Abstract
Gpr27 is a highly conserved, orphan G protein coupled receptor (GPCR) previously implicated in pancreatic beta cell insulin transcription and glucose-stimulated insulin secretion in vitro. Here, we characterize a whole-body mouse knockout of Gpr27. Gpr27 knockout mice were born at expected Mendelian ratios and exhibited no gross abnormalities. Insulin and Pdx1 mRNA in Gpr27 knockout islets were reduced by 30%, but this did not translate to a reduction in islet insulin content or beta cell mass. Gpr27 knockout mice exhibited slightly worsened glucose tolerance with lower plasma insulin levels while maintaining similar insulin tolerance. Unexpectedly, Gpr27 deletion reduced expression of Eif4e3, a neighboring gene, likely by deleting transcription start sites on the anti-sense strand of the Gpr27 coding exon. Our data confirm that loss of Gpr27 reduces insulin mRNA in vivo but has only minor effects on glucose tolerance.
Similar content being viewed by others
Introduction
Pancreatic beta-cells play a central role in regulating blood glucose homeostasis by secreting precise amounts of insulin in response to blood glucose concentrations. Beta-cell death or dysfunction contributes to the pathogenesis of both type 1 and type 2 diabetes mellitus. Since current diabetes therapies are often insufficient to manage hyperglycemia, novel strategies are needed. Therapies to improve beta cell function and survival are an attractive approach to generate new therapies for diabetes.
GPCRs have a proven track record of being viable drug targets. Thirty-four percent of currently approved therapies target 103 different GPCRs1. In diabetes, the blockbuster GLP1R agonists2 and the dipeptidyl peptidase IV inhibitors3 are prominent examples of successful drugs targeting a beta cell GPCR. Nonetheless, many GPCRs remain orphan receptors and are poorly characterized; these could be future diabetes drug targets4.
We previously identified the highly conserved GPCR Gpr27 (also known as SREB1, or super conserved receptor in brain 1) as a positive regulator of both insulin transcription and insulin secretion during a loss of function screen conducted in the MIN6 pancreatic beta cell line5. Gpr27 is predominantly expressed in the brain, pituitary, and the pancreatic beta cell5 (GTEx Portal). While this previous work indicates that Gpr27 may represent a new therapeutic target for diabetes, Gpr27 remains uncharacterized in vivo.
To explore the role of Gpr27 in glucose homeostasis in vivo, we studied a whole body Gpr27 knockout mouse. Islets from Gpr27 knockout mice had lower mRNA expression of insulin mRNA but unchanged insulin protein. Gpr27 knockout mice had reduced body mass, had modest worsening of glucose intolerance, and had decreased plasma insulin levels. This work adds to our understanding of the role of Gpr27 in regulating glucose homeostasis in vivo.
Results
Gpr27 knockout mice were generated by the Texas A&M Institute for Genomic Medicine (TIGM) by replacing the Gpr27 coding exon with an a beta galactosidase-neomycin fusion protein and a 3-phosphoglyerate kinase promoter driving puromycin resistance cassette in embryonic stem cells (Fig. 1A and Methods). Crosses of heterozygous Gpr27 mice revealed all 3 genotypes at the expected Mendelian ratios (89 wild type, 154 heterozygous, 77 knockouts at 3 weeks of age, chi-squared p value = 0.509) and displayed no gross abnormalities.
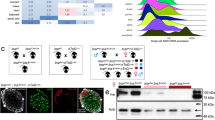
A global Gpr27 knockout mouse has reduced islet insulin and Pdx1 mRNA. (A) The Gpr27 locus contains a single exon (blue). The top panel shows polyA RNA-seq reads from sorted primary mouse beta cells16 The targeting vector (bottom) replaces the single coding exon of Gpr27 with an internal ribosomal entry site, beta-galactosidase (lacZ)-neomycin (neo) resistance cassette and polyadenylation signal (pA). This is followed by a 3-phosphoglycerate kinase promoter puromycin resistance cassette (pgk-puro-pA). (B) Gpr27 mRNA measured by RT-qPCR from isolated islets of 9 to 17-week old Gpr27 knockout animals or their wild type littermates. n = 9 wt, 11 ko. (C) Gpr27 mRNA measured by RT-qPCR from cerebral cortex, n = 3 wt, 3 ko. (D) As in 1B but for Ins1/2, Glis3, Hnf4a, Pax6 and Pdx1 mRNA. For Ins1/2, Pax6, and Hnf4a, n = 6 wt, 8 ko. For Glis3 n = 9 wt, 10 ko. For Pdx1, n = 10 wt, 11 ko. Error bars indicate SEM. p values were calculated by Student’s t-test and corrected for multiple testing by the Benjamini-Hochberg method (part D).
