Abstract
The objective of the present study was to determine the cross-protection of Ingelvac PRRS MLV against challenge with the new lineage 1 PRRSV emerged in China in pigs. Two lineage 1 PRRSV strains (FJZ03 and FJWQ16 originated from recombination event between NADC30 and JXA1-like strain). We found that pigs vaccinated with the vaccine were protected against challenge with the FJZ03 as shown by fewer days of clinical fever, reduced lung pathology scores, lower PRRS virus load in the blood and developed broadly neutralizing antibodies with high titers to FJZ03. In contrast, vaccine provided limited protection against challenge with FJWQ16 with higher fever, lower antibody titers, lower neutralizing antibodies and higher viral loads in blood. These results demonstrate PRRSV-MLV provides incomplete protection against new lineage 1 PRRSVs.
Similar content being viewed by others
Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a global viral swine disease and characterized by severe reproductive failure in sows and respiratory problems in piglets1. PRRS virus (PRRSV) is a small, enveloped, positive-sense single-stranded RNA virus belonging to the Arteriviridae family in the Nidovirales order2,3,4. The PRRSV genome is about 15 kb in length and codes for at least 16 non-structural proteins (nsps) (nsp1α, nsp1β, nsp2-6, nsp2TF, nsp2N, nsp7a, nsp7b and nsp8-12) and 8 structural proteins (GP2, E, GP5a, GP3-GP5, M and N)4,5,6,7,8,9.
PRRSV strains were divided into two distinct genotypes, type 1 (European strains, EU) and type 2 (North American strains, NA), which share only about 60% nucleotide identity at the genome level with distinct antigenicity. PRRSV emerged in China in 1996, and has become one of the country’s most serious swine diseases. In May 2006, highly pathogenic PRRS (HP-PRRSV) emerged in China and is characterized by high fever and associated with a high mortality rate, resulting in immense economic losses in the swine industry in China10. In 2013, new lineage 1 PRRSV (NADC30-like) emerged in China and spread widely in Chinese swine herds with high morbidity and mortality in sows and piglets. The NADC30-like PRRSV is now the predominant strain circulating in swineherds and have been causing great economic loss in China. Vaccination is the most effective and practical method to prevent infectious diseases and live attenuated PRRSV vaccines have been a valuable tool in PRRS disease control11. To date, several live attenuated NA-type PRRSV vaccines, such as Ingelvac PRRS® MLV, CH-1R, R98 and HP-PRRSV-derived live vaccines including JXA1-R, HuN4-F112 and TJM are commercially available in China. Among these vaccines, Ingelvac PRRS MLV (MLV) is one of the most widely used in China, and MLV strain belong to lineage 5 virus, whereas NADC30-like PRRSV is lineage 1 according to the global PRRSV classification systems12. Recently, some novel recombinant PRRSV strains (lineage 1) arising from NADC30-like PRRSV and filed or vaccine strain in China have been reported, whether there is cross-protection from the commercial vaccine against the novel recombinant PRRSV and NADC30-like PRRSV challenge is not known. Therefore, the objective of this study was to evaluate the efficacy of PRRSV vaccine MLV against heterologous NADC30-like PRRSV challenge in pigs.
Results
Complete genomic sequence analysis
Excluding poly(A) tails, complete genome sequence of FJZ03 and FJWQ16 strains was 15,016 nucleotides in length. Genome alignments revealed 81.8–96.0% identity with NA-type PRRSV strains, and identity was 92.0% between the two strains. Of note, FJZ03 and FJWQ16 had the maximum identity (97.1% and 93.0%, respectively) with NADC30. Furthermore, strains FJZ03 and FJWQ16 share 86.0–86.2% homology with the genome of vaccine strain Ingelvac PRRS MLV (MLV), however, 83.9–85.2% homology with the genome of JXA1-R vaccine (a live attenuated virus vaccine strain derived from the highly pathogenic PRRSV JXA1).
