Abstract
Pepper is an important vegetable worldwide and is a model plant for nonclimacteric fleshy fruit ripening. Drastic visual changes and internal biochemical alterations are involved in fruit coloration, flavor, texture, aroma, and palatability to animals during the pepper fruit ripening process. To explore the regulation of bell pepper fruit ripening by noncoding RNAs (ncRNAs), we examined their expression profiles; 43 microRNAs (miRNAs), 125 circular RNAs (circRNAs), 366 long noncoding RNAs (lncRNAs), and 3266 messenger RNAs (mRNAs) were differentially expressed (DE) in mature green and red ripe fruit. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that the targets of the DE ncRNAs and DE mRNAs included several kinds of transcription factors (TFs) (ERF, bHLH, WRKY, MYB, NAC, bZIP, and ARF), enzymes involved in cell wall metabolism (beta-galactosidase, beta-glucosidase, beta-amylase, chitinase, pectate lyase (PL), pectinesterase (PE) and polygalacturonase (PG)), enzymes involved in fruit color accumulation (bifunctional 15-cis-phytoene synthase, 9-cis-epoxycarotenoid dioxygenase, beta-carotene hydroxylase and carotene epsilon-monooxygenase), enzymes associated with fruit flavor and aroma (glutamate-1-semialdehyde 2,1-aminomutase, anthocyanin 5-aromatic acyltransferase, and eugenol synthase 1) and enzymes involved in the production of ethylene (ET) (ACO1/ACO4) as well as other plant hormones such as abscisic acid (ABA), auxin (IAA), and gibberellic acid (GA). Based on accumulation profiles, a network of ncRNAs and mRNAs associated with bell pepper fruit ripening was developed that provides a foundation for further developing a more refined understanding of the molecular biology of fruit ripening.
Similar content being viewed by others
Introduction
Pepper (Capsicum annuum L.) is currently the second most important vegetable worldwide, and the major pepper production areas are located in southern European countries1. Pepper is an example of a nonclimacteric fleshy fruit species, as it exhibits neither respiratory burst nor ethylene (ET) responses during the fruit ripening process2,3. Fruits of pepper are popular for their attractive color, pungency, distinct aroma and ability to be consumed at different ripening stages; moreover, pepper fruits are rich in various vitamins, carotenoids, and flavonoids and have potential health-promoting properties3. Pepper is also a model plant for fruit ripening research because its ripening process involves drastic visual changes and internal biochemical alterations involved in fruit coloration, flavor, texture, aroma, and palatability to animals4,5,6.
To date, thousands of noncoding RNAs (ncRNAs), which play important roles in different physiological and biochemical processes, have been identified in plants7,8,9. NcRNAs can be divided into different varieties, and the most common varieties are small ncRNAs (sRNAs), long ncRNAs (lncRNAs) and circular RNAs (circRNAs)10. Although numerous ncRNAs have been identified in several model plants, the functions of these ncRNAs are largely unknown. In the past few years, substantial progress has been made in deciphering the regulatory functions of sRNAs in the posttranscriptional regulation of gene expression11,12. LncRNAs can exercise their regulatory roles by sequence complementarity or homology with RNAs or DNA in either cis or trans configurations, regulating gene expression at the transcriptional or posttranscriptional level; moreover, lncRNAs can also regulate micro RNA (miRNA) activity as endogenous target mimics (eTMs) of miRNAs13,14. Thousands of circRNAs have recently been characterized, which furthers our understanding of RNA; however, although circRNAs reportedly play important roles in a range of biological processes in humans and animals15,16, knowledge of the functions of circRNAs in fruit plants is limited9,17,18.
Due to the rapid development of next-generation sequencing technology and bioinformatics, numerous ncRNAs have been identified in model plants19,20,21. To date, more than 300 conserved and novel miRNAs have been identified in different pepper varieties, and several miRNAs have been found to be involved in pepper fruit development and quality22,23,24,25. In addition, 6527 lncRNAs were found in hot pepper, and several of these lncRNAs were found during the fruit development process26. To better understand the functions of ncRNAs in bell pepper fruit ripening, deep sequencing and bioinformatic analysis were employed, leading to the identification of 43 differentially expressed (DE) miRNAs, 125 DE circRNAs, 366 DE lncRNAs, and 3266 DE mRNAs during pepper fruit ripening. In addition, the targets of the DE ncRNAs were also analyzed, and several targets were found to be involved in fruit color accumulation (such as beta-carotene hydroxylase 1/2, 15-cis-phytoene synthase, lycopene epsilon cyclase, and phytoene synthase 2), fruit flavor and aroma formation (such as glutamate-1-semialdehyde 2,1-aminomutase, alanine aminotransferase 2, and eugenol synthase 1), fruit texture (such as beta-galactosidase, cellulose synthase, and polygalacturonase (PG)) and ET and other hormone pathways (such as ACO1/4, ERF1, EIN3, ARF, gibberellic acid (GA), and auxin (IAA)). Furthermore, the networks of competing endogenous RNAs (ceRNAs) of miRNAs, circRNAs, lncRNAs and mRNAs were assessed using gene annotation to identify influenced pathways and processes.
