samplings revealed distinct and reasonable differences, thus demonstrating the potential of this study to find factors important for the establishment of the interaction between R. solani AG3-PT isolate Ben 3 with a susceptible potato cultivar and further interaction progression.
The most abundant R. solani AG3-PT transcripts in the three samplings
The interspecific interaction between R. solani AG3-PT isolate Ben 3 with the medium resistant potato cultivar 'Arkula' was initially evaluated by examining the most abundant transcripts in the three different samplings. In growing mycelium cultivated in liquid culture, transcripts of 11,206 genes of the isolate Ben3 genome were found. In total, 698 transcripts of these genes were detected with median RPKM values of 100 and higher, 37 with median RPKM values greater than 1,000. At early interaction phase (3 dpi) of tuber sprouts with the pathogen, transcripts of 10,181 genes were detected with 729 showing median RPKM values of 100 and higher, 25 median RPKM values greater than 1,000. A number of 9,939 genes of R. solani AG3-PT were transcribed with 742 showing median RPKM values of 100 and higher, 26 median RPKM values greater than 1,000 at 8 dpi (Supplementary Tables 5–7). In total, 9,679 genes of the isolate Ben3 genome were found to be expressed in all three samplings, among them are the most abundant transcripts in each of the individual sampling analyzed. Five of these highest expressed genes (Ben3g9573, Ben3g6448, Ben3g5323, Ben3g2326, and Ben3g675) are coding for R. solani specific hypothetical proteins with unknown functions. Hence, these proteins were not expected to play specific roles during the interaction with the plant, these proteins may rather be important for general growth and cellular metabolism. Among the highest abundant transcripts in each of the individual sampling are also several proteins containing lectin domains (Ben3g9146, Ben3g9350, and Ben3g8869). A number of such proteins containing a ricin-type beta-trefoil lectin domain have also been reported previously as most abundant in the R. solani AG1-IB isolate 7/3/14 during interaction with its host plant lettuce30. While specific roles of these lectin domain proteins are undetermined it has been proposed that R. solani lectins could have a function as storage protein within the mycelium37. In addition, two genes coding for proteins with thuringiensis toxin domain (Ben3g8806, and Ben3g11931) were also strongly transcribed in all three samples analyzed. Bacillus thuringiensis toxins are bacterial proteins known for their biocidal activity against insects38, but a range of further organisms are targeted as well39. The strong expression of such toxin domain homologues in R. solani indicates that these toxins may be of general importance for R. solani AG3-PT isolate Ben3 rather than playing a specific role in the fungus plant interaction. A gene coding for a septal pore cap protein (Ben3g7115) was also among the most abundant transcripts in all three samplings, indicating its contribution to hyphal homeostasis in basidiomycetous fungi40. A transcript similar to the septal pore cap protein (RSOLAG1IB_6054) was also highly abundant in R. solani AG1-IB isolate 7/3/14 transcripts found in symptomless zone of interaction with lettuce30. This protein specific for R. solani is part of the plugging material that closes the perforations within the septal pore cap of hyphal cells and prevents transport of cytoplasmic fluids between neighboring cells. Furthermore, a gene encoding a hemopexin domain protein (Ben3g6614) was also among the highest expressed genes in common to all three samplings. This domain denotes zinc-dependent metalloproteinases, which are widely recognized to play an important role in the homeostatic regulation of the extracellular environment41, but their biological functions may also extend beyond extracellular matrix degradation42. Since all these proteins showed high abundance in R. solani AG3-PT with and without contact to the host plant, they may be important for overall growth and metabolism but do not seem to be of a specific relevance in the fungal interaction with the plant.
The interaction between R. solani AG3-PT and potato
In order to find elements with relevance for supporting interaction of R. solani AG3-PT isolate Ben3 with the potato sprout, differential gene expression was performed as integrated in the ReadXplorer platform (v2.2)43. This pairwise comparison of transcriptomes of pure mycelium of isolate Ben3 with transcriptomes of either 3 dpi or 8 dpi of interaction with potato sprouts was accomplished using the DESeq2 program. Genes were assigned as differentially expressed with an adjusted P value of less than 0.05 and a minimum fold change of |2| or more. Using these criteria 592 genes could be assigned as differentially induced at 3 dpi (sum of 242 genes exclusively early induced and 350 genes induced at 3 dpi and also at 8 dpi; early up) while 520 genes are differentially reduced at 3 dpi (sum of 412 genes exclusively early reduced and 108 genes reduced at 3 dpi and also at 8 dpi; early down). At 8 dpi, 688 transcripts were found to be differentially induced (sum of 338 genes exclusively late induced and 350 genes induced at 8 dpi and also at 3 dpi; late up) and 233 are differentially reduced (sum of 125 genes exclusively late reduced and 108 genes reduced at 8 dpi and also at 3 dpi; late down). Venn diagrams were constructed to compare and visualize the differentially upregulated as well as downregulated genes at both time points during the interaction (Fig. 2). Lists of these genes together with their respective values of fold change are given in Supplementary Tables 8–9.
Venn diagrams of differentially expressed genes between the samplings. Pure mycelium of R. solani AG3-PT isolate Ben3 cultured without being attracted by a growing potato plant (Ben3) compared to Ben3 in interaction with potato sprouts at 3 dpi (early); pure mycelium of Ben3 compared to Ben3 in interaction with potato sprouts at 8 dpi (late).