High genetic diversity is a significant characteristic of PPRSV. A systematic classification of type 2 PRRSV has been conducted based on analysis of 8624 ORF5 sequences in the databases including field isolates and vaccine strains12. In this system, type 2 PRRSV was divided into nine monophyletic lineages (1–9). Therefore, this type of systematic genotyping was used for PRRSV classification in the present study.The phylogenetic analysis of ORF5 genes showed that FJZ03 and FJWQ16 formed a minor branch represented by NADC30 (lineage 1 virus) (Fig. 1). To confirm the potential recombination event, we evaluated potential recombinants using seven algorithms (RDP, GeneConv, BootScan, MaxChi, Chimera, SiScan, and 3Seq) implemented in RDP 4.80. Two inter-lineage recombination events between lineages 1 (NADC30) and 8.7 (JXA1 P45) were identified in the FJWQ16, with recombination breakpoints at position 6854–8787 (located in nsp7 (nt62)-nsp9 (nt1103), with reference to VR-2332 strain, Fig. 2). However, we note that we failed to observe any evidence of recombination in the FJZ03 by using RDP software.
Clinical observations
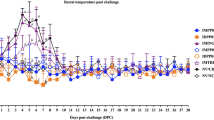
Differences in clinical expression between group 1 (MLV + FJZ03) and group 2 (MLV + FJWQ16) pigs were observed. The mean rectal temperatures was high (>40 °C) in the MLV + FJZ03 group from 1 to 4 dpc, then leveled off (Fig. 3A), whereas mean rectal temperatures was above 40 °C from 1 to 9 dpc in the MLV + FJWQ16 group 2 (Fig. 3B). All pigs in MLV + FJWQ16 group exhibited severe clinical signs including high fever, acute respiratory distress and coughing during 2–10 dpc, whereas pigs from the MLV + FJZ03 group showed no significant clinical signs throughout the experiment. The mean rectal temperature in pigs unvaccinated and challenged with FJWQ16 and those vaccinated and challenged with FJWQ16 were significantly higher (p < 0.05) than in pigs vaccinated and challenged with FJZ03 and control group at 5 dpc, 7 dpc, 8 dpc and 10 dpc (Fig. 3). Unvaccinated pigs challenged with FJZ03 or FJWQ16 developed high fever (≥40 °C) from 1 to 10 dpc and typical symptoms of PRRS such as acute respiratory distress, lethargy, anorexia, and emaciation from 2 to 10 dpc. Surprisingly, there were not statistical significant differences in rectal temperatures (except 7 dpc) between MLV + FJWQ16 and unvaccinated + FJWQ16 challenge group (Fig. 3B). Survival rates of pig in unvaccinated by challenged with FJWQ16 group was 80%, while the pigs in unvaccinated by challenged with FJZ03 group was 100% (Fig. 4). The mean respiratory scores were significantly higher (p < 0.05) in pigs unvaccinated challenged pigs and vaccinated challenged with FJWQ16 with or without vaccination pigs than pigs in other 2 groups from 3 to 14 dpc (Fig. 5). The body weight gain of pigs was monitored throughout the study. As shown in Fig. 5, by the end of the first week after challenge (on 7 dpc), pigs in unvaccinated + FJWQ16, unvaccinated + FJZ03 and MLV + FJWQ16 groups showed low weight gain, whereas in both unvaccinated + FJWQ16 and MLV + FJWQ16 groups lost weight during 7–14 dpc. Control pigs gained weight over the duration of the study. The pigs in the unvaccinated + FJWQ16 and MLV + FJWQ16 challenge groups had significantly (p < 0.05) lower body weight gain than that of MLV + FJZ03 group and control group during 7–14 dpc (Fig. 6).
Lung pathology
Macroscopic lung lesions were characterized by symptoms of interstitial pneumonia, multifocal, scattered bleeding points, tan-mottled areas with irregular and indistinct borders. The scores for gross lung lesion were shown in Table 1. In two unvaccinated groups, pigs infected with FJWQ16 showed more severe pathology than FJZ03 infected pigs (p < 0.05). Gross lung lesions in the MLV + FJWQ16 group were significantly more severe than those in MLV + FJZ03 and control groups (p < 0.05). However, no pathologic lesions were observed in control pigs. Microscopic lung lesions in unvaccinated + FJWQ16, unvaccinated + FJZ03, MLV + FJWQ16-infected pigs were characterized by interstitial pneumonia with severely alveolar septa thickening, increased numbers of lymphocytes and interstitial macrophages. Mild microscopic lung lesions were observed in MLV + FJWQ16-infected and MLV + FJZ03-infected pigs, whereas no noticeable microscopic lung lesions were observed in the control pigs.