Materials and Methods
Sample collection and preparation
Bell pepper fruit (Capsicum annuum L. cv. Suoma) in the mature green (40 d) and red ripe stages (60 d) was harvested after anthesis (d.p.a.) from a greenhouse in “Xiaotangshan” and quickly transported to the laboratory. Then, pericarp samples were collected from the bell pepper fruit (with 3 replicates per stage), frozen in liquid nitrogen and stored at −80 °C for the next experiment.
Methods of RNA extraction and detection
The RNA samples were extracted with TRIzol. RNA quality was checked by assessing the RNA integrity number (RIN) with an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA) and by measuring the RNA concentration with a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) to ensure the use of qualified samples for sequencing. The library preparation for sRNA sequencing and the library preparation for lncRNA and circRNA sequencing were the same as previously reported27.
Sequencing and ncRNA identification
The library preparations were sequenced on an Illumina Hiseq platform, and paired-end reads were generated. The raw data were filtered and all downstream analyses were based on clean data with high quality27. The known miRNAs and novel miRNAs were predicted using miRDeep2 software by comparison with known miRNAs from miRBase28. The CIRI (CircRNA Identifier) tools was used to identify circRNAs, scan SAM files twice and collect enough information to identify and characterize circRNAs, similar to previous reports29. The transcriptome was assembled using StringTie based on the reads mapped to the reference genome. The assembled transcripts were annotated using the gffcompare program, the different types of lncRNAs, including long intergenic ncRNAs (lincRNAs), intronic lncRNAs, antisense lncRNAs, and sense lncRNAs, were selected using cuffcompare previously reported27.
Quantification of expression levels and DEG analysis of ncRNAs
MiRNA expression levels were estimated for each sample. sRNAs were mapped back onto the precursor sequence, and the read count for each miRNA was obtained from the mapping results. StringTie (1.3.1) was used to calculate the fragments per kilobase of transcript per million mapped reads (FPKMs) of both lncRNAs and coding genes in each sample, and the FPKM values of genes were computed by summing the FPKM values of transcripts in each gene group. The FPKM value was calculated based on the length of the fragments and the number of reads mapped to these fragments. The expression of circRNAs was determined by the number of junction reads identified by CIRI. Differential expression analysis of two conditions/groups was performed as previously reported27.
GO and KEGG analyses and Network analysis of ceRNAs of ncRNA
Gene function was annotated as previously reported and Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R package-based Wallenius noncentral hypergeometric distribution, the KOBAS software was used to test the statistical enrichment of differentially expressed genes in KEGG pathways27,30. The ceRNAs network analysis of the ncRNAs were the same as previously reported27,31.
Results
Identification of DE and novel miRNAs, circRNAs, lncRNAs and mRNAs
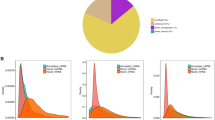
A total of 352 miRNAs were found in our libraries, of which 130 were known miRNAs and 222 were novel miRNAs. Most of the novel miRNAs were between 21 nt and 24 nt in length. We also analyzed the nucleotide bias for all the known and novel miRNAs; intriguingly, the most common nucleic acid bases were A and U, while the least common ones were G and C (Fig. 1A,B). The lengths of numerous circRNAs were between 200 nt and 400 nt, and a large number of the circRNAs were over 3000 nt in length. The majority of the circRNAs were from exons and intergenic regions. A total of 4795 novel circRNAs were found in our study, and most of them were on chromosome 8 (Fig. 1C,D). In addition, 11999 lncRNAs were found in bell pepper fruit, of which 194 were known lncRNAs and 11805 were novel lncRNAs. Interestingly, we found 139 lncRNAs that were the precursors of known and novel miRNAs, such as miR167, miR168, miR482, and miR5303 (Appendix 1). Among these lncRNAs, most were lincRNAs (10136, 84.5%), followed by antisense lncRNAs (964, 8%), sense lncRNAs (749, 6.2%) and then intronic lncRNAs (150, 1.3%) (Fig. 1E,F).