Gpr27 mRNA was undetectable in islets isolated from Gpr27 knockout mice (Fig. 1B). Gpr27 mRNA was also undetectable in the cerebral cortex of Gpr27 knockout mice (Fig. 1C). As we initially identified Gpr27 as a positive regulator of insulin transcription, we measured mature insulin mRNA by quantitative PCR (Fig. 1D). Confirming our in vitro data5, we found that insulin mRNA was reduced by ~30% in Gpr27 knockout islets. Levels of Pdx1 were also reduced by ~30% in Gpr27 knockout mice while Glis3, Pax6, and Hnf4a were not changed (Fig. 1D).
At 12 weeks of age, Gpr27 mice were 10% lighter than their wild type littermates (Fig. 2A). Despite their lower average weight, we found that Gpr27 knockout animals had slightly impaired glucose tolerance compared to their wild type littermates (two-way mixed ANOVA, time*genotype p = 0.048) that just passed our significance threshold of 0.05 (Fig. 2B). Area under the curve (AUC) analysis of blood glucose measurements confirmed this impaired glucose tolerance in Gpr27 knockout mice (p = 0.049) (Fig. 2C). Insulin tolerance testing was not significantly different between wild type and knockout animals (Fig. 2D,E). Based on our prior studies of Gpr27 in beta cell lines, we predicted Gpr27 knockout mice might have reduced insulin secretion. Indeed, plasma insulin levels were reduced in the knockout animals (two-way mixed ANOVA, time*genotype p = 0.03) (Fig. 2F), and this was due to a significant reduction in the plasma insulin measured 15 minutes after glucose challenge (p = 0.011). Islet morphology from Gpr27 knockout mice was largely normal as determined by immunofluorescence staining for insulin and glucagon (Fig. 3A). Beta cell mass and the distribution of islet sizes were unchanged. (Fig. 3B–D).
Gpr27 knockout mice have modestly impaired glucose tolerance and plasma insulin levels but normal insulin sensitivity on a chow diet. (A) Body weight at 12 weeks of age. (B) Intraperitoneal glucose tolerance test at 12 weeks of age. For time, F(2.612,211.599), p < 0.001; for time*genotype F(2.612, 3.167), p = 0.041. (C) Area under the curve for B. (D) Insulin tolerance test at 12 weeks of age. For time, F(2.139,53.5666), p < 0.001; for time*genotype F(2.139, 0.402), p = 0.686. (E) Area under the curve for D. (F) Plasma insulin at 12 weeks of age. For time, F(1,122.958), p < 0.001. For time*genotype, F(1, 5.730), p = 0.03. For B, E and D, Mauchly’s test of sphericity was violated so these were Greenhouse-Geisser corrected. Error bars show SEM. *p < 0.05. n = 8 wt, 9 ko.
Gpr27 knockout mice have normal beta cell mass and islet size. (A) Normal islet morphology in Gpr27 knockout islets. Sections were stained for insulin and glucagon and DNA. Representative of 2 mice per genotype. (B) Representative pancreas sections from wild type and Gpr27 knockout animals stained for insulin (dark brown). (C) Beta cell mass at 20–23 weeks of age, n = 4 wt, 4 ko. (D) Histogram of islet size, n = 4 wt, 4, ko. Error bars show SEM. p-value by Student’s t-test.
Islets isolated from Gpr27 mice did not display a detectable decrease in fractional insulin secretion in response to glucose (Fig. 4A) or potassium chloride stimulation (Fig. 4B). Despite the fact that insulin mRNA was reduced, there was also no detectable change in amount of insulin protein when normalized to islet number or to total protein concentration (Fig. 4C,D).