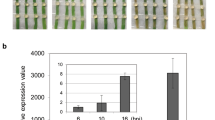
Expression differences found with this DESeq2 analysis were validated in experiments using qRT-PCR. Therefore a suitable control gene with invariant expression pattern has to be established. Glyceraldehyde-3-phosphate dehydrogenase (Ben3g7151, GAPDH), ubiquitin-conjugating enzyme E2 (EUC63156, UBC), ubiquitin-protein ligase E3 (Ben3g494, UBI), elongation factor 2 (Ben3g3364, EF-2), and beta tubulin genes (Ben3g4099; TUB1) and (Ben3g5288, TUB2) were tested and the beta tubulin gene Ben3g5288 (TUB2) showed to be the most appropriate reference in our experiments. Relative transcript levels of three candidate genes with different expression patterns were normalized on the basis of expression of this invariant control. ΔΔCq values were calculated and compared with the respective DESeq2 analysis (Table 2).
For all three candidate genes with very different expression pattern the calculated ΔΔCq values were in accordance with the respective log2fold change values computed in the DESeq2 analysis thus demonstrating the validation of expression differences.
In addition, gene ontology (GO) annotations were performed in order to assign respective functions to the particular genes. In the further analysis, a specific focus was laid on significantly induced genes to preferentially catch molecular candidates that might be important for initiation and establishing the interaction with the plant.
Differentially expressed genes (DEGs) in isolate Ben3 at 3 dpi of potato sprouts
Differentially induced genes in isolate Ben3 in interaction with potato sprouts at 3 dpi compared to pure mycelium of the isolate were selected assuming functions in initiating and supporting of the interaction process with the potato sprouts. Putative functions of the particular gene products and their distribution into the common GO aspects for biological process (BP), molecular function (MF), and cellular component (CC) are given in Fig. 3.
GO term distribution of differentially increased genes for biological process (BP), molecular function (MF), and cellular component (CC) at 3 dpi. Pure mycelium of R. solani AG3-PT isolate Ben3 cultured without being attracted by a growing potato plant (Ben3) compared to Ben3 in interaction with potato sprouts at 3 dpi (early).
At 3 dpi, differentially induced genes are mainly assigned as being involved in carbohydrate and cellular nitrogen compound metabolic processes and in transport. Almost 10% (50 genes) of the differentially upregulated genes are encoding various peptidase activities (e.g. Ben3g2070, see Table 2), while 53 genes are encoding cell wall degrading enzymes including several different hydrolases acting on glycosyl bonds, and pectate lyases (e.g. Ben3g3530, see Table 2.) or xylanases. In order to further specify these cell wall degrading enzymes, all of the DEGs were also annotated according to the Carbohydrate Active enZyme (CAZy) database (Supplementary Table 11). Secretion of a large arsenal of hydrolytic enzymes such as proteases and cell wall degrading enzymes during the course of interaction44,45 is required for necrotrophic fungal plant pathogens like R. solani to induce cell necrosis by breakdown of structural protein and carbohydrate components of the plant cell walls and to cause leakage of nutrients9,30,46,47. Thus, it is hypothesized that the induced cell wall degrading enzymes and several of the induced peptidases function in supporting interaction of R. solani AG3-PT with tissue of the potato sprout. However, secreted peptidases have also been extensively studied for their roles as effectors of gram-negative bacteria48 and also fungi47,49. Pathogen effectors are usually delivered as virulence factors into the host cells to suppress the basal defense responses and create a suitable environment for pathogen propagation50,51,52. Thus, the post-translational modification of host proteins through proteolytic processing is a widely used mechanism in regulating the plant defense response. With the current stage of knowledge it might be supposed that one or more of these induced proteases have putative roles as effectors that support the interaction of R. solani AG3-PT on the potato host. However, further functional analyses of the individual proteases are needed to clearly assign their functional role in the pathogen host interaction.
Another big group composed of 100 members of differentially upregulated genes of the interaction at 3 dpi is described as coding for hypothetical proteins. Most of these genes are R. solani specific and homologues could be found in the other five annotated genomes of R. solani AG3 Rhs1AP, R. solani AG2-2IIIB, R. solani AG8, and R. solani AG1-IA and AG1-IB. From our differential gene expression analysis, it is expected that some of these genes are involved in supporting interaction of R. solani AG3-PT on the potato sprout. It is already known that secreted effectors of fungal pathogens target host immunity using various strategies e.g. by hydrolyzing a salicylic acid precursor53, or by binding to transcription factors thus inhibiting their activity54. Further analyses are needed to reveal putative functions of the so far hypothetical proteins for their various possible roles in the interaction of R. solani AG3-PT with potato.
Interestingly, the strongest differentially increased gene (Ben3g6247) is homologous to lipid-translocating exporter (LTE) family proteins, like RTA1 from Saccharomyces. The RTA1 protein contains seven potential membrane-spanning segments55 and is predicted an integral membrane protein with function in cell resistance to xenobiotics56. Other LTE family genes may encode transporters or sensors that facilitate the excretion of biosynthetic intermediates, either directly or indirectly56. These putative functions of the LTE family proteins make the gene Ben3g6247 a favorite candidate involved in secretion of components important for the pathogen attack.