We compared the expression profiles of miRNAs, circRNAs, lncRNAs and mRNAs between mature green and red ripe bell pepper fruit and found that 43 miRNAs, 125 circRNAs, 366 lncRNAs, and 3266 mRNAs were DE (Fig. 2, Appendix 1). Among these DE RNAs, 32 miRNAs, 82 circRNAs, 186 lncRNAs, and 1188 mRNAs were upregulated, and 11 miRNAs, 43 circRNAs, 180 lncRNAs, and 2078 mRNAs were downregulated in the red ripe stage compared with the green ripe stage. The DE ncRNAs are listed in Appendix 1. The DE circRNAs were mostly found on chromosomes 1, 7, 8 and 9, and the number distributed on chromosome 7 was the largest. The DE lncRNAs were mainly distributed on chromosomes 1, 3, 6 and 10, while the DE mRNAs were chiefly distributed on chromosomes 1, 2, 3, 6 and 8; the number on chromosome 3 was the largest (Fig. 3).
DE ncRNAs target parsing involved in pepper fruit ripening
The targets of the DE miRNAs were predicted, and a total of 909 targets were found (Appendix 2). The most enriched GO terms related to biological processes included single-organism cellular process, response to oxygen-containing compound, response to abiotic stimulus, response to organic substance, and single-organism transport. However, the most relevant GO terms associated with molecular functions were protein binding, sequence-specific DNA binding transcript, identical protein binding, protein serine/threonine kinase activity, phosphotransferase activity, and divalent inorganic cation transmembrane. KEGG pathway analysis suggested that mRNAs were remarkably enriched in the pathways involved in plant-pathogen interaction, RNA degradation, protein processing in the endoplasmic reticulum, plant hormone signal transduction, and RNA transport (Appendix 2). Intriguingly, several targets of the DE miRNAs were found to be involved in the fruit ripening process, such as ACO1, RTR2, EIN3, ERF1, ARF6/8, beta-glucosidase, pectinesterase (PE), lycopene epsilon cyclase, and squamosa promoter-binding-like protein (Fig. 4, Appendix 2).
To explore the potential function of DE circRNAs, GO and KEGG pathway analyses of circRNAs were also performed. The most relevant GO terms associated with biological processes were anatomical structure formation involved in morphogenesis, cytoskeleton-dependent cytokinesis, defense response to fungus, response to jasmonic acid, external encapsulating structure organization, and the L-glutamate biosynthetic process. The most relevant GO terms associated with molecular functions were phosphatidylinositol phosphate 5-phosphatase activity and 3 iron, 4 sulfur cluster binding. However, the KEGG pathways included only spliceosome and biosynthesis of amino acids (Appendix 4). In addition, several targets of the circRNAs were also found to be involved in the fruit ripening process, such as beta-galactosidase and pectate lyase (PL) (Fig. 4, Appendix 2).
To explore the potential functions of the DE ncRNAs, we performed GO and KEGG analyses. Both the cis and trans targets of the DE lncRNAs were analyzed, and 3266 targets were found. The relevant GO terms associated with biological processes and molecular functions included many important transcription factors (TFs) and key enzymes involved in pepper fruit ripening, such as MYB, NAC, WRKY, SBP, bHLH, bZIP, ACO1/4, MADS-box protein, EIN3, ERF, 15-cis-phytoene synthase, 9-cis-epoxycarotenoid dioxygenase NCED1, carotene epsilon-monooxygenase, beta-carotene hydroxylase, beta-galactosidase, PG, beta-amylase, beta-glucosidase, catalase chitinase, peroxidase, LOX, phenylalanine ammonia-lyase, abscisic acid (ABA), IAA, ARF, and GA (Fig. 4, Appendix 2). The KEGG analysis results showed that the most frequently predicted pathways were involved in carbon metabolism, ribosomes, plant hormone signal transduction, purine metabolism, biosynthesis of amino acids, and protein processing in the endoplasmic reticulum, indicating the specific regulatory functions of lncRNAs in fruit ripening in bell pepper (Fig. 4, Appendix 2).
Comparative analysis of lncRNAs/mRNAs and functional annotation of DE mRNAs
There are many differences between lncRNAs and mRNAs, including their length, exon numbers, and open reading frame. The isoforms and interactive expression of lncRNAs and mRNAs were also examined, and the distributions of these ncRNAs on the different chromosomes are described in Appendix 4. For lncRNAs with a length <3000 nt, the number declined with increasing length (Fig. 5A), and most lncRNAs had a length <1000 nt or ≥3000 nt. The length of mRNAs showed the same trends and exhibited two peaks at approximately 400 nt and ≥3000 nt (Fig. 5B). The corresponding number of lncRNAs distributed in the range of exons were smaller than the mRNAs and mainly below 10; however, the corresponding numberof mRNAs distributed in the range of exons were ranged from 1 to 30 (Fig. 5C,D). The length of corresponding open reading frames of lncRNAs was mainly between 50 nt and 150 nt (Fig. 5E), while the length of corresponding open reading frames of mRNAs was mainly between 100 nt and 900 nt (Fig. 5F).