Ex vivo GSIS of Gpr27 knockout islets is normal. (A) Ex vivo GSIS of male 9 to 17-week old Gpr27 wild type and Gpr27 knockout islets in response to 2.8 mM and 16.7 mM glucose. n = 5 wt, 6 ko mice. (B) Ex vivo GSIS of male 9 to 17-week old Gpr27 wild type and Gpr27 knockout islets in response to 2.8 mM and 2.8 mM glucose and 40 mM KCl. n = 5 wt, 6 ko mice. (C) Total islet insulin from 25 islets of each genotype. n = 5 wt, 6 ko. (D) As in 4 C except normalized to total protein content. Error bars indicate standard error.
To confirm impaired glucose tolerance, we challenged the mice with increased insulin resistance by placing an independent cohort of Gpr27 knockout mice and wild type littermate controls on a high fat diet from 3 weeks to 12 weeks of age. After high fat feeding, the body weights of Gpr27 and wild type mice were not different (Fig. 5A). The glucose tolerance and the glucose AUC were again statistically significantly changed in the Gpr27 knockout mice (Fig. 5B,C). Insulin sensitivity exhibited a trend towards being improved in the knockout animals that did not reach our statistical significance threshold, further implying an underlying an insulin secretory defect (Fig. 5D,E). Plasma insulin levels were significantly lower in the Gpr27 knockout mice (2-way ANOVA for genotype p = 0.04) (Fig. 5F). This effect came from both the fasting and 15-minute time points as the p value for interaction (time*genotype) was not significant and the post-hoc test between genotypes at 0 and 15 minutes did not reach the significance threshold (p = 0.1 for each).
Gpr27 knockout mice are modestly hyperglycemic on a high fat diet with unchanged insulin sensitivity. (A) Body weight at 12 weeks after HFD. (B) IPGTT. For time, F(2.612, 211.599) p < 0.001. time*genotype F(2.612,3.167) p = 0.041. n = 9 wt mice, 12 ko mice. (C) Area under the curve for IPGTT. (D) Insulin tolerance test. n = 7 wt mice, 9 ko mice. For time, F(2.366,77.525), p < 0.001. For time*genotype F(2.366, 1.985), p = 0.147. (E) AUC for insulin tolerance test. SEM is shown. (F) Plasma insulin concentrations as in 2 F; Two way ANOVA: time, F(1, 9.252), p = 0.006, for genotype F(1,4.371), p = 0.04. For time*genotype F(1,0.465), p = 0.5. n = 7 ko and 4 wt. For B and F, Mauchly’s test of sphericity was violated so these were Greenhouse-Geisser corrected. Error bars show SEM.
Given the mild phenotype of the Gpr27 knockout mice, we measured the expression of the two other Gpr27 family members, Gpr85 (SREB2) and Gpr173 (SREB3). Expression of Gpr85 in islets was unchanged in the Gpr27 knockout mice (Fig. 6A) while Gpr173 was not reliably detected above background in either genotype (not shown). We also considered the possibility that the deletion of the Gpr27 coding exon could have affected neighboring gene expression. While the Prok2 (the nearest telomeric gene) is not expressed in mouse islets, the nearest centromeric gene, Eif4e3 is expressed. Surprisingly, mRNA levels of Eif4e3 were reduced in Gpr27 knockout islets (Fig. 6B) but not in Gpr27 knockout cerebral cortex (Fig. 6C). To attempt to explain this, we examined mRNA-seq data from wild type primary mouse beta cells and observed splicing events between the antisense strand of the Gpr27 exon and the Eif4e3 gene (Fig. 6D). We observed similar splice junctions in 2 human RefSeq EIF4E3 transcripts (NM_173359 and NM_001134649), each beginning with an exon located on the anti-sense strand of the GPR27 coding exon. Taken together, these data suggest that the deletion of the Gpr27 exon has the unintended consequence of removing the first exon of a subset of Eif4e3 transcripts. A less likely possibility would be that Gpr27 mRNA expression is required for Eif4e3 transcription. To test this, we knocked down Gpr27 mRNA using an shRNA in MIN6 cells (Fig. 6E). We found that Gpr27 knockdown did not reduce Eif4e3 mRNA (Fig. 6F), suggesting that the reduction of Eif4e3 in Gpr27 knockout islets is likely due to the deletion of the Gpr27 exon itself.
