Abstract
Neutrophils are formidable defenders. Their vast numbers, constant production, high cytotoxicity and capacity to produce extracellular traps, underlie their ability to efficiently protect in a microorganism-rich world. However, neutrophils are much more than immune sentinels, as evidenced by the expanding repertoire of functions discovered in the context of tissue homeostasis, regeneration or chronic pathologies. In this Perspective, we discuss general functional features of the neutrophil compartment that may be relevant in most, if not all, physiological scenarios in which they participate, including specialization in naïve tissues, transcriptional noise in the bloodstream as a potential strategy for diversification and functional bias in inflammatory sites. We intentionally present the reader with more questions than answers and propose models and approaches that we hope will shed new light onto the biology of these fascinating cells and spark new directions of research.
Similar content being viewed by others
Main
For over a century, neutrophils (microphages) have been recognized as the most rapid and aggressive responders among immune cells to an insult, be it sterile or infectious. However, as an immune lineage, neutrophils are rather unconventional. They are built as a large army of non-proliferative cells that constantly patrol the body in search of microorganisms, follow strict circadian patterns, live for only a handful of hours and must be readily eliminated to avoid collateral damage to the host (recently reviewed in refs. 1,2,3). Paradoxically, the strategies that they have evolved to protect the host and to additionally contribute to other aspects of organismal physiology remain largely enigmatic. Here, we embrace discussion of what these strategies may be, and examine principles of their functional organization and education throughout the body.
Architecture of the neutrophil compartment
Under physiological conditions, transit times of newly formed neutrophils from the bone marrow to the circulation are estimated to last 2.6 d in mice and 6 d in humans4,5,6. Once released to the blood, the estimated half-life of mature neutrophils in circulation is less than a day5. Their transit to the bloodstream is accompanied by substantial changes at the chromatin and transcriptional levels, which appear to occur unevenly during maturation, and enable the acquisition of immune functionalities7,8,9. These properties are further expanded as the cells move to tissues both during inflammatory responses and in the steady state10,11,12. We posit that global understanding of these migratory and functional trajectories is needed to capture the biological architecture of neutrophils across tissues and physiological scenarios. Experimental-based modeling of this biological architecture is lacking, and this void in knowledge generates confusion when trying to understand unique aspects of neutrophil biology, including short lifespan and plasticity.
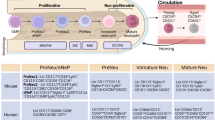
Although neutrophils are conspicuously poor in RNA content, application of single-cell transcriptomics to neutrophils has provided insights into their transcriptional heterogeneity during maturation at an unprecedented resolution, which should enable the generation of models explaining their phenotypic and functional organization (Fig. 1). For example, using mRNA as a molecular readout allowed the description of a transcriptional continuum termed ‘neutrotime’ for neutrophils from the bone marrow, blood and spleen13. This model proposes that a single developmental spectrum dominates neutrophil heterogeneity under steady-state conditions (Fig. 1a). Building on this view, studies have extended the characterization of neutrophil heterogeneity to other body compartments, including the lung, intestine and the skin10. Even in naïve conditions, neutrophils adopt distinct phenotypic and functional properties (Fig. 1b). These functions enable, for example, support of B cell maturation in the spleen or vascular regeneration in the lung10,14,15. It is logical to predict that this enormous diversity emanates from local, rapid reprogramming of neutrophils under tissue-derived signals (Fig. 1b).
a, During development in the bone marrow, myeloid progenitors undergo sequential maturation to generate mature neutrophils that are released to the circulation. This process can be modeled as a single developmental continuum under steady-state conditions. b, Extending analyses to peripheral tissues reveals additional degrees of heterogeneity as neutrophils adapt to the different environments, and acquire distinct phenotypic and functional properties. These changes likely rely on tissue-derived signals and localization in specific anatomic niches (hubs), such as perivascular areas in the lungs (environmental model), or alternatively, these are already imprinted in circulating clones of neutrophils with predefined programs (pre-committed model). c, During emergency granulopoiesis, increased neutrophil production driven by a stress (for example, infection or cancer) may use existing developmental trajectories (1) or create new ones (2) to increase population size, without altering the overall architecture of the neutrophil compartment. Alternatively, the stress could alter the normal architecture of granulopoiesis to generate entirely new populations (3).
An alternative model is that clonally distinct granulocytic progenitors in the bone marrow or mature neutrophils differentially primed in the circulation are endowed with distinct migratory capabilities or differentiation properties that ultimately govern the acquisition of tissue-associated phenotypes once the cells nest in the appropriate tissue microenvironments (Fig. 1b). It remains to be tested, however, whether neutrophils exist in different genetic states (transcriptional or epigenetic) in the circulation in ways that enable divergent responses to infectious, inflammatory or environmental cues. In support of this model, single-cell studies have identified three transcriptionally distinct populations of mature neutrophil in the blood of healthy mice that are predicted to arise from distinct maturing bone marrow neutrophils16. Similarly, studies in neutrophils from human blood have characterized discrete phenotypic subsets, including a population of CD177+ neutrophils that is overrepresented in the blood of patients with anti-neutrophil cytoplasmic antibody-dependent vasculitis17,18,19 or a population expressing OLFM4 (ofactomedin-4)20. Likewise, in functional terms, it is relevant to note that only a fraction of neutrophils are able to release neutrophil extracellular traps (NETs) upon stimulation21. Although this phenotypic diversity does not provide definitive proof of lineage independence between these subsets, it does raise the possibility that the circulating neutrophil compartment is organized clonally (Fig. 1b), a possibility that needs to be formally explored with the help of novel barcoding tools coupled with single-cell RNA sequencing22,23, or through more challenging clonal studies in vitro and in vivo24. Regardless of which mechanism(s) dominate(s) in vivo, the relevance of understanding how neutrophils attain diversity is clear if we are to develop therapeutics that target the right population, be it progenitors in the marrow or fully differentiated neutrophils in peripheral tissues.