For DE mRNAs, the most relevant GO terms associated with molecular functions were protein binding, sequence-specific DNA binding transcription factor activity, protein kinase activity, serine-type endopeptidase activity, protein serine/threonine kinase activity, transmembrane receptor protein kinase activity, flavonoid 3′-monooxygenase activity, and chlorophyll binding. KEGG pathway analysis indicated that the most frequently predicted pathways were involved in plant hormone signal transduction, carbon metabolism, ribosome, biosynthesis of amino acids, phenylpropanoid biosynthesis, starch and sucrose metabolism (Fig. 6). We found that many DE mRNAs were involved in pepper fruit ripening-related processes and could be divided into five different groups: the first group included enzymes associated with fruit texture such as PE, PL, beta-glucosidase, pectin acetylesterase, PG, and pectin methyltransferase; the second group included enzymes involved in fruit pigment accumulation such as 15-cis-phytoene desaturase, 9-cis-epoxycarotenoid dioxygenase NCED1, beta-carotene hydroxylase 1, phytoene synthase (PSY), lycopene epsilon cyclase, and carotenoid cleavage dioxygenase, and some of these enzymes were also found to be upregulated during chili pepper development32; the third group included components associated with fruit flavor or aroma such as 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, bifunctional phosphatase IMPL2, delta(7)-sterol-C5(6)-desaturase, and glutamate decarboxylase; and the fourth group included components associated with plant hormone-related processes such as ACO, MADS-box, EIN3, ERF, ABA, IAA, GA, and ARF. In addition, a fifth group included several TFs, such as MYB, bHLH, WRKY, and NAC (Appendix 4).
Network analysis of the ncRNAs involved in fruit ripening
As reported previously, circRNAs can act as “miRNA sponges”, and lncRNAs can be the precursors or targets of miRNAs. To explore coordinated regulatory functions, analysis of circRNA-mRNA-miRNA, miRNA-mRNA-lncRNA and circRNA-miRNA-mRNA-lncRNA networks was performed. In the circRNA-mRNA-miRNA network analysis, 9 circRNAs, 35 miRNAs and 84 mRNAs were found to be involved in the network; however, in the miRNA-mRNA-lncRNA network analysis, 34 miRNAs, 4 lncRNAs and 84 mRNAs were found in the network. Furthermore, in the circRNA-miRNA-mRNA-lncRNA network analysis, 9 circRNAs, 35 miRNAs, 124 lncRNAs and 256 mRNAs were involved in network regulation. Intriguingly, several biological processes were involved in the various fruit ripening processes, such as ethylene biosynthesis and signal transduction (ACO1, ACO4 and ERF039), fruit color formation (bifunctional 15-cis-phytoene synthase and lycopene epsilon cyclase), fruit texture (PG, pectin acetylesterase, and beta-galactosidase), and fruit flavor and aroma (alpha-farnesene synthase and anthocyanin 5-aromatic acyltransferase), indicating that their specific networks were involved in the coordinated regulation of the fruit ripening processes (Fig. 7, Appendix 5).
LncRNAs, circRNAs and mRNAs can interact with miRNAs through microRNA response elements (MREs) within ceRNA networks9,33,34. We developed candidate ceRNA relationships based on miRNA-target relationships and obtained 271 pairs of ceRNA relationships, including 148 pots. Then, we extracted three comprehensive ceRNA networks from the ceRNA relationship pairs, including 10 mRNAs and 20 circRNAs. More importantly, we found two important pathways, namely, the ERF TF, which plays important roles in ET signal transduction, and eugenol synthase 1, which may participate in pepper flavor formation, in the ceRNA regulatory network (Appendix 6).
Discussion
Pepper is a typical nonclimacteric fruit and has emerged as a model for research on fruit ripening35. Fruit ripening is a highly coordinated biological process and involves numerous morphological and physiological processes, including pigment accumulation, nutritional changes, texture variation and metabolism of flavor and aromatic substances36,37. With the advancement of sequencing technology, increasing numbers of ncRNAs are being found in model fruit plants9,38. Several miRNAs and lncRNAs have been identified in different kinds of pepper varieties, but their functions are largely unknown. Here, in our research, we identified 222, 4795 and 11085 novel miRNAs, circRNAs and lncRNAs, respectively, which enrich the ncRNA database. Furthermore, 43 miRNAs, 125 circRNAs, 366 lncRNAs, and 3266 mRNAs were DE during fruit ripening, and several ncRNAs’ targets were found to be associated with fruit color, texture, flavor, aroma, hormones and several TFs, indicating that they have specific regulatory roles in the ripening of bell pepper fruit.