Detection of virus in the serum
Genomic copies of the vaccine virus were detected in the sera from the vaccinated and challenged pigs at 4 dpi, and had a low serum viral load 28 days post immunization. In the pigs from the MLV + FJZ03 group, high level of the mean serum viral load was detected (104.7 copies/mL) on 4 dpc, which peaked at 106.0 copies/mL on 7 dpc, and then decreased (Fig. 7). After challenge with FJWQ16, the pigs in the MLV + FJWQ16 group had a high mean serum viral titer on 4 dpc (105.5 copies/mL), which peaked (106.5 copies/mL) on 7 dpc and remained high until 14 dpc (Fig. 7). In the unvaccinated + FJZ03 and unvaccinated + FJWQ16 groups, viral load was detected starting on 4 dpc (105.2 copies/mL, 106.1 copies/mL, respectively), which peaked on 7 dpc (106.7 copies/mL, 107.2 copies/mL, respectively) and remained high until 14 dpc (Fig. 7). Vaccinated and challenged pigs had a significantly (p < 0.05) lower number of genomic copies of type 2 PRRSV in the blood than the unvaccinated and challenged pigs from 7 to 11 dpc (Fig. 7).
Anti-PRRSV IgG antibodies
In all unvaccinated pigs, mean S/P ratio for every pig was 0.0 at 0 dpc. After FJZ03 and FJWQ16 challenge, all serum samples from unvaccinated + FJZ03 and unvaccinated + FJWQ16 group became PRRSV-positive with S/P ratio ≥ 0.4 occurred at 7 dpc, and by 14 dpc all pigs developed antibodies with a mean S/P ratio for the FJZ03 and FJWQ16 groups of 1.23 ± 0.12, and 1.48 ± 0.14, respectively. In all vaccinated pigs, seroconversion occurred on 14 dpi and the mean S/P ratio was 0.86 ± 0.15 for the MLV + FJZ03 group at 0 dpc, and 1.63 ± 0.18 at 14 dpc after FJZ03 challenge. At 0 dpc, the mean S/P ratio was 0.90 ± 0.15 for the MLV + FJWQ16 group, 1.78 ± 0.16 at 14 dpc after FJWQ16 challenge. Anti-PRRSV antibody titers were not detected in the negative control group throughout the experiment.
PRRSV-specific neutralizing antibodies
Neutralizing antibodies (NA) were detected in pigs vaccinated with MLV on 28 dpi. Pigs vaccinated with MLV developed high NA titers for FJZ03 (>4, Fig. 8), but only low titers for FJWQ16 (≤2, Fig. 8). Furthermore, the NA titers to FJZ03 increased in vaccinated pigs following challenge (>8), but the titers to FJWQ16 maintained at a low level (≤6) and never reached protective levels (Fig. 8). NAs were not detected in any pigs in the control group throughout the experiment.
Discussion
PRRS has been causing substantial economic losses in the swine industry, especially, the HP-PRRSV, since 2006, in China10. Recently, new lineage 1 PRRSV (NADC30-like) emerged in China and is now a severe threat to Chinese pig population. The PRRSV-MLV vaccine can induce a protective immune response, but it may not have broad cross-protection against all isolates13,14,15,16. Although six PRRSV vaccines are currently marketed in China, Ingelvac PRRS MLV is one of the most widely used in China. In the present study, FJWQ16 and FJZ03 are lineage 1 virus (NADC30-lile PRRSV), while vaccine strain MLV is lineage 5 based on ORF5 analysis. However, whether there is cross-protection from the vaccine MLV (lineage 5) against heterologous NADC30-like PRRSV (lineage 1) challenge is not known. Therefore, the aim of the present study was to investigate the efficacy of Ingelvac PRRS MLV in a challenge study with the NADC30-like PRRSVs.