Early phase of necrotrophic interaction is associated with cell death of the plant host and production of various secondary metabolites and the accumulation of reactive oxygen species21. It has been shown that antioxidant processes and respective gene expression were correlated to necrotic tissues in several R. solani pathosystems (potato sprout-R. solani AG3; soybean hypocotyl-R. solani AG4 and soybean leaves-R. solani AG1-IA)27. At early phase of interaction of the potato host with isolate Ben3 (3 dpi) no strong evidence for induction of antioxidant processes in pathogen hyphae could be observed on transcriptional level, because there was no strong increase in glutathione S-transferase gene expression or upregulation of other genes known to be involved in the scavenging of reactive oxygen species. Therefore, it could be postulated that in our system to colonize the potato sprout with R. solani AG3-PT, tissue analysis at 3 dpi resembles an early stage of the plant pathogen interaction maybe prior infection of sprout tissue. No visible symptoms were observed at this time point.
Differentially expressed genes in isolate Ben3 at 8 dpi of potato sprouts
In order to find transcripts that are important in an advanced stage of the interaction, differentially induced genes between pure mycelium of isolate Ben3 and Ben3 attracted to potato sprouts at 8 dpi were screened. Respective functional annotations of the gene products and their distribution into the common GO features are shown in Fig. 4. At 8 dpi, differentially induced genes are mainly assigned as being involved in carbohydrate and macromolecular metabolic processes and in transport. At this later stage of interaction, the upregulated 152 genes (> 22%) are encoding various cell wall degrading enzymes (e.g. Ben3g3530, see Table 2). In addition, all of the DEGs were also annotated according to the Carbohydrate Active enZyme (CAZy) database (Supplementary Table 11). This increased expression of genes coding for cell wall hydrolytic enzymes nicely demonstrates the increasing pathogenic activity of the isolate Ben3 as well as the importance of breakdown of cell wall components to access the nutrients during the course of interaction, thus confirming the described virulence strategy of a necrotrophic pathogen21,57. This destructive tactic was accompanied by an induction of expression of genes encoding integral components of membranes. These 154 mostly uncharacterized integral membrane proteins and putative transporters were presumably involved in the uptake of nutrients and degradation products of the hydrolase activities. At this stage of interaction the predominant importance of genes coding for peptidases seemed to be decreasing, but still 33 genes encoding peptidases were differentially upregulated (e.g. Ben3g2070, see Table 2). This could also be explained by the fact that whole sprouts were harvested, including the sprouts with up to 8 days of interaction with the challenging pathogen and the subsequent emerging sprouts with a shorter interaction period. In general, this is also represented in the 350 genes that were in common differentially increased at 3 and 8 dpi (Fig. 2).
GO term distribution of differentially increased genes for biological process (BP), molecular function (MF), and cellular component (CC) at 8 dpi. Pure mycelium of R. solani AG3-PT isolate Ben3 cultured without being attracted by a growing potato plant (Ben3) compared to Ben3 in interaction with potato sprouts at 8 dpi (late).
In addition, another big group of DEGs at 8 dpi is composed of 98 genes with unknown function. Since these genes are Rhizoctonia specific without any regions of similarity to sequences with assigned functions, it can only be speculated about their role in supporting the pathogen plant interaction. Further functional comparisons of these genes and e.g. their differential expression in the respective pathosystems might give hints to their putative function.
In the here described experimental system to colonize the potato sprout with R. solani AG3-PT isolate Ben3 lesions become first visible at 8 dpi. It has been shown in other experiments25,27 that necrotrophic interaction and cell death in the plant host was correlated with respective gene expression in the plant and in the fungus. Samsatly and co-workers27 demonstrated by using quantitative RT-PCR, that the expression of antioxidant genes coding for glutathione S-transferase and catalase were significantly increased in R. solani AG3 five days after inoculation of detached potato sprouts. But such strongly increased expression of these antioxidant genes could not be found in the here described experiments. Reasons for these differences could be due to inoculation using isolates of R. solani AG3 exhibiting differences in pathogenicity. However, another distinction is the fact that Samsatly and co-workers27 performed experiments with detached sprouts in a limited in vitro system while our experimental setup fully reflects the conditions of the in vivo environment with growing sprouts on cultured seed tubers. Further investigations together with the concomitant transcriptome analysis in the potato host will finally increase the understanding of a mutual relationship in the host pathogen interaction in an environment resembling the natural situation.
Differentially expressed genes in isolate Ben3 comparing 3 and 8 dpi of potato sprouts
To differentiate between R. solani AG3-PT transcripts that were mainly relevant at early time point and those becoming more important at advanced stages of the interaction differentially expressed genes between 3 and 8 dpi of the interaction were also analyzed with DESeq2. Using the above mentioned criteria with adjusted P-values of less than 0.05 and a minimum fold change of |2| or more 173 genes could be assigned as differentially reduced expressed between 3 and 8 dpi while 400 genes are differentially increased expressed at the later timepoint. Complete lists of these genes together with their respective baseMean values and values of fold change are given in Supplementary Table 10, lists of the 20 most differentially expressed genes between 3 and 8 dpi are presented in Tables 3 and 4. While 10 out of the 20 most differentially reduced expressed genes between 3 and 8 dpi are coding for proteins involved in protein degradation and nitrogen uptake and assimilation (Table 3), the majority of genes differentially increased expressed between 3 and 8 dpi are involved in polysaccharide degradation with focus on copper-dependent lytic polysaccharide monooxygenases for cleavage of cellulose chains with oxidation of various carbons (Table 4).