Color change is visible and is used as an indicator of fruit ripening; red peppers accumulate higher levels of total carotenoids during the ripening process39,40. The expression levels of some carotenoid biosynthetic genes encoding key carotenoid biosynthetic enzymes, such as PSY, phytoene desaturase (PDS), and capsanthin-capsorubin synthase (CCS), are higher in red pepper fruits than in pepper fruits of other colors; however, some of these genes are not expressed in peppers with lower levels of total carotenoids39,41,42. We found that one miRNA’s (unconservative_Chr05_14982) target is lycopene epsilon cyclase (Capana09g000177), which is a key enzyme in carotenoid synthesis and showed opposite expression patterns43. We also found that two lncRNAs (MSTRG.81557.3 and MSTRG.81557.4) target 15-cis-phytoene synthase, which is a key enzyme in carotenoid synthesis and showed the same expression44. Several lncRNAs (MSTRG.139116.1, MSTRG.139115.1, MSTRG.139111.1, and MSTRG.139117.1) target carotene epsilon-monooxygenase, which is a key enzyme in the lutein pathway45. In addition, we found that one miRNA (unconservative_Chr02_5911) targets glutamate-1-semialdehyde 2,1-aminomutase (Capana03g001893), which is an enzyme involved in tomato flavor synthesis46; one circRNA (Chr06:21090976|21092632) targets alanine aminotransferase 2 (Capana06g001160); and several lncRNAs target anthocyanin 5-aromatic acyltransferase (Capana10g000432), which contributes to pepper aroma formation47. One ncRNA can target several different genes, and different miRNAs, lncRNAs and circRNAs can also have the same target; however, the method through which they regulate their target genes synergistically to control pigment accumulation during pepper ripening is still largely unknown and needs further research.
Fruit ripening and softening involve numerous modifications to cell-wall polysaccharides, and during the bell pepper fruit ripening process, a dramatic change in the texture of the walls also occurs. Several enzymes, such as PG, pectin methyl esterase (PME), PL, and β-galactosidase, are involved in fruit ripening and softening48. We found that two known miRNAs (miR157d-3p and miR396h) target beta-glucosidase, which is an important enzyme in the fruit ripening process49 and that a novel miRNA (unconservative_Chr05_14982) targets cellulose synthase, which takes part in fruit cell wall formation50. Furthermore, several circRNAs and lncRNAs target beta-galactosidase, which plays important roles in the bell pepper ripening process48. In addition, several lncRNAs and circRNAs target PG, PL and PE, which are key enzymes in the fruit softening process9.
Fruit ripening is also affected by various hormones, such as ET, IAA, and ABA35,36,51. For example, IAA is known to retard tomato ripening by regulating ET and ABA34. We found that several DE ncRNA targets are important enzymes or TFs in the fruit ripening process, which is consistent with the results of previous studies23,26. Several lncRNAs and miRNAs target ACO1 and ACO4, which are important enzymes in ET biosynthesis, and several miRNAs (miR164a-5p, miR167b-3p, miR168a-3p, miR396b, miR482a and unconservative_Chr06_17217) target ETR2, EIN3 and ERFs, which are important regulators in the ET signal transduction pathway36,52,53. Previous studies reported that ARFs were targets of the can-miR160 and can-miR160 families in pepper, similar to our study23. In addition, DE ncRNA targets were also found to be involved in the GA, ABA and IAA pathways. Further, numerous TFs, such as bHLH, NAC, MYB, bZIP, and WRKY, have also been reported to participate in the regulation of fruit ripening in tomato and pepper fruit34,49. Our study revealed that several lncRNAs and miRNA targets were the bHLH, TCP, bZIP, MYB and WRKY TFs and squamosa promoter-binding-like protein, which is an important part of the fruit ripening regulatory network.
CircRNAs were found to act as “miRNA sponges” and were found to be enriched with functional miRNA binding sites54,55. Several lncRNAs can form internal structures for certain biological outputs, such as sRNA production, as can primary miRNAs (pri-miRNAs); lncRNAs can also compete with miRNAs for binding sites on shared target mRNAs; furthermore, lncRNAs can bind to miRNAs to “communicate” with other RNA targets56,57,58,59. CircRNAs, lncRNAs and mRNAs can act as ceRNAs in gene regulation9. To date, there has been no report on the ncRNA networks and ceRNAs in bell pepper fruit. Here, we analyzed circRNA-mRNA-miRNA, miRNA-mRNA-lncRNA and circRNA-miRNA-mRNA-lncRNA networks and found that several vital enzymes and TFs were involved in the coordinated regulatory networks, such as ACO1/4, ARF8, PSY1, WRKY, ERF039, lycopene epsilon cyclase, beta-galactosidase, and PG, pectin acetylesterase 9, all of which are important regulators involved in fruit ripening3,6,35,60. In addition, the ceRNA networks were parsed, revealing that two important regulators, ethylene-responsive transcription factor RAP2-7 and eugenol synthase 1, may participate in the pepper fruit ripening process35,61. These findings provide a theoretical basis for deciphering novel mechanisms of fruit ripening and functionally characterizing ceRNA networks in future studies.