Ingelvac PRRS MLV has been shown to provide complete protection from infection by VR-2332, however, offers incomplete protection against newly emerging heterologous strains13,14. There is relatively little data pertaining to the immune responses and efficacy of MLV vaccine against challenges with NADC30-like PRRSV strains. In the present study, we found different levels of protection against challenge with NADC30-like PRRSVs following vaccination with the MLV. PRRSV viremia plays a key factor in the development of respiratory diseases16. The efficacy of vaccine is usually assessed by the reduction in viral loads after challenge with a virulent PRRSV17,18,19. Hence, the reduction of PRRSV viral loads in the blood and lungs is an important parameter to determine the efficacy of PRRSV vaccines17,18,19. In the present study, vaccine reduced PRRSV viral loads significantly in vaccinated and challenged groups compared with unvaccinated and challenged groups, however, in the vaccinated and challenged group (MLV + FJWQ16), the viral load remained high from 4 to 14 dpc. The high levels PRRSV viremia in MLV + FJWQ16 group result in significant respiratory diseases. Severity of the interstitial pneumonia is correlated with the amount of viral load20, in addition, PRRSV virulence was associated with higher titers of virus in vivo21. MLV + FJWQ16 group had high overall viral serum titers at 8 days after challenge, vaccination of pigs with MLV reduced only partially the severity of FJWQ16-induced lung lesions and the amount of PRRSV antigen within the interstitial pneumonia. Overall, compared to control and MLV + FJZ03 group, the MLV + FJWQ16 group had more days with high fever, exhibiting higher levels of viremia and significant body weight lost after FJWQ16 challenge, indicating that pigs vaccinated with MLV were not protected from FJWQ16 challenge. Although the use of PRRSV-MLV reduced body temperatures and viral load in the present study, there was a significant difference between MLV + FJZ03 and MLV + FJWQ16 groups. We have no clear explanation why MLV has different results of protection against FJZ03 and FJWQ16 challenge. It may be attributed to antigenic differences between the vaccine and the challenge strains, that is, MLV vaccine is less effective in inducing cross-protection against viral isolates that are significantly different from the vaccine strains. These results in present study agree with previous findings that commercial modified live PRRS vaccine is partially protect heterologous PRRS challenge13,14,22,23,24. It may be due to remarkable antigenic variation between the vaccine strains and challenging PRRSV strains25.
Neutralizing antibodies might play a significant role in protection against PRRSV17,26. In this study, neutralizing antibody titers to FJZ03 were present at protective titers (>8) in the vaccinated pigs after challenge with FJZ03. However, the titers to FJWQ16-specific neutralizing antibodies in pigs vaccinated with MLV never reached protective levels, indicating that vaccination with MLV does not induce broadly neutralizing antibodies to FJWQ16.
Although commercial vaccines provide limited protection to NADC30-like PRRSV infection, previous studies have focused on only one NADC30-like PRRSV strain24,27,28. The aim of the present study was to investigate the efficacy of the commercially available PRRSV type 2 vaccine MLV in a challenge study with two different NADC30-like PRRSV isolates. Phylogenetic and molecular evolutionary analyses indicated that FJWQ16 originated from recombination event between NADC30 and JXA1-like PRRSV strain (JXA1 P45), whereas FJZ03 was not observed any evidence of recombination between PRRSV. Additionally, recombinant PRRSVs (JL580 and FJ1402) between NADC30-like PRRSV and HP-PRRSV were shown to have higher pathogenicity than NADC30-like PRRSV. In the present study, recombinant strain FJWQ16 has higher pathogenicity than FJZ03 and agree with previous studies28,29. Furthermore, the JXA1-R vaccine virus was shown to have reverted to virulence under field conditions30. This JXA1 P45 strain maybe related to the virulence reversion of the JXA1-R vaccine virus circulating in swineherds. Thus, it is critical to monitor of PRRSV evolution in China. FJWQ16 replicated in serum at a higher titer than did FJZ03, and one piglet in the FJWQ16 group died, whereas the piglets in the FJZ03 group and control group survived throughout the experiment. Recently, the recombination between NADC30-like PRRSV and Chinese field strains/vaccine strains have been reported in many studies; these recombinant viruses showed more virulent than NADC30-like PRRSV, such as strain JL580, 14LY01-FJ, 14LY02-FJ 15LY01-FJ, and 15LY02-FJ from the recombination between NADC30 and HP-PRRSVs, which has been confirmed as highly pathogenic. In the present study, a recombinant type 2 PRRSV FJWQ16 between NADC30-like PRRSV and JXA1-like PRRSV was shown to have higher pathogenicity (20% mortality rate) than FJZ03, NADC30, and NADC30-like strain28,29,31. These results showed that genomic recombination is an important mechanism that plays a role in conditioning pathogenicity and antigenicity of the virus.