It is known that nitrogen metabolism and nitrogen-regulated gene expression in the plant pathogenic fungi is of great importance for the establishment of the disease in the host plant58. However, nitrate is the less preferred nitrogen source compared to ammonium and L-glutamine with respect to nutrient utilization in fungi at least during infection of leaves59. In fungi, this preferred nutrient utilization is regulated via nitrogen metabolite repression and ensures the transcription of ammonium and urea active permease encoding genes. Besides the strong transient expression of genes encoding ammonium and nitrogenous compounds transporting permeases (Ben3g6147, Ben3g6767, Ben3g4369, and Ben3g7775) at the early interaction stage at 3 dpi, R. solani AG3-PT isolate Ben3 exerted also high induction and expression of genes involved in nitrate uptake and assimilation (Ben3g6360, Ben3g6359, and Ben3g6361). Whether this is a distinctive feature of the isolate Ben3 or a characteristic of soil-borne plant pathogenic fungi would need further investigations.
The majority of genes with differentially increased expression between 3 and 8 dpi are involved in cell wall degradation coding for hydrolases acting on glycosyl bonds, and pectate lyases. This was expected and again demonstrates the increasing importance of breakdown of cell wall components designating the virulence strategy of a necrotrophic pathogen21.
Conclusions
The results of the molecular responses of R. solani AG3-PT isolate Ben 3 during the interaction with a medium resistant host genotype of potato resemble the typical necrotrophic interaction. Specific induction of gene expression at the initial stages of interaction and during the course of interaction with the host plant was related to distinct functions. Expression induction of exporters facilitating excretion and strong upregulation of genes coding for peptidases and cell wall digesting enzymes was significant for the initial stage, while an additional increase of expression of cell wall hydrolyzing enzymes and genes encoding various integral membranes proteins with transporter function was linked to interaction progression. Interestingly, several Rhizoctonia specific yet unknown genes were differentially expressed, but further functional analysis will be needed to resolve their role in the R. solani AG3-PT potato interaction.
The used transcriptome sequencing approach allows addressing the expression response of the fungus as well as the plant simultaneously. In a following step the transcriptional answers of the potato sprouts towards the interaction with R. solani AG3-PT will give valuable information about the plants’ defense response. In addition, the same system to colonize the potato sprout with R. solani AG3-PT isolate Ben3 can be used with potato varieties showing differences in degree of field resistance to R. solani AG3-PT or to analyze the response of different R. solani AG3 isolates. A comparison of the interaction responses will lead the way to new candidates that can be used for the development of breeding strategies for new less susceptible varieties.
Methods
Plant material
The potato cultivar 'Arkula' (Norika GmbH, Sanitz, Germany) was used in this study. It has been demonstrated previously that 'Arkula' is susceptible to R. solani AG3-PT25, and is described as medium resistance to stem canker as stated in the European Cultivated Potato Database (https://www.europotato.org). To study interactions with R. solani AG3-PT, the use of pathogen free plant material is a prerequisite, thus potato mini-tubers were produced from in vitro plantlets (kindly provided by Norika GmbH). Mini-tubers were pre-sprouted in the dark using a cycle of changing temperatures (8 °C 1 week, 4 °C 3 weeks, 8 °C 3 days, 20–22 °C until sprouting) and a treatment with 10 mg/L gibberellic acid for 20 min. One mini-tuber was planted into each pot (pot size: 12 × 12 × 20 cm) containing a quartz sand/grit mixture (Euroquarz, Dorsten, Germany) and cultivated in a growth chamber (York, Mannheim, Germany) at 16/8 h day/night cycle at 18/15 °C day/night temperature and 400 μmol m-2 s-1 light and a relative humidity of 80%. The tubers were poured with B’cuzz Hydro A + B nutrient solution twice a week (Atami B.V., Rosmalen, The Netherlands), which had been adjusted to provide an EC of 2.1 dS m-1 and a pH of 5.8. If required, tubers were additionally watered with osmotic water. Each treatment (potato tubers without and with R. solani AG3-PT inoculation) included three replicates with ten plants per replicate, which were arranged in a completely randomized block design.
Pathogen inoculation and sampling
The R. solani AG3-PT isolate Ben3 (kindly provided by Marianne Benker, North Rhine-Westphalia–Plant Protection Service, Germany) used in this study was cultured on Petri dishes on potato dextrose agar (PDA, Merck, Germany) at 20–22 °C for 5 days.
About 27 days after transferring mini-tubers into pots, when their sprouts had reached a length of 3–4 cm, an agar plug (Ø 10 mm) grown with isolate Ben3 was placed on the tuber next to the first emerging sprout25. Afterwards tubers were completely covered with quartz sand and each pot was sprayed with osmotic water to maintain humidity and promote fungal growth.
At both sampling dates [3 and 8 days post inoculation (dpi)], three biological replicate pools composed of all emerging sprouts from 10 plants per replicate were used. Subsequently, sprout samples were shock frozen in liquid nitrogen and stored at − 80 °C. All sprouts were examined regarding the incidence of necrotic lesions, while these lesions become first visible at the second sampling date.
For transcriptome analysis of the control isolate Ben3 mycelium without being attracted by a growing potato plant, the fungus was grown in liquid culture as follows: R. solani AG3-PT plugs (5 day old agar plugs of Ø 10 mm) were taken from the margin of a Petri dish culture and placed in 100 ml Erlenmeyer flasks in 30 ml of B’cuzz Hydro A + B nutrient solution, which was also used to culture the plants. The mycelium was grown without shaking for 8 days at the same time and conditions alongside the infected plants. Fungal hyphae were harvested and immediately shock frozen in liquid nitrogen and stored at − 80 °C prior to RNA extraction.