Beyond homeostasis, many common pathological perturbations such as cancer or infections result in enhanced granulopoiesis, which causes neutrophilia by mobilizing immature neutrophils into blood and inflamed sites (Fig. 1c). This phenomenon, collectively referred to as emergency granulopoiesis, is believed to be orchestrated by diffusible, systemic signals such as pathogen-associated molecular patterns and cytokines like granulocyte colony-stimulating factor that generally target the bone marrow25,26,27,28. Whether the molecular and evolutionary mechanisms driving enhanced granulopoiesis differ depending on the infectious or noninfectious nature of the initiating stimuli, and if it entails expansion of putative clones with different properties remains undefined. Stress-driven granulopoiesis may simply expand the existing pool of neutrophils or generate new sources of heterogeneity when compared with healthy states (Fig. 1c). For example, studies in mice have identified a type of circulating neutrophil enriched in expression of interferon-stimulated genes that expands during bacterial infection16. It is worth noting that, although this interferogenic subset is primed for augmented functionality during infections, it already exists in the blood of healthy mice, hinting that the genetic architecture of the granulopoietic compartment is conserved but functionally adaptable16 (Fig. 1c). In settings such as cancer, phenotypic and functional changes in neutrophils can be remarkable, and tumor-induced granulopoiesis is known to strongly modulate neutrophil properties (reviewed in ref. 26). In severe cases of coronavirus disease 2019 (COVID-19), a similarly disruptive scenario, analyses of circulating leukocytes have showed the presence of dysfunctional neutrophils, including CD274 (PD-L1)-expressing neutrophils29, loss of the interferon-driven program30 and the appearance of immature cells with plasmablast-like features31, altogether suggesting anomalous neutrophil maturation and release under extreme stress. Along this line, studies of human blood neutrophils have revealed context-dependent transcriptional programs that associate with changes in functional output32. This series of observations raise the possibility that stress alters the normal architecture of granulopoiesis to generate entirely new populations, such as Siglec F+ neutrophils, which appear during acute inflammatory stress and in cancer33,34 (Fig. 1c). In summary, we emphasize the importance of understanding the principles driving the adaptation of neutrophils to different environments, stresses or disease from the perspective of the global architecture of the neutrophil compartment. This holistic approach, we believe, will provide a more robust platform to understand the true physiological purpose of these cells.
The circulating neutrophil
Although neutrophils are first responders to insults, they are not mere eradicators of pathogens. They combine an extraordinary capacity to patrol the organism with a built-in arsenal of enzymes and antimicrobial peptides and a kamikaze-type behavior that contains pathogenic spread, with a remarkable plasticity that can be molded by the different environments that they visit35, as discussed in more detail below. In the circulation, multiple flavors of neutrophils have been reported that associate with diseases or inflammatory scenarios, but this has largely relied on discrete parameters such as cell density or expression of surface proteins (as reviewed in ref. 36). In particular cases, cell-surface markers identify circulating populations with distinct functions, as is the case of VEGFR1+ neutrophils, which are efficiently recruited to hypoxic tissues to promote angiogenesis37. This illustrates the notion that, already in the circulation, neutrophils are a mix of cells in different phenotypic and functional states32,36. However, the actual degree of heterogeneity of neutrophils in blood, its purpose and underlying mechanisms remain unclear.
A critical limitation in addressing these gaps in knowledge is the lack of genetic and molecular tracers to unequivocally call for heterogeneity of neutrophils beyond surface markers. For example, while it is relatively simple to catalog T lymphocyte subsets based on their origin, transcriptional profile and genetic drivers38, we lack this degree of molecular resolution when studying neutrophils. Indeed, while single-cell sequencing technologies have unveiled a broad range of transcriptional states for neutrophils, these states have not been formally ‘promoted’ to subsets in the absence of reliable proteins and genes to trace the different cells39. In addition, these states exist as a (transcriptional) continuum rather than as independent clusters, with few or no markers to differentiate them16,32,40 (Fig. 2a), which makes classification even more challenging.
a, Profiling of blood neutrophils by single-cell transcriptomics fails to find diversity using standard computational approaches, but functionally distinct populations could exist inside this transcriptional cloud (red, green, blue and yellow). b, Genetic and phenotypic heterogeneity of blood neutrophils could be imprinted through a series of orthogonal mechanisms that act during granulopoiesis and in the circulation. c, Different sources of heterogeneity, including somatic mutations, circadian aging, migration across vessels or circulating metabolites influence neutrophils at different stages of maturation to generate diverse transcriptional and functional outputs (cells of different colors). BM, bone marrow; UMAP, uniform manifold approximation and projection.
Despite these limitations, studies analyzing the transcriptomic and proteomic profiles of blood neutrophils have used standard computational approaches to score subsets, maturation trajectories and gene signatures. While not always comparable, they demonstrated consistency in some aspects, for example, in the identification of type I interferon-inducible genes in a subset of neutrophils, particularly in response to stimulation16,32,40. Unfortunately, we still lack globally accepted methods for high-dimensional clustering, which, combined with the absence of correlation between transcriptomic and proteomic datasets, hinders the isolation of such subsets for functional studies. Hence, single-cell studies remain largely descriptive and fail to shed definitive light into the functional organization of circulating neutrophils. We propose that building reference frameworks that include neutrophils from many different contexts (including healthy or diseased tissues) will be critical to understand what the different states mean41.