In summary, although a commercial MLV vaccine could shorten the period of fever, reduce levels of vermeil in sera and mortality rate, it was not able to reduce the clinical symptoms and lung lesions of the challenged pigs. These findings indicate that MLV vaccine provided incomplete cross-protection against newly emerging heterologous NADC30-like PRRSV and recombinant PRRSV infection. It is thus urgent to develop novel, safe and effective vaccines for controlling NADC30-like PRRSV.
Materials and Methods
Animal ethics
The animal study and all pig experimental procedures were performed in accordance with the guidelines of South China Agricultural University Institutional Animal Care and Use Committee (SCAU-AEC-2014-10) and approved by animal Ethics committee of South China Agricultural University.
Virus and vaccine
Type 2 PRRSV FJZ03 strain (GenBank ID: KP860909) was isolated from lung samples of an aborted fetus in 2013 and FJWQ16 strain (GenBank ID: KX758249) from a herd with high morbidity and mortality in sows and piglets respiratory disease, in 2015. PRRSV was isolated from the positive sera and tissues using MARC-145 cells and real-time RT-PCR analysis was applied on the supernatants to confirm the growth of the virus. The virus-infected cells in 6-well plates were determined by indirect fluorescent antibody assay (IFA). The cell cultures were subjected to the extraction of total RNA using a viral nucleic extraction kit (TIANGEN, China). Nine overlapped fragments covering the whole genome were amplified by RT-PCR as previously described32. Recombinant clones were sequenced by Ruibo Life Technologies Corporation (Beijing, China).
Modified live virus (MLV) vaccines based off VR-2332 (Ingelvac PRRS MLV) from Boehringer Ingelheim Vetmedica, Inc. Strains FJZ03 and FJWQ16 have been passaged 3 times in MARC-145 cells for use in the swine study.
Complete genomic sequence analysis
To identify the evolutionary relationship of FJZ03 and FJWQ16, 38 representative PRRSV strains in GenBank were utilized in sequence alignments and phylogenetic analyses (Table 2). Multiplex sequence alignments were performed using CLUSTAL X (version 1.83). The phylogenetic tree was constructed using the NJ method with MEGA 6.0, the maximum composite likelihood model and the bootstrap confidence value of 1000 replicates33. The ORF5 gene sequences were classified according to the global PRRSV classification systems12.
The recombination event was confirmed using a recombination detection program (RDP v.4.80)34 as described in Ramos et al.35. Potential recombination events were tested by seven different algorithms (RDP, GeneConv, BootScan, MaxChi, Chimera, SiScan, and 3Seq) with Bonferroni correction and the highest acceptable p value 0.01. Recombination breakpoints was further analyzed by the Genetic Algorithm for Recombination Detection (GARD) and SimPlot software v.3.5.136,37.
Animal study design and clinical observation
Twenty-five 21-day-old pigs confirmed to be free of PRRSV, PCV2, PRV, and CSFV were used for this study. Pigs were allowed to acclimate for one week before initiation of the experiments. All pigs randomly divided into 5 groups (5 pigs/group) and were raised separately in different isolation rooms with individual ventilation. The pigs in groups 1 (MLV + FJZ03 challenge group) and 2 (MLV + FJWQ16 challenge group) were vaccinated intramuscularly with a single dose of MLV according to manufacturer’s directions (Ingelvac PRRS® MLV) on day 0. The pigs in group 3 (unvaccinated + FJZ03 challenge group), group 4 (unvaccinated + FJWQ16 challenge group) and group 5 (unvaccinated unchallenged, control) were mock vaccinated with PBS on the same day. Twenty-eight days post immunization (dpi) (0 day post challenge, dpc), groups 1 and 3 challenged with FJZ03 (2 × 105 TCID50/pig, 2 mL), groups 2 and 4 challenged with FJWQ16 (2 × 105 TCID50/pig, 2 mL) by intranasal (1 mL) and intramuscular (1 mL) routes, respectively. The pigs in group 5 received PBS (2 mL) and served as the negative control group.
