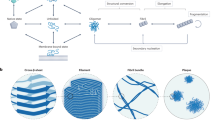
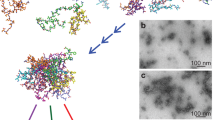
Abstract
The study of the aggregation of soluble proteins into highly ordered, insoluble amyloid fibrils is fundamental for the understanding of neurodegenerative disorders. Here, we present a method for the observation of single amyloid fibrils that allows the investigation of fibril growth, secondary nucleation or fibril breakup that is typically hidden in the average ensemble. Our approach of thermophoretic trapping and rotational diffusion measurements is demonstrated for single Aβ40, Aβ42 and pyroglutamyl-modified amyloid-β variant (pGlu3-Aβ3–40) amyloid fibrils.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
A dataset for demonstration can be downloaded at https://doi.org/10.5281/zenodo.1414296. The full dataset that supports the findings of this study is available from the corresponding author upon reasonable request.
Code availability
The source code and the files for the software used in this study are contained in the Supplementary Software, and a maintained version can be downloaded at https://github.com/molecular-nanophotonics/thermophoretic-trap-for-protein-aggregation-studies.
References
Knowles, T. P. J., Vendruscolo, M. & Dobson, C. M. Nat. Rev. Mol. Cell Biol. 15, 384–396 (2014).
Iadanza, M. G., Jackson, M. P., Hewitt, E. W., Ranson, N. A. & Radford, S. E. Nat. Rev. Mol. Cell Biol. 19, 755–773 (2018).
Linse, S. Biophys. Rev. 9, 329–338 (2017).
Blackley, H. et al. J. Mol. Biol. 298, 833–840 (2000).
Pinotsi, D. et al. Nano Lett. 14, 339–345 (2014).
Ban, T., Yamaguchi, K. & Goto, Y. Acc. Chem. Res. 39, 663–670 (2006).
Tycko, R. Annu. Rev. Phys. Chem. 62, 279–299 (2011).
Knowles, T. P. J. et al. Proc. Natl Acad. Sci. USA 108, 14746–14751 (2011).
Wolff, M. et al. Sci. Rep. 6, 22829 (2016).
Braun, M. & Cichos, F. ACS Nano 7, 11200–11208 (2013).
Braun, M., Bregulla, A. P., Günther, K., Mertig, M. & Cichos, F. Nano Lett. 15, 5499–5505 (2015).
Piazza, R. & Parola, A. J. Phys. Condens. Matter 20, 153102 (2008).
Würger, A. Rep. Prog. Phys. 73, 126601 (2010).
Bregulla, A. P., Würger, A., Günther, K., Mertig, M. & Cichos, F. Phys. Rev. Lett. 116, 188303 (2016).
Xue, C., Lin, T. Y., Chang, D. & Guo, Z. R. Soc. Open Sci. 4, 160696 (2017).
Reichl, M., Herzog, M., Götz, A. & Braun, D. Phys. Rev. Lett. 112, 198101 (2014).
Scheidt, H. A., Adler, J., Krueger, M. & Huster, D. Sci. Rep. 6, 1–7 (2016).
Cohen, A. E. & Moerner, W. E. Proc. Natl Acad. Sci. USA 103, 4362–4365 (2006).
Watkins, L. P. & Yang, H. J. Phys. Chem. B 109, 617–628 (2005).
Yang, H. J. Chem. Phys. 129, 074701 (2008).
Fränzl, M. et al. Protocol Exchange https://doi.org/10.1038/protex.2019.031 (2019).
Luk, V. N., Mo, G. C. & Wheeler, A. R. Langmuir 24, 6382–6389 (2008).
Acknowledgements
F.C. and D.H. acknowledge financial support by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) through the Collaborative Research Center TRR 102 ‘Polymers under multiple constraints: restricted and controlled molecular order and mobility’ (funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), project number 189853844–SFB TRR 102) and project CI 33/14-1. J.P. and M.M. acknowledge financial support by the BMBF (contract no. 03WKCL01G) and the DFG via the Cluster of Excellence ‘cfaed’ (contract no. EXC 1056/1). We thank A. Kramer for helping to revise the manuscript.
Author information
Authors and Affiliations
Contributions
M.F., T.T. and F.C. designed the experiments. M.F. and T.T. performed the experiments. M.F., T.T. and F.C. analyzed the data. J.A. prepared the amyloid samples and carried out fibrillation kinetics measurements. J.P., M.F., T.T., F.C. and M.M. developed the trap preparation procedure. D.H. and F.C. provided the experimental equipment. M.F., T.T., J.A., D.H. and F.C. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information: Allison Doerr was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–12 and Supplementary Protocol
Supplementary Software
The repository contains a collection of Python scripts and Jupyter Notebooks for the tracking and data analysis of a single amyloid fibril in a thermophoretic trap.
Supplementary Video 1
Aβ40 fibril of length L = 1.5 μm confined inside the thermophoretic trap with an incident laser heating power of Pheat = 1 mW. A sample image from the movie is displayed in Fig. 1b of the main text. The dashed circle indicates the inner diameter (10 μm) of the trap. The exposure time and inverse framerate correspond to 30 ms.
Supplementary Video 2
Tracked position and orientation of the Aβ40 fibril shown in Supplementary Video 1 overplayed with the original video. The dashed circle indicates the inner diameter (10 μm) of the trap. The exposure time and inverse framerate correspond to 30 ms.
Supplementary Video 3
Fragmentation of a trapped Aβ40 fibril of length L = 1.1 μm corresponding to the image sequence Fig. 2f. The dashed circle indicates the inner diameter (10 μm) of the trap. The exposure time and inverse framerate correspond to 30 ms.
Rights and permissions
About this article
Cite this article
Fränzl, M., Thalheim, T., Adler, J. et al. Thermophoretic trap for single amyloid fibril and protein aggregation studies. Nat Methods 16, 611–614 (2019). https://doi.org/10.1038/s41592-019-0451-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41592-019-0451-6
This article is cited by
-
CRISPR-powered optothermal nanotweezers: Diverse bio-nanoparticle manipulation and single nucleotide identification
Light: Science & Applications (2023)
-
Hypothermal opto-thermophoretic tweezers
Nature Communications (2023)
-
Low-temperature optothermal nanotweezers
Nano Research (2023)
-
Light-driven single-cell rotational adhesion frequency assay
eLight (2022)
-
Hydrodynamic manipulation of nano-objects by optically induced thermo-osmotic flows
Nature Communications (2022)