RNA extraction
Prior to RNA extraction from inoculated sprout tissue the samples were ground using a mixer mill (2 min, 30/s; Retsch MM400, Haan, Germany) with two grinding balls (7 mm, 3 mm; Askubal, Korntal-Münchingen, Germany) under constant cooling in liquid nitrogen. Total RNA was extracted from 70 to 90 mg of ground sprout material using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) including DNase treatment (QIAGEN). After measuring the quantity of extracted RNA with the NanoDrop ND-1000 spectral photometer, quality and intactness of the RNA was analyzed using the bioanalyzer (Agilent Technologies Deutschland GmbH, Waldbronn, Germany).
RNA of fungal pellet was extracted using the same material and method.
Sequencing
Sequencing of the prepared stranded cDNA libraries performed on an Illumina HiSeq 1500 platform (Illumina Inc., San Diego, U.S.A.). In total, 36 cDNA libraries were sequenced in six runs. The cDNA libraries were single-end sequenced in rapid mode with 1 × 50 cycles. Data analysis and base calling were accomplished with in-house software based on CASAVA 1.8.247. In total 276 Gb data were obtained for 36 resulting libraries, with an average of 7.6 Gb mappings per library. The sequencing raw data for all libraries has been made available on the EBI ArrayExpress server, with the accession E-MTAB-7137, https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-7137.
Transcript mapping and expression analysis
Read mapping was carried out as described recently30. In brief, the obtained reads were quality filtered (> Q30) and subsequently mapped to the R. solani AG3-PT Ben3 draft genome33 [EMBL: FXZJ01000001-FXZJ01001390] using tophat260. Two mismatches were allowed to handle possible sequencing errors and allelic variants of the diploid R. solani Ben3 genome. For transcript abundance analysis, resulting data were imported and analyzed using the ReadXplorer platform (v2.2)43. Reads per kilobase per million mapped reads (RPKM) values were calculated, using the single best match options (single perfect match mapping and single best match mapping of only uniquely mapped reads) for each of the separate libraries61. To determine the most abundant transcripts per sample, means of the RPKM values were recorded of the three biological replicates. As threshold, only transcripts with more than three raw reads in at least two of the three biological replicates were included and defined as being expressed.
For DESeq2 calculations62 the genes were counted as differentially expressed with an adjusted P-value of less than 0.0563 and a minimum fold change of 2 or more. Selected genes based on differential expression and RPKM results were annotated in detail using Blast2GO version 5.064 applying default settings with an expect value of 1 × 10–6 and the fungiDB taxa 4751.
Validating expression differences with RT-qPCR
RNA was extracted and quality controlled as described above. Single-stranded cDNA synthesis was carried out with 1 µg of total RNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories GmbH, Feldkirchen, Germany) in a 25 µl reaction following manufacturer´s instructions. Subsequently cDNA was diluted tenfold. RT-qPCR was performed using 96-well reaction plates on a Thermal Cycler CFX96 C1000 Touch (Bio-Rad). The thermal profile was 95 °C for 5 min, 40 cycles of 95 °C for 15 s and 60 °C for 1 min, followed by dsDNA melting curve analysis to ensure amplicon specificity. Each reaction was done in a 10 µL volume containing 200 nM of each primer, 3 µL of cDNA (1:10) and 5 µL of Sensi Fast SYBR NO ROX Kit (Bioline GmbH, Luckenwalde, Germany). Data collection and analysis was performed using CFX Manager Software 3.0 (Bio-Rad). At least three biological replicates were measured in duplicates, uninfected control plants and also non-template controls were included.
Relative transcript levels were normalized on the basis of expression of an invariant control. Therefore oligonucleotide primer sets of putative reference genes were tested with various infected plant samples and with pure mycelium grown without plant contact: Glyceraldehyde-3-phosphate dehydrogenase (Ben3g7151, GAPDH), Rs-GAPDHf: CATCATTCCATCGTCCACTG, Rs-GAPDHr: GAGGCAGATTTCTCCAATCG; Ubiquitin-conjugating enzyme E2 (EUC63156, UBC), Rs-UBCf: TAATCCAGGCGAGAGCAAGT, Rs-UBCr: CCCGCGATAGTTTAATCGAC; Ubiquitin-protein ligase E3 (Ben3g494, UBI), Rs-UBIf: AACGATTCTGGAGGGTTGTG, Rs-UBIr: GGTCATCGGACGTAGCATCT; Elongation factor 2 (Ben3g3364, EF-2), Rs-EF-2f: GTCTTCTCGGAAGAGCAACG, Rs-EF-2r: ACTCCCAGTGGTCAAAGACG; Beta tubulin (Ben3g4099; TUB1), Rs-TUB1f: ATGAAGGAAGTCGAGGAGCA, Rs-TUB1r: GCGGTAGAGTTGCCGATAAA; Beta tubulin (Ben3g5288, TUB2), Rs-TUB2f: AACTCGGCATCCTTCGTAGA, Rs-TUB2r: AGCAAATTGTCCATGGCTTC. While primer sets of EF-2 and TUB1 were not suitable because of amplification products in the uninfected plant background, primer sets of all other tested references gave reliable amplifications with efficiencies of close to 2. Reference gene stability was calculated with the CFX Manager software 3.0 (Bio-Rad) and with NormFinder65, selecting TUB2 as the best invariant control.