While speculative at this point, the existence of circulating neutrophils with distinct phenotypic and transcriptional profiles suggests that they undergo a type of education that may be important to prime responses against different types of pathogens, as recently hinted in mice by ex vivo studies using Candida24. An extreme case of diversity among circulating neutrophils is seen during systemic stress, for example, cancer or bone marrow transplantation, in which immature forms manifest differential production of reactive oxidative species and neutrophil extracellular traps, but higher production of cytokines32. However, because mobilization from medullary reservoirs cannot account for the diversity seen in the blood of healthy individuals, we propose that heterogeneity is attained by mechanisms acting at different stages of the neutrophil life cycle, which converge to generate a pool of functionally distinct neutrophils in the circulation, each of which is primed for a different function (Fig. 2b). In this regard, it is conceivable that granulocytic precursors undergo divergent developmental trajectories, somatic mutations, or simply undergo desynchronized release into the bloodstream. Because circulating neutrophils rapidly engage into changes in phenotype, transcription and function driven by circadian rhythm and microbiota (referred to as ‘aging’), this desynchronized release would drive the accumulation of functionally heterogeneous (out-of-phase) neutrophils in blood21,42,43 (Fig. 2c).
Clonal diversification emerging from medullary progenitors is an intriguing, yet unproven mechanism of diversification as phenotypic ‘clones’ of neutrophils have not been detected in blood using traditional methods. Lineage-determining transcription factors, such as PU.1, C/EBPα and C/EBPβ7, drive sequential expression of lineage-specific genes while at the same time silencing lineage-foreign genes, and can additionally modify the surrounding chromatin (a property of pioneer transcription factors)44. Collaboration of these lineage-determining transcription factors with a heterogeneous set of transcription factors with tissue-restricted45 and/or stimulus-dependent46 activity is likely to generate diverging epigenetic landscapes and push the different granulocytic progenitors along distinct transcriptional programs, as shown for macrophages45. Such a model aligns well with findings in the context of trained immunity, in which signals during infection or cancer generate persistent changes in neutrophil function47,48. It also agrees with data showing that granulopoiesis organizes locally and clonally in the bone marrow of both mice and humans49,50,51. Related to this spatial diversification, analyses of human bulk and single-cell transcriptomic datasets of neutrophils show that the extent and type of transcriptional responses to different challenges (growth factors, transplantation of hematopoietic stem cells, cancer and viral infection) differ both with maturation stage and with tissue of residence32. Finally, migration trajectories can be another source of diversification. Neutrophils migrating to inflamed tissues can migrate back into the circulation, a process referred to as reverse migration that causes phenotypic alterations (for example, in the expression of the adhesion receptor ICAM-1, the cytokine receptor VEGFR1 and the chemokine receptor CXCR1)52, increased reactive oxygen species production and resistance to apoptosis52,53. Thus, maturation, tissue residence or emigration and clonal organization of neutrophils are potential sources of phenotypic and functional diversity in the circulation, a possibility that challenges current views and should influence how we study the heterogeneity and physiology of these cells.
Transcriptional noise
Human and mouse neutrophils undergo gradual changes of the genetic, epigenetic and transcriptional material as they mature in the marrow and transition to blood8,25,54,55,56. A conclusion of recent analyses is that lineage specification occurs early at the granulocyte–monocyte progenitor stage and involves gradual loss of proliferative and metabolic activity (including mitochondrial content) and granule protein synthesis8,25,54, while at the same time allowing the gain of sensing and migratory receptors (such as chemokine and Toll-like receptors; Fig. 3a). These dynamics are proposed to protect the bone marrow by uncoupling the synthesis of cytotoxic elements from that of the receptors that can promote their release8,25. In addition, activation of multiple effector and transcriptional programs occurs in the transition from the marrow to the circulation, and is concomitant with a remarkable drop in transcript content8,10. This implies that, as neutrophils transition to blood, many genes become transcriptionally active but contribute less to the global transcriptional output8. At the molecular level, this suggests the opening of multiple chromatin regions with variable gene expression across the neutrophil pool present in blood, without commitment to specific immune programs. These observations are consistent with studies of human neutrophils that showed remarkable variability in the gene expression and DNA methylation across individuals57. It also aligns with the correlation between the abundance of variable genes in a population and the susceptibility to infection and autoimmune disease58. At the cellular level, neutrophil clusters extend across large areas in the transcriptional space when compared with other circulating leukocytes29, suggesting high variance in the genes that are expressed across individual neutrophils. Overall, these findings suggest that circulating neutrophils have adopted a strategy of biological noise akin to that identified in unicellular organisms or in uncommitted pluripotent cells that exist in oligo-stable states59 (Box 1). In some ways, these properties suggest that the granulocytic compartment, when considered as a whole, features adaptable, stem-like properties, despite the limited plasticity of each of its cellular constituents.
a, Trajectory of neutrophil maturation with different stages shown over a UMAP plot, with the abundance of transcripts and transcriptional noise at each stage shown at bottom. b, Transcriptional noise of a group refers to the compound Euclidean distance of each cell to the transcriptional centroid of the corresponding group/cluster. c, Transcriptional rates and noise across immature neutrophils in the marrow, and mature neutrophils in blood and peripheral tissues, shown in a linear bubble plot. Note that transcriptional noise increases as rates decrease, as cells mature to reach the blood. These properties appear to be maintained in tissues.
This unconventional transcriptional strategy of circulating neutrophils raises new questions such as where this biological noise originates, and does it persist when circulating cells move into tissues or under pathological scenarios. Ad hoc analyses of existing single-cell datasets16 for this Perspective confirmed the loss of transcript abundance throughout maturation, which starts in the bone marrow and is lowest in the blood. These changes are concomitant with increases in transcriptional noise (Fig. 3b), defined here as the average Euclidean distance of individual cells in the transcriptional space to a representative cell with average expression of all genes (centroid) present in a given population60 (for example, granulocyte–monocyte progenitors or mature blood neutrophils; Fig. 3a). Importantly, both transcript abundance and transcriptional noise are predicted to persist as circulating neutrophils move into tissues (Fig. 3c), suggesting that this may be a general property of neutrophils in extramedullary tissues. The consistent finding of high transcriptional noise of neutrophils in blood and tissues further suggests that the stochastic nature of this biological property may be evolutionarily selected to enhance the antimicrobial diversification of the neutrophil pool, for example, by generating primed states against different types of pathogens24.