Rectal temperature was recorded daily from 0 to 14 dpc and blood samples were collected on 0, 4, 7, 11, and 14 dpc for virus titration. The pigs were monitored daily for clinical respiratory disease as previously described38, pigs were monitored every day for clinical signs and scored daily for clinical respiratory disease severity using scores ranging from 0 to 6 (0 = normal, 6 = severe). All of the pigs were euthanized on 14 dpc. Lungs were collected from each pig at necropsy and the macroscopic lesions in the lungs were recorded using a scoring system as previously described38, the scoring system is based on the approximate volume that the dorsal and ventral surfaces of each lung lobe accounts for the entire lung: the right anterior lobe, right middle lobe, cranial part of the left anterior lobe, and the caudal part of the left anterior lobe were assigned each 10% of the total lung volume, the accessory lobe were assigned 5%, and the right and left caudal lobes each contribute 27.5%. Macroscopic lung lesions were given a score in a blinded fashion by two veterinary pathologists. Lung were collected and fixed in 10% neutral-buffered formalin and routinely processed for histological examination. Microscopic lung lesions were evaluated in a blinded fashion by two veterinary pathologists as described previously39.
Quantification of PRRSV RNA
To attain a relative quantity of viral RNA, TaqMan fluorescent quantitative RT-PCR (RT-qPCR) was performed on all serum samples as described previously40. The PCR products of conserved regions within ORF7 for type 2 PRRSV strains (180 base pair) was cloned with the PMD-19T (Takara, Korea) and transformed into DH5a competent cells (TIANGEN, China). Plasmid DNA was extracted by using a plasmid purification kit (TIANGEN, China) and quantified by the Thermo Scientific Varioskan Flash multimode reader. Real-time RT-PCR using Taqman probes was performed to generate a standard curve by known amounts of the serially diluted ORF7-based plasmid standards (101–108 copies/μL). Specific primers for qPCR in this study was performed as described40, PRRSV F: 5′-ACAACGGCAAGCAGCAGAA-3′ and PRRSV R: 5′-GAGCGATGATCTTACCCAGCAT-3′ and the PRRSV probe: 5′-FAM-CTGGGYARGATYATCGCCCAGCA-BHQ1-3′. The concentrations in the tested samples were obtained by the Ct values plotted against the known concentration of the ORF7-based plasmid standards.
Serology
Serum samples were analyzed by ELISA using the PRRS Virus Antibody Test Kit 2XR (IDEXX Laboratories Inc., Westbrook, ME, USA). The serology test was performed by the manufacturer’s instructions. Samples are considered positive for antibodies to PRRSV when sample/positive (S/P) > 0.4.
Virus neutralization test in serum
Virus neutralization test was performed in the serum as described previously41. Briefly, a 100-μl aliquot of each diluted sample was mixed with an equal volume of FJZ03 or FJWQ16 containing 200 TCID50. Each mixture was transferred to MARC-145 monolayers in 96-well plates after incubation at 37 °C for 1 h. The presence of virus-infected cells in each well was determined by IFA. The neutralizing antibody (NA) titers of the sera against the different PRRSV were calculated using the Reed-Muench method42. Animals were considered to be protected from viremia when a titer of greater than eight43.
Statistical analysis
The numerical data is expressed as the mean ± S.D. and was analyzed using SPSS 16.0 software. Differences between groups were assessed using ANOVA. Differences were considered statistically significant when p < 0.05.
References
Wensvoort, G. et al. Lelystad virus, the cause of porcine epidemic abortion and respiratory syndrome: a review of mystery swine disease research at Lelystad. Veterinary Microbiology 33, 185–193 (1992).