ΔCq was calculated as the difference between control and target products (ΔCq = Cqgene − CqTUB2). Differences in relative expression levels between the treated samples were calculated as ΔΔCq = ΔCq (Ben3) − ΔCq (early or late). Oligonucleotide primer sets used for RT-qPCR are as follows: pectate lyase (Ben3g3530), Rs-3530f: TCCAACGTTATTGCAAACGA, Rs-3530r: GGCTCGTCACCATTGCTATT; extracellular metalloprotease (Ben3g2070), Rs-2070f: TTTCCCCTCCGACTATGCTA, Rs-2070r: CCCTGGAAGGTGTGGTAGAG; C2H2-type zinc-finger protein (Ben3g4553), Rs-4553f: CCCTCATGTGTGTGAGCACT, Rs-4553r: TGGACTGCGTCCTTCCTACT.
References
Yang, G. & Li. C. General description of Rhizoctonia species complex. In Plant Pathology, ed C.J. Cumagun (2012).
Ogoshi, A. Ecology and pathogenicity of anastomosis and intraspecific groups of Rhizoctonia solani Kuhn. Annu. Rev. Phytopathol. 25, 125–143 (1987).
Garcia, V. G., Onco, M. A. P. & Susan, V. R. Review. Biology and systematics of the form genus Rhizoctonia. Span. J. Agric. Res. 4, 55–79 (2006).
Ajayi-Oyetunde, O. O. & Bradley, C. A. Rhizoctonia solani: taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant Pathol. 67, 3–17 (2018).
Carling, D. E., Kuninaga, S. & Brainard, K. A. Hyphal anastomosis reactions, rDNA-internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani anastomosis group-2 (AG-2) and AG-BI. Phytopathology 92, 43–50. https://doi.org/10.1094/PHYTO.2002.92.1.43 (2002).
Sharon, M., Kuninaga, S., Hyakumachi, M., Naito, S. & Sneh, B. Classification of Rhizoctonia spp. using rDNA-ITS sequence analysis supports the genetic basis of the classical anastomosis grouping. Mycoscience 49, 93–114. https://doi.org/10.1007/s10267-007-0394-0 (2008).
Wibberg, D. et al. Development of a Rhizoctonia solani AG1-IB specific gene model enables comparative genome analyses between phytopathogenic R. solani AG1-IA, AG1-IB, AG3 and AG8 isolates. PLoS ONE 10, e0144769. https://doi.org/10.1371/journal.pone.0144769 (2015).
Banville, G. J. Yield losses and damage to potato plants caused by Rhizoctonia solani Kuhn. Am. Potato J. 66, 821–834 (1989).
Campion, C., Chatot, C., Perraton, B. & Andrivon, D. Anastomosis groups, pathogenicity and sensitivity to fungicides of Rhizoctonia solani isolates collected on potato crops in France. Eur. J. Plant Pathol. 109, 983–992. https://doi.org/10.1023/B:EJPP.0000003829.83671.8f (2003).
Woodhall, J. W., Lees, A. K., Edwards, S. G. & Jenkinson, P. Characterization of Rhizoctonia solani from potato in Great Britain. Plant Pathol. 56, 286–295. https://doi.org/10.1111/j.1365-3059.2006.01545.x (2007).
Lehtonen, M. J., Somervuo, P. & Valkonen, J. P. Infection with Rhizoctonia solani induces defense genes and systemic resistance in potato sprouts grown without light. Phytopathology 98, 1190–1198. https://doi.org/10.1094/PHYTO-98-11-1190 (2008).
Tsror, L. Biology, epidemiology and management of Rhizoctonia solani on potato. J. Phytopathol. 158, 649–658. https://doi.org/10.1111/j.1439-0434.2010.01671.x (2010).
Fiers, M. et al. Genetic diversity of Rhizoctonia solani associated with potato tubers in France. Mycologia 103, 1230–1244. https://doi.org/10.3852/10-231 (2011).
Kuninaga, S., Carling, D. E., Takeuchi, T. & Yokosawa, R. Comparison of rDNA-ITS sequences between potato and tobacco strains in Rhizoctonia solani AG-3. J. Gen. Plant Pathol. 66, 2–11. https://doi.org/10.1007/PL00012917 (2000).
Wilson, P. S., Ketola, E. O., Ahvenniemi, P. M., Lehtonen, M. J. & Valkonen, J. P. T. Dynamics of soilborne Rhizoctonia solani in the presence of Trichoderma harzianum: effects on stem canker, black scurf and progeny tubers of potato. Plant Pathol. 57, 152–161. https://doi.org/10.1111/j.1365-3059.2007.01706.x (2008).
Hide, G. A. & Horrocks, J. K. Influence of stem canker (Rhizoctonia solani Kuhn) on tuber yield, tuber size, reducing sugars and crisp color in cv Record. Potato Res. 37, 43–49 (1994).
Stevenson, W. R., Loria, R., Franc, G. D. & Weingartner, D. P. Compendium of potato diseases 2nd edn. (American Phytopathological Society Press, New York, 2001).
Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. https://doi.org/10.1146/annurev.phyto.43.040204.135923 (2005).
Horbach, R., Navarro-Quesada, A. R., Knogge, W. & Deising, H. B. When and how to kill a plant cell: infection strategies of plant pathogenic fungi. J. Plant Physiol. 168, 51–62. https://doi.org/10.1016/j.jplph.2010.06.014 (2011).
Laluk, K. & Mengiste, T. Necrotroph attacks on plants: wanton destruction or covert extortion?. Arabidopsis Book 8, e0136. https://doi.org/10.1199/tab.0136 (2010).
Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. https://doi.org/10.1146/annurev-phyto-081211-172955 (2012).
Kouzai, Y. et al. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 217, 771–783. https://doi.org/10.1111/nph.14849 (2018).
Zhang, X. Y. et al. Histological observation of potato in response to Rhizoctonia solani infection. Eur. J. Plant Pathol. 145, 289–303. https://doi.org/10.1007/s10658-015-0842-1 (2016).
Kankam, F. et al. Isolation, purification and characterization of phytotoxins produced by Rhizoctonia solani AG-3, the cause agent of potato stem canker. Am. J. Potato Res. 93, 321–330 (2016).
Genzel, F., Franken, P., Witzel, K. & Grosch, R. Systemic induction of salicylic acid-related plant defences in potato in response to Rhizoctonia solani AG3PT. Plant Pathol. 67, 337–348 (2018).
Rioux, R. et al. Comparative analysis of putative pathogenesis-related gene expression in two Rhizoctonia solani pathosystems. Curr. Genet. 57, 391–408. https://doi.org/10.1007/s00294-011-0353-3 (2011).
Samsatly, J., Copley, T. R. & Jabaji, S. H. Antioxidant genes of plants and fungal pathogens are distinctly regulated during disease development in different Rhizoctonia solani pathosystems. PLoS ONE 13, e0192682. https://doi.org/10.1371/journal.pone.0192682 (2018).
Yang, F., Li, W. S. & Jorgensen, H. J. L. Transcriptional reprogramming of wheat and the hemibiotrophic pathogen Septoria tritici during two phases of the compatible interaction. PLoS ONE 8, e81606. https://doi.org/10.1371/journal.pone.0081606 (2013).
Wibberg, D. et al. Transcriptome analysis of the phytopathogenic fungus Rhizoctonia solani AG1-IB 7/3/14 applying high-throughput sequencing of expressed sequence tags (ESTs). Fungal Biol. Uk 118, 800–813. https://doi.org/10.1016/j.funbio.2014.06.007 (2014).
Verwaaijen, B. et al. The Rhizoctonia solani AG1-IB (isolate 7/3/14) transcriptome during interaction with the host plant lettuce (Lactuca sativa L.). PLoS ONE 12, e0177278. https://doi.org/10.1371/journal.pone.0177278 (2017).
Verwaaijen, B. et al. A comprehensive analysis of the Lactuca sativa, L. transcriptome during different stages of the compatible interaction with Rhizoctonia solani. Sci. Rep. 9, 7221. https://doi.org/10.1038/s41598-019-43706-5 (2019).
Copley, T. R., Duggavathi, R. & Jabaji, S. The transcriptional landscape of Rhizoctonia solani AG1-IA during infection of soybean as defined by RNA-seq. PLoS ONE 12, e0184095. https://doi.org/10.1371/journal.pone.0184095 (2017).
Gkarmiri, K. et al. Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica. BMC Genom. 16, 630. https://doi.org/10.1186/s12864-015-1758-z (2015).
Wibberg, D. et al. Draft genome sequence of the potato pathogen Rhizoctonia solani AG3-PT isolate Ben3. Arch Microbiol 199, 1065–1068. https://doi.org/10.1007/s00203-017-1394-x (2017).
Cubeta, M. A. et al. Draft genome sequence of the plant-pathogenic soil fungus Rhizoctonia solani anastomosis group 3 strain Rhs1AP. Genome Announc. https://doi.org/10.1128/genomeA.01072-14 (2014).
Patil, V. U., Girimalla, V., Sagar, V., Bhardwaj, V. & Chakrabarti, S. K. Draft genome sequencing of Rhizoctonia solani anastomosis group 3 (AG3-PT) causing stem canker and black scurf of potato. Am. J. Potato Res. 95, 87–91. https://doi.org/10.1007/s12230-017-9606-0 (2018).
Kellens, J. T. C. & Peumans, W. J. Developmental accumulation of lectin in Rhizoctonia solani - a potential role as a storage protein. J. Gen. Microbiol. 136, 2489–2495. https://doi.org/10.1099/00221287-136-12-2489 (1990).
Palma, L., Munoz, D., Berry, C., Murillo, J. & Caballero, P. Bacillus thuringiensis toxins: an overview of their biocidal activity. Toxins 6, 3296–3325. https://doi.org/10.3390/toxins6123296 (2014).
Ohba, M., Mizuki, E. & Uemori, A. Parasporin, a new anticancer protein group from Bacillus thuringiensis. Anticancer Res. 29, 427–433 (2009).
van Driel, K. G. A. et al. Septal pore cap protein SPC18, isolated from the basidiomycetous fungus Rhizoctonia solani, also resides in pore plugs. Eukaryot. Cell 7, 1865–1873. https://doi.org/10.1128/Ec.00125-08 (2008).
Nagase, H. & Woessner, J. F. Jr. Matrix metalloproteinases. J. Biol. Chem. 274, 21491–21494. https://doi.org/10.1074/jbc.274.31.21491 (1999).
Dufour, A., Sampson, N. S., Zucker, S. & Cao, J. Role of the hemopexin domain of matrix metalloproteinases in cell migration. J. Cell Physiol. 217, 643–651. https://doi.org/10.1002/jcp.21535 (2008).
Hilker, R. et al. ReadXplorer–visualization and analysis of mapped sequences. Bioinformatics 30, 2247–2254. https://doi.org/10.1093/bioinformatics/btu205 (2014).