Transcriptional noise appears to be independent of granulopoiesis because it emerges when cells have left the medullary compartment (Fig. 3c). Indeed, egress of neutrophils from the marrow into blood coincides with major chromatin remodeling, whereby multiple regions become accessible or inaccessible in a short period of time8,10,12. What drives this ‘noisy’ pattern in the periphery is not determined yet. One possibility is that rapid epigenetic remodeling causes stochastic decondensation of the neutrophil chromatin. In turn, stochastic changes in chromatin density could lead to intermittent access of transcription factors to regulatory regions thereby promoting ‘bursts’ of gene expression (Box 1), a known driver of transcriptional noise61. A fair question is, however, how accurate and physiologically relevant this noise is, given the low transcriptional rates of neutrophils. It is tempting to speculate that it is precisely because of these remarkably low transcriptional rates that small changes in gene expression can have a strong impact in their transcriptional programs and functional imprinting. Thus, transcriptional noise may represent an unappreciated mechanism that controls neutrophil dynamics and functional diversification in blood. Nevertheless, there remains much work to do to understand the molecular footprint, biological trigger and physiological significance of this property of neutrophils. To this end, it will be critical to solve technical and analytical limitations associated with the low transcript content in single-cell technologies.
Functional states of inflammatory neutrophils
A recurring challenge in the field is how to classify neutrophils in ways that inform useful (patho)physiology. This is not trivial because neutrophils appear to transit through so-called ‘states’ rather than durable subsets that can be traced by defined markers or by driver transcription factors1,39. In the absence of such markers, single-cell transcriptomics have taken center stage despite the poor transcriptional nature of mature neutrophils, and have been extremely useful to describe even small differences across tissues, disease or even within the same tissue10,16,41,62,63. However, transcriptional profiling is only useful to describe durable properties, and we lack reliable criteria to annotate functions associated with most transcriptional profiles. Hence, describing the functional states of circulating neutrophils becomes extremely challenging, yet this could transform our understanding of how they respond to infections or to sterile inflammation.
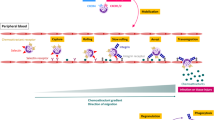
There is no single or simple answer to the question of whether there are reliable ways to describe the functional states of circulating neutrophils, and clearly new methodology and concepts will need to be developed to address this challenge. Orthogonal approaches may be particularly helpful in better defining seemingly identical cells (as defined by transcription). Multiparametric analyses using mass cytometers or high-dimensional spectral or conventional flow cytometers combined with computational algorithms54,64 may capture rapid changes in surface or intracellular proteins as neutrophils sense diverse stimuli or transit across tissues10. An alternative is to use live imaging, the oldest single-cell technology available65. In principle, this should allow measuring the response of individual cells in their native environment, as they ‘read’ chemical and physical cues that cannot be mimicked ex vivo and are lost when tissues are dissociated to extract cells. Because cells in different functional states should respond differently in these environments, by capturing multiple parameters of their physical response under a microscope, it may be possible to build ‘behavioral’ landscapes that categorize cells based on function, rather than on their molecular fingerprint. Based on this tenet, we were able to categorize neutrophils recruited to inflamed vessels into at least three functional states, one of which strongly associated with pathological inflammation66. Critical for this approach is to score morphological and kinetic variables that can be readily obtained using three-dimensional imaging over time, but we propose that combining these with other readable features (molecular or physical) will enable more accurate depictions of diversity and function of neutrophils or any other cell type. Moving forward, it will be important to link these behavioral states with the genetic traits of neutrophils, as this will allow to define the molecular principles of such states, for example, to understand why different neutrophils select alternative routes to migrate from capillaries to tissues67, to perform reverse transendothelial migration52 or to engage in ‘swarming’ behaviors in response to injury or vaccination68.
The functional states of inflammatory neutrophils identified by behavioral profiling raises new questions, for example, if the functional diversity originates from cell-extrinsic or cell-intrinsic cues (Fig. 4). The finding that approximately 20% of neutrophils can transit between behaviors66 suggests that external cues are relevant, but probably do not account for the type of response. Determining whether neutrophils primed for specific behaviors exist in the circulation, and the molecular basis of such priming, is an exciting future challenge.
a,b, Inflammatory neutrophils display at least three behavioral states (B1–B3), as identified by live imaging inside inflamed vessels. This functional diversification may be the result of different environmental signals (flat cells of different colors) acting on a unique neutrophil cell state at the inflamed site (a), or alternatively, be the result of already biased neutrophil populations exposed to similar inflammatory signals inside the vessels (b). These preexisting states are predicted to have distinct protein composition and/or genetic backgrounds (genetic modifications or epigenetic marks). In all cases, individual neutrophils manifest certain capacity to transition among the different behavioral states.
Adaptation to tissues
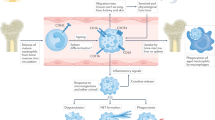
Although neutrophils are best understood in blood and in humans, as they are the most abundant circulating leukocyte type, they can also be found as a marginated pool in contact with the inner side of vessel walls in the naïve spleen, liver, bone marrow and lung, and in the parenchymal space of the intestine and skin, among many other tissues11,69. Specifically, lung neutrophils interact with endothelial cells, with which they are in close contact and circulate across the tightly matted intersecting pulmonary capillaries10,11,70. Likewise, most of the neutrophils identified in the healthy liver following blood perfusion remain intravascularly, whereas spleen and bone marrow neutrophils are mainly located in the red pulp and perivascular spaces, respectively10,11,71. In the intestine, neutrophils localize around isolated lymphoid follicles11 and in the skin they can be found, in low numbers, in the dermis72. Neutrophil margination, sequestration and infiltration to specific locations take place through specialized mechanisms. For example, hyaluronan from hepatic sinusoids and veins mediates hepatic sequestration of neutrophils through CD44, migration to the lymph nodes relies on the chemokine receptor CCR7 (ref. 73) and recruitment or mobilization neutrophils in the lungs is mediated by CXCR4 (refs. 10,74). These tissue-specific mechanisms of recruitment suggest that phenotypic differences within the circulating neutrophil pool (such as differential expression of chemokine receptors or integrins) may favor their recruitment to, and localization within the different tissues75, and partly explain their subsequent specification in those tissues10.