References
Palma, J. M., Corpas, F. J., Del Río, L. A. & López-Huertas, E. Pepper fruit as a model to study the metabolism of antioxidants, ROS and RNS. Free Radic Biol Med 75, 1–39, https://doi.org/10.1016/j.freeradbiomed.2014.10.786 (2014).
Liu, M., Pirrello, J., Chervin, C., Roustan, J. P. & Bouzayen, M. Ethylene control of fruit ripening: revisiting the complex network of transcriptional regulation. Plant Physiology 169, 2380–2390, https://doi.org/10.1104/pp.15.01361 (2015).
Corpas, F. J. et al. Nitro-oxidative metabolism during fruit ripening. J Exp Bot 69, 3449–3463, https://doi.org/10.1093/jxb/erx453 (2018).
Paran, I. & van der Knaap, E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot 58, 3841–3852, https://doi.org/10.1093/jxb/erx453 (2007).
Giovannoni, J., Nguyen, C., Ampofo, B., Zhong, S. & Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annual Review of Plant Biology 68, 61–84, https://doi.org/10.1146/annurev-arplant-042916-040906 (2017).
Klie, S. et al. Conserved changes in the dynamics of metabolic processes during fruit development and ripening across species. Plant Physiol 164, 55–68, https://doi.org/10.1104/pp.113.226142 (2014).
Li, Z. F., Zhang, Y. C. & Chen, Y. Q. miRNAs and lncRNAs in reproductive development. Plant Sci 238, 46–52, https://doi.org/10.1016/j.plantsci.2015.05.017 (2015).
Chekanova, J. A. Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol 27, 207–216, https://doi.org/10.1016/j.pbi.2015.08.003 (2015).
Wang, Y. et al. Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol Plant 161, 311–321, https://doi.org/10.1111/ppl.12600 (2017).
Dou, C. et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci Rep 6, 21499, https://doi.org/10.1038/srep21499 (2016).
Liu, H., Yu, H., Tang, G. & Huang, T. Small but powerful: function of microRNAs in plant development. Plant Cell Rep 37, 515–528, https://doi.org/10.1007/s00299-017-2246-5 (2018).
Deng, P., Muhammad, S., Cao, M. & Wu, L. Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol J 16, 965–975, https://doi.org/10.1111/pbi.12882 (2018).
Kornienko, A. E., Guenzl, P. M., Barlow, D. P. & Pauler, F. M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol 11, 59, https://doi.org/10.1186/1741-7007-11-59 (2013).
Dykes, I. M. & Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 15, 177–186, https://doi.org/10.1016/j.gpb.2016.12.005 (2017).
Greene, J. et al. Circular RNAs: Biogenesis, Function and Role in Human Diseases. Front Mol Biosci 4, 38, https://doi.org/10.3389/fmolb.2017.00038 (2017).
Wilusz, J. E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA e1478, https://doi.org/10.1002/wrna.1478 (2018).
Zuo, J., Wang, Q., Zhu, B., Luo, Y. & Gao, L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun 479, 132–138, https://doi.org/10.1016/j.bbrc.2016.07.032 (2016).
Tan, J., Zhou, Z., Niu, Y., Sun, X. & Deng, Z. Identification and Functional Characterization of Tomato CircRNAs Derived from Genes Involved in Fruit Pigment Accumulation. Sci Rep 7, 8594, https://doi.org/10.1038/s41598-017-08806-0 (2017).
Ye, C. Y., Chen, L., Liu, C., Zhu, Q. H. & Fan, L. Widespread noncoding circular RNAs in plants. New Phytol 208, 88–95, https://doi.org/10.1111/nph.13585 (2015).
Chen, G. et al. Genome-Wide Identification of Circular RNAs in Arabidopsis thaliana. Front Plant Sci 8, 1678, https://doi.org/10.3389/fpls.2017.01678 (2017).
Liu, D., Mewalal, R., Hu, R., Tuskan, G. A. & Yang, X. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic Res 4, 17031, https://doi.org/10.1038/hortres.2017.31 (2017).
Joy, N. & Soniya, E. V. Identification of a miRNA candidate reflects the possible significance of transcribed microsatellites in the hairpin precursors of black pepper. Funct Integr Genomics 12, 387–395, https://doi.org/10.1007/s10142-012-0267-2 (2012).
Hwang, D. G. et al. The hot pepper (Capsicum annuum) microRNA transcriptome reveals novel and conserved targets: a foundation for understanding MicroRNA functional roles in hot pepper. PLoS One 8, e64238, https://doi.org/10.1371/journal.pone.0064238 (2013).
Asha, S., Sreekumar, S. & Soniya, E. V. Unraveling the complexity of microRNA-mediated gene regulation in black pepper (Piper nigrum L.) using high-throughput small RNA profiling. Plant Cell Rep 35, 53–63, https://doi.org/10.1007/s00299-015-1866-x (2016).