Benfield, D. A. et al. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR- 2332). The Journal of Veterinary Diagnostic Investigation 4, 127–133 (1992).
Wensvoort, G. Lelystad virus and the porcine epidemic abortion and respiratory syndrome. Veterinary Research 24, 117–124 (1993).
Meulenberg, J. J. PRRSV, the virus. Veterinary Research 31, 11–21 (2000).
Fang, Y. & Snijder, E. J. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Research 154, 61–76 (2010).
Fang, Y. et al. Efficient-2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proceedings of the National Academy of Sciences of the United States of America 109, E2920–E2928 (2012).
Firth, A. E. et al. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. Journal of General Virology 92, 1097–1106 (2011).
Li, Y. et al. Transactivation of programmed ribosomal frameshifting by a viral protein. Proceedings of the National Academy of Sciences of the United States of America 111, E2172–E2181 (2014).
Wu, W. H. et al. The 2b protein as a minor structural component of PRRSV. Virus Research 114, 177–181 (2005).
Tian, K. et al. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2, e526 (2007).
Mengeling, W. L. et al. Comparative safety and efficacy of attenuated single-strain and multistrain vaccines for porcine reproductive and respiratory syndrome. Veterinary Microbiology 93, 5–38 (2003).
Shi, M. et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. Journal of Virology 84, 8700–8711 (2010).
Li, X. et al. Comparison of host immune responses to homologous and heterologous type II porcine reproductive and respiratory syndrome virus (PRRSV) challenge in vaccinated and unvaccinated pigs. BioMed Research International 2014, 416727 (2014).
Meng, X. J. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Veterinary Microbiology 74(4), 309–329 (2000).
Wei, Z. et al. Immunization of pigs with a type 2 modified live PRRSV vaccine prevents the development of a deadly long lasting hyperpyrexia in a challenge study with highly pathogenic PRRSV JX143. Vaccine 31, 2062–2066 (2013).
Lager, K. M. et al. Efficacy of type 2 PRRSV vaccine against Chinese and Vietnamese HP-PRRSV challenge in pigs. Vaccine 32, 6457–6462 (2014).
Labarquea, G. G., Nauwyncka, H. J., van Woenselb, P. A., Visserb, N. & Pensaerta, M. B. Efficacy of an American and a European serotype PRRSV vaccine after challenge with American and European wild-type strains of the virus. Veterinary Research 31, 97 (2000).
van Woensel, P. A., Liefkens, K. & Demaret, S. Effect on viraemia of an American and a European serotype PRRSV vaccine after challenge with European wild-type strains of the virus. Veterinary Record 142, 510–512 (1998).
Roca, M. et al. Effect of challenge with a virulent genotype II strain of porcine reproductive and respiratory syndrome virus on piglets vaccinated with an attenuated genotype I strain vaccine. Veterinary Journal 193, 92–96 (2012).
Han, K., Seo, H. W., Oh, Y., Kang, I., Park, C. & Chae, C. Comparison of the virulence of European and North American genotypes of porcine reproductive and respiratory syndrome virus in experimentally infected pigs. Veterinary Journal 195, 313–318 (2013).
Brockmeier, S. L. et al. Genomic sequence and virulence comparison of four Type 2 porcine reproductive and respiratory syndrome virus strains. Virus Research 169(1), 212–221 (2012).
Murtaugh, M. P. & Genzow, M. Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 29, 8192–8204 (2011).
Kimman, T. G., Cornelissen, L. A., Moormann, R. J., Rebel, J. M. & Stockhofe-Zurwieden, N. Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27, 3704–3718 (2009).
Zhou, L. et al. Efficacy evaluation of three modified-live virus vaccines against a strain of porcine reproductive and respiratory syndrome virus NADC30-like. Veterinary Microbiology 207, 108–116 (2017).
Kvisgaard, L. K., Hjulsager, C. K., Kristensen, C. S., Lauritsen, K. T. & Larsen, L. E. Genetic and antigenic characterization of complete genomes of type 1 porcine reproductive and respiratory syndrome viruses (PRRSV) isolated in Denmark over a period of 10 years. Virus. Research 178, 197–205 (2013).