Gpr27 knockout islets have reduced Eif4e3 mRNA. (A) As in Fig. 1B but for Gpr85 mRNA. (B) As in 6A except for Eif4e3 mRNA. (C) As in Fig. 1C except for Eif4e3 mRNA. (D) mRNA-seq coverage and junctions from sorted primary mouse beta cells showing splicing between the Gpr27 coding exon and the Eif4e3 gene. Data plotted from16 using17. (E) Human Refseq EIF4E3 transcripts showing two that contain overlap with the GPR27 coding exon. Note the similar splicing structure to 6D. (F) MIN6 cells were infected with the indicated adenoviruses. Expression of Gpr27 was measured by RT-qPCR. (G) As in E, but Eif4e3 was measured. For F and G, n = 4. p values were determined by Student’s t-test. (H) IPGTT of 13-week old Eif4e3 knockout mice versus control mice. n = 8 male knockout mice vs 320 control mice. Error bars indicate standard error. Data plotted from IMPC6.
Finally, we examined data from the International Mouse Phenotyping Consortium on Eif4e3 knockout mice to ask if downregulation of Eif4e3 could partially explain the modestly worsened glucose tolerance of Gpr27 knockout animals. Eif4e3 knockout mice had a trend to lower plasma glucose that did not meet their significance threshold (28.38 mmol/L vs 25.17 mmol/L in males, 25.05 mmol/L vs 17.95 mmol/L in females, combined p-value = 0.007)6 and an unchanged glucose tolerance (Fig. 6G). On the other hand, the Eif4e3 knockout male mice had lower body weights (39.8 grams vs 34.0 grams, p = 0.000928 in males)6, suggesting that Eif4e3 downregulation might explain lower body weights in the Gpr27 knockout mice.
Discussion
Herein we provide a description of the in vivo consequences of Gpr27 deletion. We found that Gpr27 loss reduces insulin mRNA in vivo, confirming our previous findings in vitro in MIN6 cells5. This reduced insulin transcript may be caused by a reduction in mRNA levels of Pdx1 as we found in MIN6 cells after knockdown of Gpr27. However, we found only a non-significant trend towards reduction in levels of Hnf4a and Glis3 in Gpr27 knockout islets in contrast to the clear reductions in these genes that we found in Gpr27 knockdown MIN6. One possible reason for this difference is that our experiments in cultured cells silenced Gpr27 for only 72 hours5 while the Gpr27 knockout beta cells have been without the gene for months and may have compensated for the loss of Gpr27. Alternatively, this could be due to differences between cultured beta cells and primary beta cells.
We were initially surprised to find that mRNA transcripts of Eif4e3, a gene 25 kb away from Gpr27, were reduced in islets but not cerebral cortex of Gpr27 knockout animals. We believe that the most likely reason for this is that unannotated Eif4e3 transcriptional start sites are located on the antisense strand of the Gpr27 coding exon. Although the mouse Refseq annotation does not describe such transcripts, we find evidence of them in mRNA-seq data from mouse beta cells. Confirming this, we find very similar overlapping GPR27/EIF4E3 transcripts are present in the human Refseq annotation. We speculate that these alternative transcriptional start sites of Eif4e3 may be more specific to the islet in mouse versus human, possibly explaining why Eif4e3 levels are unchanged in the brain of Gpr27 knockout mice and why the mouse Refseq annotation lacks these transcripts (as islet expression data may not have been included). Our observations suggest that future attempts to manipulate the Gpr27 exon (e.g. by a Cre-lox strategy or CRISPR) must be done with caution since they may also affect Eif4e3. Notably, the alternative transcription start sites change only the 5′ untranslated region of Eif4e3, suggesting that the primary effect would be on the Eif4e3 expression level and not on the protein sequence.
We cannot completely rule of the possibility that the phenotype of our Gpr27 knockout mice is due to local downregulation of Eif4e3 by the knockout allele. However, we do not believe this is likely since silencing (by RNA interference) of Gpr27 does not affect Eif4e3 transcription but does reduce insulin and Pdx1 mRNA5 and Eif4e3 knockout mice have unchanged glucose tolerance.