Liu, Z. et al. Identification and characterization of novel microRNAs for fruit development and quality in hot pepper (Capsicum annuum L.). Gene 608, 66–72, https://doi.org/10.1016/j.gene.2017.01.020 (2017).
Ou, L. et al. Noncoding and coding transcriptome analysis reveals the regulation roles of long noncoding RNAs in fruit development of hot pepper (Capsicum annuum L.). Plant Growth Regul 83, 141–156, https://doi.org/10.1007/s10725-017-0290-3 (2017).
Zuo, J. et al. Analysis of the Coding and Non-Coding RNA Transcriptomes in Response to Bell Pepper Chilling. Int J Mol Sci. 19, 2001, https://doi.org/10.3390/ijms19072001 (2018).
Friedlander, M. R., Mackowiak, S. D., Li, N., Chen, W. & Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40, 37–52, https://doi.org/10.1093/nar/gkr688 (2012).
Gao, Y., Wang, J. & Zhao, F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome biology 16, 4, https://doi.org/10.1186/s13059-014-0571-3 (2015).
Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinform 21, 3787–3793, https://doi.org/10.1093/bioinformatics/bti430 (2005).
Li, J. H., Liu, S., Zhou, H., Qu, L. H. & Yang, J. H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42, 92–97, https://doi.org/10.1093/nar/gkt1248 (2014).
Martínez-López, L. A., Ochoa-Alejo, N. & Martínez, O. Dynamics of the chili pepper transcriptome during fruit development. BMC Genomics 15, 143, https://doi.org/10.1186/1471-2164-15-143 (2014).
Salzman, J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet 32, 309–316, https://doi.org/10.1016/j.tig.2016.03.002 (2016).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388, https://doi.org/10.1038/nature11993 (2013).
Cheng, Y. et al. Putative WRKYs associated with regulation of fruit ripening revealed by detailed expression analysis of the WRKY gene family in pepper. Sci Rep 6, 39000, https://doi.org/10.1038/srep39000 (2016).
Lee, S., Chung, E. J., Joung, Y. H. & Choi, D. Non-climacteric fruit ripening in pepper: increased transcription of EIL-like genes normally regulated by ethylene. Funct Integr Genomics 10, 135–146, https://doi.org/10.1007/s10142-009-0136-9 (2010).
Ma, N. et al. Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol 14, 351, https://doi.org/10.1186/s12870-014-0351-y (2014).
Hou, Y. et al. Comparative Analysis of Fruit Ripening-Related miRNAs and Their Targets in Blueberry Using Small RNA and Degradome Sequencing. Int J Mol Sci 18, E2767, https://doi.org/10.3390/ijms18122767 (2017).
Li, Z., Wang, S., Gui, X. L., Chang, X. B. & Gong, Z. H. A further analysis of the relationship between yellow ripe-fruit color and the capsanthin-capsorubin synthase gene in pepper (Capsicum sp.) indicated a new mutant variant in C. annuum and a tandem repeat structure in promoter region. PLoS One 8, e61996, https://doi.org/10.1371/journal.pone.0061996 (2013).
Li, Y. et al. Comparative Transcriptome Analysis of Genes Involved in Anthocyanin Biosynthesis in Red and Green Walnut (Juglans regia L.). Molecules 23, E25, https://doi.org/10.3390/molecules23010025 (2017).
Ha, S. H., Kim, J. B., Park, J. S., Lee, S. W. & Cho, K. J. A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin sythase gene is not a prerequisite for the formation of a yellow pepper. J Exp Bot 58, 3135–3144, https://doi.org/10.1093/jxb/erm132 (2007).
Lado, J., Zacarías, L. & Rodrigo, M. J. Regulation of Carotenoid Biosynthesis During Fruit Development. Subcell Biochem 79, 161–198, https://doi.org/10.1007/978-3-319-39126-7_6 (2016).
Ahrazem, O., Rubio-Moraga, A., López, R. C. & Gómez-Gómez, L. The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron’s apocarotenoid precursors. J Exp Bot 61, 105–119, https://doi.org/10.1093/jxb/erp283 (2010).
Nisar, N., Li, L., Lu, S., Khin, N. C. & Pogson, B. Carotenoid metabolism in plants. Mol Plant 8, 68–82, https://doi.org/10.1016/j.molp.2014.12.007 (2015).
Yuan, H., Zhang, J., Nageswaran, D. & Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic Res 2, 15036, https://doi.org/10.1038/hortres.2015.36 (2015).
Araújo, W. L. et al. Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24, 2328–2351, https://doi.org/10.1105/tpc.112.099002 (2012).
D’Auria, J. C. Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9, 331–340, https://doi.org/10.1016/j.pbi.2006.03.016 (2006).