Upon injury and microbial infection, neutrophil recruitment to tissues is accompanied by rapid removal to allow return to homeostasis76. It is intriguing, however, that clearance sites and dynamics in tissues under homeostatic conditions remain virtually unknown and that apoptotic neutrophils are rarely found in intact tissues77. Estimations of neutrophil half-lives have predominantly been restricted to the bone marrow and blood, in part because they were believed to be cleared from the circulation by macrophages in the spleen and liver76,78,79,80. This notion was challenged by the discovery that neutrophils populate most healthy organs11. Use of an inducible, neutrophil-specific reporter mouse model (Ly6GCreERT mice) to track synchronous waves of neutrophils released from the marrow allowed estimation of neutrophil lifetimes in tissues to show equal or even extended lifetimes compared with those in blood10, a finding that suggested that neutrophils persist long enough to integrate environmental cues and to acquire functional and phenotypic diversity within tissues10. These local environments, which we refer to as niches here, may control the heterogeneity, function and identity of tissue neutrophils, much like those instructing diversity in tissue-resident macrophages81. Understanding the nature of such niches across tissues, aided for example by imaging reporter models or spatial transcriptomics, will be instrumental in establishing their cellular identity and understanding their relevance. The finding that tissue-associated neutrophils acquire specialized functions during homeostasis10 expands and complements the roles for other myeloid subsets (that is, resident macrophages) in organ homeostasis82 in ways that remain to be defined. In other words, it is unclear to what extent the tissue-specific contribution of the different myeloid lineages is redundant and how this varies across different physiological scenarios.
In the particular case of the lung, CXCL12-producing vessels mediate the retention of neutrophils in specific perivascular areas where they are reprogramed to support vascular growth10. Angiogenic features of neutrophils were reported in the context of ischemia and hypoxia, liver injury and irradiation37,83,84, but not in naïve tissues. Whether the lung functions as a unique reservoir of angiogenic neutrophils that can be mobilized on demand to distant tissues needs further exploration. In line with the idea that neutrophils can be deployed from reservoirs outside the marrow, it was reported that neutrophils recruited to the liver following sterile injury can reenter the vasculature and serially traffic through the lung and into the bone marrow for clearance84, whereas during skin infection they can migrate to the draining lymph nodes through lymphatic vessels where they are phagocytosed by dendritic cells85. In another relevant example, neutrophils that infiltrate the heart following myocardial infarction migrate to the bone marrow to deliver interleukin-1β and promote granulopoiesis86, whereas in the steady state they enter this organ to suppress mesenchymal activity and promote circadian release of hematopoietic precursors87. Finally, it is intriguing that classical niche signals, such as the chemokine CXCL12, not only act as attractants, but also induce chromatin remodeling to enable nuclear compaction and facilitate navigation of neutrophils in complex three-dimensional environments88. These and other examples of migration in vivo52 suggest that neutrophils engage into unconventional traffic patterns to distant tissues to accomplish specific tasks. We propose that the neutrophil compartment can be envisioned as a ‘migratory tissue’ that infiltrates virtually every organ to be educated, execute functions or deliver messages from one organ to the next.
Co-option of neutrophils by disease
Neutrophil heterogeneity is considered a disease-modifying factor and studies in murine models of cancer have been particularly revealing of the enormous diversity and association of certain transcriptional states with disease outcome. Proposed or demonstrated functions range from clearly pro-tumoral (for example, promoting tumor angiogenesis, tumor cell dissemination and metastatic seeding) to antitumoral and antimetastatic26,89,90. Illustrative of this remarkable diversity, 12 different transcriptional states have been reported in the context of human liver cancer62. Heterogeneity has also been described in several pathological conditions including stroke, myocardial infarction, autoimmune disease or infections such as COVID-19 (refs. 29,91,92,93). A conclusion that can be extracted from this recent body of work is that neutrophils adapt to the altered physiology around them in the same way that they adapt to (non-pathological) tissue microenvironments, but whether this reflects the co-option of homeostatic programs or instead involves entirely new mechanisms needs further exploration. Studies in macrophages, however, have suggested that pathological states of the tissue, such as cancer, reprogram these cells by mimicking the physiology of fetal growth (reviewed in ref. 94). It is therefore conceivable that similar maladaptive processes are active during neutrophil reprogramming by disease.
A related question is to what extent the diversification of neutrophils seen in pathology is stochastic (that is, unpredictable) or follows deterministic, targetable programs. Strategies to address this question have been challenging for two main reasons. First, the short lifespan and postmitotic nature of neutrophils precludes expansion, analysis and rigorous fate mapping of these cells. Second, the sensitivity of the cells to manipulation, as well as their low transcriptional rate precludes clear isolation and transcriptional profiling in most scenarios, especially when using single-cell transcriptomics62,95. In the context of polymicrobial sepsis or lung inflammation, engineered nanoparticles were used to specifically target tissue-toxic neutrophils and to improve host survival96, but these strategies were directed against broad functional properties rather than specific transcriptional programs. The recent discovery of a set of key transcription factors that control the functional adaptation of neutrophils during sterile inflammation revealed a deterministic path that could be exploited to blunt inflammation by specifically intercepting the involved factors, such as JunB12. Thus, the identification of transcriptional hubs that control the full spectrum of diversity will be fundamental to modulate the function of neutrophil subsets, for example, to promote antitumoral states in individuals with cancer, or pro-angiogenic states to improve healing in older or diabetic individuals. Clearly, a better understanding of how the neutrophil compartment organizes and adapts to the surrounding environment has a massive potential for therapeutics, by directing the generation of diagnostic or prognostic tools.