While we did observe worsened glucose tolerance in the Gpr27 knockout mice on a chow diet, the p-values were close to our predetermined statistical threshold and the magnitude of the phenotype was quite mild and likely biologically inconsequential. To minimize the chance of a type 1 error, we confirmed worsened glucose tolerance on an independent cohort of mice fed a high fat diet. In both cases, worsened glucose tolerance in combination with reduced plasma insulin after glucose challenge and unchanged insulin tolerance suggest the Gpr27 global knockout mice have an insulin secretory defect. However, we did not observe a defect in insulin secretion in ex vivo islets from Gpr27 knockout mice. We speculate that one possibility for the normal ex vivo GSIS could be the lack of a hypothetical Gpr27 ligand ex vivo. Alternatively, the defect in GSIS in Gpr27 deficient islets could be below our limit of detection, especially given the modest effect on glucose tolerance. Finally, given Gpr27’s expression in the brain, we cannot rule out a primary brain effect of Gpr27 loss that then causes reduced GSIS in the periphery.
Gpr27 remains an orphan GPCR. One possible explanation for the phenotype in Gpr27 knockout mice is that there are as of yet unidentified endogenous ligands that stimulate Gpr27, and loss of Gpr27 blocks these signals. Alternatively, Gpr27 may simply have basal activity that is required for insulin production and insulin secretion7. Several groups have now described small molecules that may activate Gpr27. Through a chemical screen, Dupuis and Hanson identified two small molecules that can trigger Gpr27 association with β arrestin-2 in 293T cells8. Plasmalogens, a form of glycerophospholipid, may also signal through Gpr27, although direct binding of plasmalogens to Gpr27 has not been demonstrated9. Additionally, two inverse agonists of Gpr27 have been described10. The Gpr27 knockout mouse will be a useful resource for testing the specificity of these putative ligands.
The G-protein coupling of Gpr27 has been controversial. While our initial study showed that Gpr27 over-expression in 293T cells increased IP1 suggesting Gαq coupling, this finding was not reproduced by Dupuis and Hanson who found no evidence of Gαq, Gαs, or Gαi coupling8. On the other hand, Martin and Aronstam found evidence of constitutive Gαi coupling in 293T cells7. Thus, it is not clear how Gpr27 signals even in these heterologous over-expression systems. However, impaired insulin secretion in Gpr27 knockout mice implies that Gpr27 might normally signal to enhance insulin secretion, which would implicate either Gαs or Gαq11. Additional studies are needed to further interrogate the signaling mechanism and pathway of Gpr27.
In summary, we describe a whole-body knockout mouse for Gpr27 and show reduced insulin mRNA but only modest effects on glucose homeostasis and an unexpected effect on Eif4e3 gene expression.
Methods
Animals
129SvEvBrd embryonic stem cells were transfected with a targeting vector containing 5′ and 3′ homology arms to the Gpr27 locus where the entire coding sequence of Gpr27 was replaced with an IRES-lacZ-neomycin and a bovine growth hormone polyadenylation signal followed by the 3-phosphoglycerate kinase promoter driving a puromycin resistance gene and another bovine growth hormone polyadenylation signal. Cas9 was not used. A clone containing proper integration by homologous recombinaton was confirmed by PCR and southern blot (Supplemental Fig. 1). This clone was injected into blastocysts12,13 and germ line transmission was achieved. We obtained heterozygous mice from the Texas A&M Institute for Genomic Medicine and bred the mice to C57B6J for >5 generations in our mouse facility. The mouse line is currently available from Taconic (https://www.taconic.com/knockout-mouse/gpr27-targeted). The sequence of the targeting vector and the final genomic sequence of the targeted allele are in Supplemental Data.