Xu, X. H., He, Q., Chen, C. & Zhang, C. L. Differential communications between fungi and host plants revealed by secretome analysis of phylogenetically related endophytic and pathogenic fungi. PLoS ONE 11, e0163368. https://doi.org/10.1371/journal.pone.0163368 (2016).
Krishnan, P., Ma, X., McDonald, B. A. & Brunner, P. C. Widespread signatures of selection for secreted peptidases in a fungal plant pathogen. BMC Evol. Biol. 18, 7. https://doi.org/10.1186/s12862-018-1123-3 (2018).
Lowe, R. G. T. et al. Extracellular peptidases of the cereal pathogen Fusarium graminearum. Front. Plant Sci. 6, 962. https://doi.org/10.3389/fpls.2015.00962 (2015).
Wibberg, D. et al. Genome analysis of the sugar beet pathogen Rhizoctonia solani AG2-2IIIB revealed high numbers in secreted proteins and cell wall degrading enzymes. BMC Genom. 17, 245. https://doi.org/10.1186/s12864-016-2561-1 (2016).
Xia, Y. J. Proteases in pathogenesis and plant defence. Cell. Microbiol. 6, 905–913. https://doi.org/10.1111/j.1462-5822.2004.00438.x (2004).
Franceschetti, M. et al. Effectors of filamentous plant pathogens: commonalities amid diversity. Microbiol. Mol. Biol. Rev. https://doi.org/10.1128/MMBR.0006616 (2017).
Boller, T. & He, S. Y. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. https://doi.org/10.1126/science.1171647 (2009).
Cui, H., Xiang, T. & Zhou, J. M. Plant immunity: a lesson from pathogenic bacterial effector proteins. Cell Microbiol. 11, 1453–1461. https://doi.org/10.1111/j.1462-5822.2009.01359.x (2009).
Dou, D. & Zhou, J. M. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. https://doi.org/10.1016/j.chom.2012.09.003 (2012).
Liu, T. et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5, 4686. https://doi.org/10.1038/ncomms5686 (2014).
Qin, J. et al. The plant-specific transcription factors CBP6Og and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. Elife 7, e34902. https://doi.org/10.7554/eLife.34902 (2018).
Soustre, I., Letourneux, Y. & Karst, F. Characterization of the Saccharomyces cerevisiae RTA1 gene involved in 7-aminocholesterol resistance. Curr Genet 30, 121–125. https://doi.org/10.1007/s002940050110 (1996).
Manente, M. & Ghislain, M. The lipid-translocating exporter family and membrane phospholipid homeostasis in yeast. FEMS Yeast Res. 9, 673–687. https://doi.org/10.1111/j.1567-1364.2009.00513.x (2009).
Kawahara, Y. et al. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE 7, e49423. https://doi.org/10.1371/journal.pone.0049423 (2012).
Bolton, M. D. & Thomma, B. P. H. J. The complexity of nitrogen metabolism and nitrogen-regulated gene expression in plant pathogenic fungi. Physiol. Mol. Plant Pathol. 72, 104–110. https://doi.org/10.1016/j.pmpp.2008.07.001 (2008).
Fernandez, J., Marroquin-Guzman, M. & Wilson, R. A. Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu. Rev. Phytopathol. 52, 155–174. https://doi.org/10.1146/annurev-phyto-102313-050135 (2014).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. https://doi.org/10.1186/gb-2013-14-4-r36 (2013).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. https://doi.org/10.1038/nmeth.1226 (2008).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. https://doi.org/10.1093/bioinformatics/bti610 (2005).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496 (2004).
Acknowledgements
We gratefully acknowledge the technical assistance with sequencing from Anika Winkler (Center for Biotechnology, Bielefeld University, Bielefeld, Germany) and from Sabine Breitkopf, Angelika Fandrey and Sieglinde Widiger (Leibniz Institute of Vegetable and Ornamental Crops, Großbeeren, Germany). The bioinformatics support of the German Network for Bioinformatics Infrastructure (de.NBI) is gratefully acknowledged. This work was financially supported by the Federal Ministry of Food and Agriculture (BMEL) of the Federal Republic of Germany and the Ministry of Science, Research and Cultural Affairs (MWFK) of the State of Brandenburg, and by the Federal Office for Agriculture and Food (BLE, Grant No. 2814700911).
Author information
Authors and Affiliations
Contributions
R.G. contributed conception and design of the study; F.G. performed the experiments; B.V. and D.W. carried out the sequencing and transcript mapping; R.Z. performed expression analysis and data interpretation; R.Z. wrote the manuscript. All authors read and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zrenner, R., Genzel, F., Verwaaijen, B. et al. Necrotrophic lifestyle of Rhizoctonia solani AG3-PT during interaction with its host plant potato as revealed by transcriptome analysis. Sci Rep 10, 12574 (2020). https://doi.org/10.1038/s41598-020-68728-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-68728-2
This article is cited by
-
Comprehensive antifungal investigation of green synthesized silver nanoformulation against four agriculturally significant fungi and its cytotoxic applications
Scientific Reports (2024)
-
Diversity and Pathogenicity of Anastomosis Groups of Rhizoctonia solani Isolates Associated with Potato Diseases in Northern Sinaloa, Mexico
Potato Research (2023)
-
Black scurf of potato: Insights into biology, diagnosis, detection, host-pathogen interaction, and management strategies
Tropical Plant Pathology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.