Osorio, F. A. et al. Passive transfer of virus specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology 302, 9–20 (2002).
Bai, X. et al. Commercial vaccines provide limited protection to NADC30-like PRRSV infection. Vaccine 34, 5540–5545 (2016).
Zhang, Q. et al. Pathogenicity and antigenicity of a novel NADC30-like strain of porcine reproductive and respiratory syndrome virus emerged in China. Veterinary Microbiology 197, 93–101 (2016).
Zhao, K. et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. Journal of Virology 89, 10712–10716 (2015).
Jiang, Y. F. et al. Characterization of three porcine reproductive and respiratory syndrome virus isolates from a single swine farm bearing strong homology to a vaccine strain. Veterinary Microbiology 179, 242–249 (2015).
Liu, J. K. et al. Emergence of a novel highly pathogenic porcine reproductive and respiratory syndrome virus in China. Transboundary and Emerging Diseases 64, 2059–2074 (2017).
Wei, C. H. et al. Molecular characterization of NADC30-like PRRSV isolate FJLY01 from Fujian. Journals Journal of Northwest A&F University 45, 51–60+67 (2017).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729 (2013).
Martin, D. P. et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26, 2462–2463 (2010).
Ramos, N., Mirazo, S., Castro, G. & Arbiza, J. Molecular analysis of Porcine Circovirus Type 2 strains from Uruguay: evidence for natural occurring recombination. Infection Genetics and Evolution 19, 23–31 (2013).
Lole, K. S. et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. Journal of Virology 73, 152–160 (1999).
Kosakovsky Pond, S. L., Posada, D., Gravenor, M. B., Woelk, C. H. & Frost, S. D. Automated phylogenetic detection of recombination using a genetic algorithm. Molecular Biology and Evolution 23, 1891–1901 (2006).
Halbur, P. G. et al. Comparative pathogenicity of nine US porcine reproductive and respiratory syndrome virus (PRRSV) isolates in a five-week-old cesarean-derived, colostrum-deprived pig model. Journal of Veterinary Diagnostic Investigation 8, 11–20 (1996).
Halbur, P. G. et al. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Veterinary Pathology 32, 648–660 (1995).
Lu, W. H. et al. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3) and increased pathogenicity after genomic recombination with vaccine variant. Veterinary Microbiology 175, 332–3340 (2015).
Leng, C. L. et al. Highly pathogenic porcine reproductive and respiratory syndrome virus GP5 B antigenic region is not a neutralizing antigenic region. Veterinary Microbiology 159, 273–281 (2012).
Reed, L. J. & Muench, H. A simple method of estimating fifty percent endpoint. The American Journal of Hygiene 27, 493–497 (1938).
Lopez, O. J. et al. Protection against Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) infection through passive transfer of PRRSV-neutralizing antibodies is dose dependent. Clinical and Vaccine Immunology 14, 269–275 (2007).
Acknowledgements
This work was supported by Leading Project Foundation of Science Department of Fujian Province (2018N0022), Fujian Natural Science Foundation, Fujian Province, China (2016J01168), The Fujian Provincial Department of Education Project (JAT170571), and Industry-University-Research Project of Longyan University (LC2017005 and LC2016010).
Author information
Authors and Affiliations
Contributions
J.L. and X.Y. designed the study. C.W., J.F., Y.L., A.C., and X.Z. conducted the experiments. C.W., A.D., L.M., X.Y. and J.L. analyzed the data. All authors have read and approved the submitted manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wei, C., Dai, A., Fan, J. et al. Efficacy of Type 2 PRRSV vaccine against challenge with the Chinese lineage 1 (NADC30-like) PRRSVs in pigs. Sci Rep 9, 10781 (2019). https://doi.org/10.1038/s41598-019-47239-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47239-9
This article is cited by
-
Glycyrrhiza polysaccharides inhibits PRRSV replication
Virology Journal (2023)
-
Efficacy of a live attenuated highly pathogenic PRRSV vaccine against a NADC30-like strain challenge: implications for ADE of PRRSV
BMC Veterinary Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.