Mice were group-housed in a colony maintained with a standard 12 h light/dark cycle and given food and water ad libitum. Experiments were conducted according to the Guide for the Care and Use of Laboratory Animals, as adopted by the National Institutes of Health, and with approval of the UCSF Institutional Animal Care Use Committee (IACUC). Genotyping for the mutant allele was performed with the following primers: puroLR = GAACCAGCTGATTACCCTGTTATCCCTAC and Gpr27-R = CGCAAAGGTAATGCCACTTGAGG. Genotyping for the wild type allele was performed with the following primers: Gpr27-F = CTGAAAGGCATTGGTTTGTGAAGC and Gpr27-R = CGCAAAGGTAATGCCACTTGAGG. Male mice age 9–15 weeks were used for all experiments unless otherwise specified in figure legends. For high fat diet, mice were placed on Research Diets D12492 (5.21 Kcal/g, 20% protein, 20% carbohydrates, 60% fat) from 3 weeks until 12 weeks of age. For all other experiments, animals were fed PicoLab Mouse Diet 20 (3.75 Kcal/g, 23.2% protein, 55.2% carbohydrates, 21.6% fat). Intraperitoneal glucose tolerance test, insulin tolerance test, and plasma insulin collection were performed as previously described14. Briefly, mice were fasted individually for 16 hours prior to 2 g/kg glucose injection or 0.1 mU/kg insulin. Glucose (Freestyle Lite, Abbott) and insulin (mouse insulin ELISA, Mercodia) were measured from blood or plasma taken from the tail.
Islet RT-qPCR
Pancreatic islets were isolated as previously described15 and cultured overnight in RPMI with 10% fetal bovine serum, 1% penicillin/streptomycin, 25 mM HEPES. Total RNA was extracted by Trizol (ThermoFisher) or Direct-zol RNA MiniPrep Kit (Zymo 11–330), treated with DNase I (Turbo DNase, Ambion), and reverse transcribed with Superscript III (ThermoFisher). Gene expression was calculated utilizing the delta delta CT method relative to Gusb. Probes utilized were: Ins1/25, Gpr275, Glis3 (Mm00443081_m1, ThermoFisher), Hnf4a (Mm01247712_m1, ThermoFisher), Pax6 (Mm00615386_m1, ThermoFisher), Pdx1 (Mm00435565_m1, ThermoFisher), Eif4e3 (Mm01182452_m1, ThermoFisher), Gpr85 (Mm00460767_s1, ThermoFisher), Gpr173 (Mm02620389_s1, ThermoFisher), Gusb5.
Gpr27 knockdown
Control or Gpr27 shRNA adenovirus5 were used to infect MIN6 cells at an MOI of 50, cultured for 3 days and analyzed for Gpr27 and Eif4e3 expression as above.
Ex vivo glucose stimulated insulin secretion (GSIS)
Islets were rested for 24 hours after isolation RPMI (Gibco) with HEPES, penicillin, streptomycin, and glutamine supplementation. For static GSIS, 15–20 islets were hand-picked and equilibrated in Krebs-Ringer Bicarbonate HEPES buffer (KRBH; 137 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 2.5 mM CaCl2, 25 mM NaHCO3, 20 mM HEPES) containing 2.8 mM glucose for 1 hour at 37 °C. Equilibration buffer was then removed and islets were sequentially stimulated with KRBH containing glucose and compounds as indicated in figure panels. Islets were lysed in RIPA buffer for determination of total islet insulin content. Islet lysate protein content was determined using the Pierce 660 Protein Assay with Ionic Detergent Compatibility Reagent (ThermoFisher).
Immunofluorescence imaging
Pancreata were fixed in Z-FIX (Anatech) and paraffin embedded. Five µm sections were deparaffinized and subjected to antigen retrieval using Citrate unmasking solution (Vector H3300). Primary antibodies used: guinea-pig anti-insulin (1:250, Dako A0564), rabbit anti-glucagon (1:250, Immunostar 20076). Secondary antibodies: anti-guinea pig Alexa Fluor 488 (1:500, ThermoFisher A11073), anti-rabbit Alexa Fluor 555 (1:500, ThermoFisher A31572) Images were captured with a Zeiss Apotome widefield microscope.
Beta cell mass and distribution of islet size
Pancreatic sections as above were taken at approximately every 100 microns through the whole pancreas, fixed and cut as done for the immunofluorescence except only the guinea-pig anti-insulin antibody was used for the primary antibody at 1:1000. The ABC Kit (Elite EK-6100) and DAB Chromogen System (Dako) were used to develop. A Keyence BZ-X710 at 2X magnification was used to capture images. Beta cell area was measured with photoshop for each section and % beta cell area was measured. Beta cell mass was calculated by the product of % beta cell area * pancreas wet weight.