The challenges ahead
Despite advances in the past few years, we still know far too little about the mechanisms that underlie the remarkable antimicrobial efficacy, adaptability and contribution of neutrophils to disease. Looking forward, we narrow down the main challenges of the field in three areas. The first is to better define the global architecture of the neutrophil compartment in mammals, an effort that will provide a much-needed reference for the myriad of studies reporting an ever-expanding list of phenotypes and transcriptional states, particularly in the context of cancer41. The second challenge is to understand which specific environments (niches) within a tissue enable the functional reprograming of neutrophils from patrolling behaviors toward more refined, tissue-tailored tasks. We believe that defining the anatomical context where such reprogramming takes place will be fundamental to delineate how pathologies co-opt neutrophils to their benefit. The third, broader challenge entails addressing the question of why mammals invest such enormous resources in producing vast quantities of short-lived cells, most of which will never encounter a microorganism in their lifetime. Indeed, other myeloid lineages (such as macrophages) have opted for lesser numbers, longer lifespans and elective proliferation, yet are able to effectively execute immune defense and tissue homeostasis. It is conceivable that the massive production of short-lived neutrophils enables the appearance of populations that are simultaneously useful in the never-ending fight against pathogens but potentially harmful to the host and therefore they must be eliminated rapidly, but this remains speculative. Identifying why neutrophils evolved such unique strategies may unlock the key to understanding why they are such formidable defenders, and to bridle them for clinical use.
References
Burn, G. L., Foti, A., Marsman, G., Patel, D. F. & Zychlinsky, A. The neutrophil. Immunity 54, 1377–1391 (2021).
Ley, K. et al. Neutrophils: new insights and open questions. Sci. Immunol. 3, eaat4579 (2018).
Aroca-Crevillén, A., Adrover, J. M. & Hidalgo, A. Circadian features of neutrophil biology. Front. Immunol. 11, 576 (2020).
Dancey, J. T., Deubelbeiss, K. A., Harker, L. A. & Finch, C. A. Neutrophil kinetics in man. J. Clin. Invest. 58, 705–715 (1976).
Lahoz-Beneytez, J. et al. Human neutrophil kinetics: modeling of stable isotope labeling data supports short blood neutrophil half-lives. Blood 127, 3431–3438 (2016).
Pillay, J. et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood 116, 625–627 (2010).
Ai, Z. & Udalova, I. A. Transcriptional regulation of neutrophil differentiation and function during inflammation. J. Leukoc. Biol. 107, 419–430 (2020).
Grassi, L. et al. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 24, 2784–2794 (2018).
Theilgaard-Mönch, K. et al. The transcriptional program of terminal granulocytic differentiation. Blood 105, 1785–1796 (2005).
Ballesteros, I. et al. Co-option of neutrophil fates by tissue environments. Cell 183, 1282–1297 (2020).
Casanova‑Acebes, M. et al. Neutrophils instruct homeostatic and pathological states in naive tissues. J. Exp. Med. 215, 2778–2795 (2018).
Khoyratty, T. E. et al. Distinct transcription factor networks control neutrophil-driven inflammation. Nat. Immunol. 22, 1093–1106 (2021).
Grieshaber-Bouyer, R. et al. The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat. Commun. 12, 2856 (2021).
Fine, N. et al. Distinct oral neutrophil subsets define health and periodontal disease states. J. Dent. Res. 95, 931–938 (2016).
Puga, I. et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat. Immunol. 13, 170–180 (2012).
Xie, X. et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 21, 1119–1133 (2020).
Hu, N. et al. Coexpression of CD177 and membrane proteinase 3 on neutrophils in antineutrophil cytoplasmic autoantibody-associated systemic vasculitis: anti-proteinase-3-mediated neutrophil activation is independent of the role of CD177-expressing neutrophils. Arthritis Rheum. 60, 1548–1557 (2009).
Silvestre-Roig, C., Hidalgo, A. & Soehnlein, O. Neutrophil heterogeneity: implications for homeostasis and pathogenesis. Blood 127, 2173–2181 (2016).
Verheugt, F. W. A., Borne, A. E. G. K., Noord-Bokhorsl, J. C., Elven, E. H. & Engelfriet, C. P. Serological, immunochemical and immuoncytological properties of granulocyte antibodies. Vox Sang. 35, 294–303 (1978).
Clemmensen, S. N. et al. Olfactomedin 4 defines a subset of human neutrophils. J. Leukoc. Biol. 91, 495–500 (2012).
Adrover, J. M. et al. Programmed ‘disarming’ of the neutrophil proteome reduces the magnitude of inflammation. Nat. Immunol. 21, 135–144 (2020).
Bowling, S. et al. An engineered CRISPR–Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cells 181, 1410–1422 (2020).
Weinreb, C., Rodriguez-Fraticelli, A., Camargo, F. D. & Klein, A. M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, eaaw3381 (2020).
Scherer, A. K. et al. Predestined neutrophil heterogeneity in homeostasis varies in transcriptional and phenotypic response to Candida. Preprint at bioRxiv https://doi.org/10.1101/2022.11.01.514676 (2022).
Evrard, M. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking and effector functions. Immunity 48, 364–379 (2018).
Hedrick, C. C. & Malanchi, I. Neutrophils in cancer: heterogeneous and multifaceted.Nat. Rev. Immunol. 22, 173–187 (2022).
Manz, M. G. & Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014).
van Grinsven, E. et al. Immature neutrophils released in acute inflammation exhibit efficient migration despite incomplete segmentation of the nucleus. J. Immunol. 202, 207–217 (2019).
Schulte-Schrepping, J. et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440 (2020).
Combes, A. J. et al. Global absence and targeting of protective immune states in severe COVID-19. Nature 591, 124–130 (2021).
Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020).