Ogasawara, S., Abe, K. & Nakajima, T. Pepper beta-galactosidase 1 (PBG1) plays a significant role in fruit ripening in bell pepper (Capsicum annuum). Biosci Biotechnol Biochem 71, 309–322, https://doi.org/10.1271/bbb.60179 (2007).
Biles, C. L., Bruton, B. D., Russo, V. & Wall, M. M. Characterisation of β-galactosidase isozymes of ripening peppers. J Sci Food Agric 75, 237–243, https://doi.org/10.1002/(sici)1097-0010(199710)75:23.3.CO;2-t (1997).
Le Gall, H. et al. Cell Wall Metabolism in Response to Abiotic Stress. Plants (Basel) 4, 112–166, https://doi.org/10.3390/plants4010112 (2015).
Navarro, J. M., Flores, P., Garrido, C. & Martínez, V. Changes in the contents of antioxidant compounds in pepper fruits at different ripening stages, as affected by salinity. Food Chem 96, 66–73, https://doi.org/10.1016/j.foodchem (2006).
Dong, T. et al. A non-climacteric fruit gene CaMADS-RIN regulates fruit ripening and ethylene biosynthesis in climacteric fruit. PLoS One 9, e95559, https://doi.org/10.1371/journal.pone.0095559 (2014).
Cherian, S., Figueroa, C. R. & Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: a cross-dissection of climacteric versus non-climacteric fruit. J Exp Bot 65, 4705–4722, https://doi.org/10.1093/jxb/eru280 (2014).
Li, L. J. et al. Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin Ther Targets 21, 639–648, https://doi.org/10.1080/14728222.2017.1319938 (2017).
Zhang, H. et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget 8, 61687–61697, https://doi.org/10.18632/oncotarget.18671 (2017).
Ebert, M. S. & Sharp, P. A. Emerging roles for natural microRNA sponges. Curr Biol 20, 858–861, https://doi.org/10.1016/j.cub.2010.08.052 (2010).
Yoon, J. H., Abdelmohsen, K. & Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol 34, 9–14, https://doi.org/10.1016/j.semcdb.2014.05.015 (2014).
Ma, X., Shao, C., Jin, Y., Wang, H. & Meng, Y. Long non-coding RNAs: a novel endogenous source for the generation of Dicer-like 1-dependent small RNAs in Arabidopsis thaliana. RNA Biol 11, 373–390, https://doi.org/10.4161/rna.28725 (2014).
Ballantyne, M. D., McDonald, R. A. & Baker, A. H. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther 99, 494–501, https://doi.org/10.1002/cpt.355 (2016).
Zuo, J. et al. SRNAome and degradome sequencing analysis reveals specific regulation of sRNA in response to chilling injury in tomato fruit. Physiol Plant 160, 142–154, https://doi.org/10.1111/ppl.12509 (2017).
Aragüez, I. & Valpuesta, V. Metabolic engineering of aroma components in fruits. Biotechnol J 8, 1144–1158, https://doi.org/10.1002/biot.201300113 (2013).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31772022), the Natural Science Foundation of Beijing (6182016), the National Key Research and Development Program of China (2016YFD0400901), the China Agriculture Research System Project (CARS-23), Special innovation ability construction fund of Beijing Academy of Agricultural and Forestry Sciences (20180404 and 20180705), the Young Investigator Fund of Beijing Academy of Agricultural and Forestry Sciences (201709), Beijing Academy of Agriculture and Forestry fruit and vegetable preservation and processing innovation team (201602), and the International Cooperation Fund Project of Beijing Academy of Agricultural and Forestry Sciences. The authors acknowledge James Giovannoni (USDA/Cornell) and Lance Courtney (Cornell) for useful discussions during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
J.H.Z. conceived and designed the experiments and wrote the manuscript; Y.X.W., B.Z.Z., Y.B.L., Q.W. and L.P.G. participated in the related experiments and analyzed the data.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zuo, J., Wang, Y., Zhu, B. et al. Network analysis of noncoding RNAs in pepper provides insights into fruit ripening control. Sci Rep 9, 8734 (2019). https://doi.org/10.1038/s41598-019-45427-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45427-1
This article is cited by
-
Characterization of leucine aminopeptidase (LAP) activity in sweet pepper fruits during ripening and its inhibition by nitration and reducing events
Plant Cell Reports (2024)
-
Identification and functional prediction of CircRNAs of developing seeds in high oleic acid sunflower (Helianthus annuus L.)
Acta Physiologiae Plantarum (2023)
-
Identification and functional deciphering suggested the regulatory roles of long intergenic ncRNAs (lincRNAs) in increasing grafting pepper resistance to Phytophthora capsici
BMC Genomics (2021)
-
Genome-wide identification and characterization of long non-coding RNAs involved in flag leaf senescence of rice
Plant Molecular Biology (2021)
-
Noncoding RNAs: functional regulatory factors in tomato fruit ripening
Theoretical and Applied Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.