Statistical analysis
For IPGTT and plasma insulin, a mixed two-way ANOVA with repeated measures was implemented by SPSS (IBM). For multiple testing of individual time points, we used Benjamini-Hochberg post-hoc testing. For AUC and body weight, we used a two-tailed Student’s t-test.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schioth, H. B. & Gloriam, D. E. Trends in GPCR drug discovery: new agents, targets and indications. Nat. Rev. Drug. Discov. 16, 829–842 (2017).
Ahren, B. GLP-1 receptor agonists for type 2 diabetes- A rationale drug development. J Diabetes Investig (2018).
Nauck, M. A., Meier, J. J., Cavender, M. A., Abd El Aziz, M. & Drucker, D. J. Cardiovascular Actions and Clinical Outcomes With Glucagon-Like Peptide-1 Receptor Agonists and Dipeptidyl Peptidase-4 Inhibitors. Circulation. 136, 849–870 (2017).
Persaud, S. J. Islet G-protein coupled receptors: therapeutic potential for diabetes. Curr. Opin. Pharmacol. 37, 24–28 (2017).
Ku, G. M., Pappalardo, Z., Luo, C. C., German, M. S. & McManus, M. T. An siRNA screen in pancreatic beta cells reveals a role for Gpr27 in insulin production. PLoS Genet. 8, e1002449 (2012).
Dickinson, M. E. et al. High-throughput discovery of novel developmental phenotypes. Nature 537, 508–514 (2016).
Martin, A. L., Steurer, M. A. & Aronstam, R. S. Constitutive Activity among Orphan Class-A G Protein Coupled Receptors. PLoS One 10, e0138463 (2015).
Dupuis, N. et al. Activation of the Orphan G Protein-Coupled Receptor GPR27 by Surrogate Ligands Promotes beta-Arrestin 2 Recruitment. Mol. Pharmacol. 91, 595–608 (2017).
Hossain, M. S., Mineno, K. & Katafuchi, T. Neuronal Orphan G-Protein Coupled Receptor Proteins Mediate Plasmalogens-Induced Activation of ERK and Akt Signaling. PLoS One 11, e0150846 (2016).
Yanai, T. et al. Identification and molecular docking studies for novel inverse agonists of SREB, super conserved receptor expressed in brain. Genes. Cell 21, 717–27 (2016).
Jain, S. et al. Chronic activation of a designer G(q)-coupled receptor improves beta cell function. J. Clin. Invest. 123, 1750–62 (2013).
Doetschman, T. et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 330, 576–8 (1987).
Thomas, K. R. & Capecchi, M. R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell 51, 503–12 (1987).
Pappalardo, Z. et al. A Whole-Genome RNA Interference Screen Reveals a Role for Spry2 in Insulin Transcription and the Unfolded Protein Response. Diabetes 66, 1703–1712 (2017).
Szot, G. L., Koudria, P. and Bluestone, J. A. Murine pancreatic islet isolation. J Vis Exp. 255 (2007).
Ku, G. M. et al. Research resource: RNA-Seq reveals unique features of the pancreatic beta-cell transcriptome. Mol. Endocrinol. 26, 1783–92 (2012).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–6 (2011).
Acknowledgements
This work was supported by NIH R01 DK107650 and K08 DK087945 and P30 DK063720 (a Pilot and Feasibility Grant to GK and core laboratories). GMK was supported by a Career Development Award from JDRF (5-CDA-2014-199-A-N). We would like to thank Michael S. German and Gerold M. Grodsky for helpful discussions; Vi Dang, Vinh Nguyen and Greg Szot from the UCSF Mouse Islet Production Core for providing pancreatic islets. The UCSF Islet Isolation Facility is supported by Diabetes Research Center (DRC) grant NIH P30 DK063720. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Author information
Authors and Affiliations
Contributions
G.M.K. conceived of the work. T.G.H., D.G.C., N.Y., Y.Z. and G.M.K. performed experiments and analyzed the data. G.M.K. wrote the manuscript. T.G.H. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chopra, D.G., Yiv, N., Hennings, T.G. et al. Deletion of Gpr27 in vivo reduces insulin mRNA but does not result in diabetes. Sci Rep 10, 5629 (2020). https://doi.org/10.1038/s41598-020-62358-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62358-4
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.