Montaldo, E. et al. Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat. Immunol. https://doi.org/10.1038/s41590-022-01311-1 (2022).
Engblom, C. et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science 358, eaal5081 (2017).
Ryu, S. et al. Siglec-F–expressing neutrophils are essential for creating a profibrotic microenvironment in renal fibrosis. J. Clin. Invest. 132, e156876 (2022).
Christoffersson, G. & Phillipson, M. The neutrophil: one cell on many missions or many cells with different agendas? Cell Tissue Res. 371, 415–423 (2018).
Silvestre-Roig, C., Fridlender, Z. G., Glogauer, M. & Scapini, P. Neutrophil diversity in health and disease. Trends Immunol. 40, 565–583 (2019).
Christoffersson, G. et al. VEGF-A recruits a proangiogenic MMP-9-delivering neutrophil subset that induces angiogenesis in transplanted hypoxic tissue. Blood 120, 4653–4662 (2012).
Zhu, X. & Zhu, J. CD4 T helper cell subsets and related human immunological disorders. Int. J. Mol. Sci. 21, 8011 (2020).
Deniset, J. F. & Kubes, P. Neutrophil heterogeneity: bona fide subsets or polarization states? J. Leukoc. Biol. https://doi.org/10.1002/JLB.3RI0917-361R (2018).
Wigerblad, G. et al. Single-cell analysis reveals the range of transcriptional states of circulating human neutrophils. J. Immunol. 209, 772–782 (2022).
Ballesteros, I., Cerezo‐Wallis, D. & Hidalgo, A. Understanding NSCLC, one cell at a time. Cancer Cell https://doi.org/10.1038/nrc3060 (2022).
Adrover, J. M. et al. A neutrophil timer coordinates immune defense and vascular protection. Immunity 50, 390–402 (2019).
Zhang, D. et al. Neutrophil ageing is regulated by the microbiome. Nature 525, 528–532 (2015).
Iwafuchi-Doi, M. & Zaret, K. S. Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Kalafati, L. et al. Innate immune training of granulopoiesis promotes anti-tumor activity. Cell 183, 771–785 (2020).
Moorlag, S. J. C. F. M. et al. BCG vaccination induces long-term functional reprogramming of human neutrophils. Cell Rep. 33, 108387 (2020).
Wu, Q. & Lucas, D. Resilient anatomy and local microplasticity of naïve and stress hematopoiesis. Preprint at bioRxiv https://doi.org/10.1101/2022.05.23.492315 (2020).
Zhang, J. et al. In situ mapping identifies distinct vascular niches for myelopoiesis. Nature 590, 457–462 (2020).
Osman, A. et al. Paired bone marrow and peripheral blood samples demonstrate lack of widespread dissemination of some CH clones. Blood Adv. 13, 269–274 (2022).
Nourshargh, S., Renshaw, S. A. & Imhof, B. A. Reverse migration of neutrophils: where, when, how and why. Trends Immunol. 37, 273–286 (2016).
Buckley, C. D. et al. Identification of a phenotypically and functionally distinct population of long-lived neutrophils in a model of reverse endothelial migration. J. Leukoc. Biol. 79, 303–311 (2006).
Kwok, I. et al. Combinatorial single-cell analyses of granulocyte–monocyte progenitor heterogeneity reveals an early uni-potent neutrophil progenitor. Immunity 53, 303–318 (2020).
Zhu, Y. P. et al. CyTOF mass cytometry reveals phenotypically distinct human blood neutrophil populations differentially correlated with melanoma stage. J. Immunother. Cancer 8, e000473 (2020).
Zhu, Y. P. et al. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Rep. 24, 2329–2341 (2018).
Ecker, S. et al. Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types. Genome Biol. 18, 18 (2017).
Naranbhai, V. et al. Genomic modulators of gene expression in human neutrophils. Nat. Commun. 6, 7545 (2015).
Rosales-Alvarez, R. E., Rettkowski, J., Herman, J. S., Cabezas-Wallscheid, N. & Grün, D. Gene expression noise dynamics unveil functional heterogeneity of ageing hematopoietic stem cells. Preprint at bioRxiv https://doi.org/10.1101/2022.08.04.502776 (2022).
Enge, M. et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330 (2017).
Fraser, L. T. C. R., Dikdan, R. J., Dey, S., Singh, A. & Tyagi, S. Reduction in gene expression noise by targeted increase in accessibility at gene loci. Proc. Natl Acad. Sci. USA 118, e2018640118 (2021).
Xue, R. et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature https://doi.org/10.1038/s41586-022-05400-x (2022).
Hackert, N. S., Radtke, F. A., Exner, T., Lorenz, H. & Grieshaber-Bouyer, R. Human and murine neutrophils share core transcriptional programs in both homeostatic and inflamed contexts. Preprint at bioRxiv https://doi.org/10.1101/2022.11.13.516246 (2022).
Simoni, Y., Chng, M. H. Y., Li, S., Fehlings, M. & Newell, E. W. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr. Opin. Immunol. 51, 187–196 (2018).
Mihlan, M., Safaiyan, S., Stecher, M., Paterson, N. & Lämmermann, T. Surprises from intravital imaging of the innate immune response. Annu. Rev. Cell Dev. Biol. 38, 467–489 (2022).
Crainiciuc, G. et al. Behavioural immune landscapes of inflammation. Nature 601, 415–421 (2022).
Woodfin, A. et al. The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nat. Immunol. 12, 761–769 (2011).
Pizzagalli, D. U. et al. Characterization of the dynamic behavior of neutrophils following influenza vaccination. Front. Immunol. 10, 2621 (2019).
Liew, P. X. & Kubes, P. The Neutrophil’s role during health and disease. Physiol. Rev. 99, 1223–1248 (2019).
Yipp, B. G. et al. The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci. Immunol. 2, eaam8929 (2017).
Deniset, J. F., Surewaard, B. G., Lee, W. Y. & Kubes, P. Splenic Ly6Ghigh mature and Ly6Gint immature neutrophils contribute to eradication of S. pneumoniae. J. Exp. Med. 214, 1333–1350 (2017).
Ng, L. G. et al. Visualizing the neutrophil response to sterile tissue injury in mouse dermis reveals a three-phase cascade of events. J. Invest. Dermatol. https://doi.org/10.1038/jid.2011.179 (2011).
Beauvillain, C. et al. CCR7 is involved in the migration of neutrophils to lymph nodes. Blood 117, 1196–1204 (2011).
Devi, S. et al. Neutrophil mobilization via plerixaformediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J. Exp. Med. 210, 2321–2336 (2013).
He, W. et al. Circadian expression of migratory factors establishes lineage-specific signatures that guide the homing of leukocyte subsets to tissues. Immunity 49, 1175–1190 (2018).
Bratton, D. L. & Henson, P. M. Neutrophil clearance: when the party is over, clean-up begins. Trends Immunol. 32, 350–357 (2011).
A-Gonzalez, N. et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214, 1281–1296 (2017).
Suratt, B. T. et al. Neutrophil maturation and activation determine anatomic site of clearance from circulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L913–L921 (2001).
Saverymuttu, S. H., Peters, A. M., Keshavarzian, A., Reavy, H. J. & Lavender, J. P. The kinetics of 111Indium distribution following injection of 111Indium labelled autologous granulocytes in man. Br. J. Haematol. 61, 675–685 (1985).
Furze, R. C. & Rankin, S. M. The role of the bone marrow in neutrophil clearance under homeostatic conditions in the mouse. FASEB J. 22, 3111–3119 (2008).
Guilliams, M., Thierry, G. R., Bonnardel, J. & Bajenoff, M. Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Bowers, E. et al. Granulocyte-derived TNFα promotes vascular and hematopoietic regeneration in the bone marrow. Nat. Med. 24, 95–102 (2018).
Wang, J. et al. Visualizing the function and fate of neutrophils in sterile injury and repair. Science 358, 111–116 (2017).
Özcan, A. et al. CCR7-guided neutrophil redirection to skin-draining lymph nodes regulates cutaneous inflammation and infection. Sci. Immunol. 7, eabi9126 (2022).
Sreejit, G. et al. Retention of the NLRP3 inflammasome-primed neutrophils in the bone marrow is essential for myocardial infarction-induced granulopoiesis. Circulation 145, 31–44 (2022).
Casanova-Acebes, M. et al. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 (2013).
Calì, B. et al. Atypical CXCL12 signaling enhances neutrophil migration by modulating nuclear deformability. Sci. Signal. 15, eabk2552 (2022).
Cerezo‐Wallis, D. & Ballesteros, I. Neutrophils in cancer, a love–hate affair. FEBS J. 289, 3692–3703 (2022).
Coffelt, S. B., Wellenstein, M. D. & de Visser, K. E. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446 (2016).
Cuartero, M. I. et al. N2 neutrophils, novel players in brain inflammation after stroke: modulation by the PPARγ agonist rosiglitazone. Stroke 44, 3498–3508 (2013).
Ng, L. G., Ostuni, R. & Hidalgo, A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 19, 255–265 (2019).
Vafadarnejad, E. et al. Dynamics of cardiac neutrophil diversity in murine myocardial infarction. Circ. Res. 127, e232–e249 (2020).
Sharma, A., Blériot, C., Currenti, J. & Ginhoux, F. Oncofetal reprogramming in tumour development and progression. Nat. Rev. Cancer 22, 593–602 (2022).
Bronte, V. et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150 (2016).
Bachmaier, K. et al. Albumin nanoparticle endocytosing subset of neutrophils for precision therapeutic targeting of inflammatory tissue injury. ACS Nano 16, 4084–4101 (2022).
Raj, A., Peskin, C. S., Tranchina, D., Vargas, D. Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, 1707–1719 (2006).
Ibáñez-Solé, O., Ascensión, A. M., Araúzo-Bravo, M. J. & Izeta, A. Lack of evidence for increased transcriptional noise in aged tissues. eLife 11, e80380 (2022).
Acknowledgements
We thank members of the laboratory of A.H., P. Frenette, D. Lucas, L. Ng, G. Fernandez-Calvo, F. Sanchez-Cabo, M. Casanova-Acebes and J. M. Adrover for past and present discussion and inspiration on questions presented here. This manuscript has been possible through grants R01AI165661 from NIH/NIAD, RTI2018-095497-B-I00 from MCIN, HR17_00527 from Fundación La Caixa, the Transatlantic Network of Excellence (TNE-18CVD04) from the Leducq Foundation, and FET-OPEN (no. 861878) from the European Commission. M.P.-S. is supported by the EMBO ALTF (no. 1142-2020) long-term fellowship and from MICINN (RYC2021-033511-I). J.S. and I.B. are supported by fellowships from MICINN (PRE2019-089130 and RYC2020-029563-I, respectively). The CNIC is supported by the MCIN and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (CEX2020-001041-S).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Hongbo Luo and Renato Ostuni for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Palomino-Segura, M., Sicilia, J., Ballesteros, I. et al. Strategies of neutrophil diversification. Nat Immunol 24, 575–584 (2023). https://doi.org/10.1038/s41590-023-01452-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-023-01452-x
This article is cited by
-
Neutrophils seeking new neighbors: radiotherapy affects the cellular framework and the spatial organization in a murine breast cancer model
Cancer Immunology, Immunotherapy (2024)
-
Evaluation of the innate immune response of caprine neutrophils against Mycobacterium avium subspecies paratuberculosis in vitro
Veterinary Research (2023)
-
Neutrophil diversity and plasticity: Implications for organ transplantation
Cellular & Molecular Immunology (2023)
-
Whole-genome screens reveal regulators of differentiation state and context-dependent migration in human neutrophils
Nature Communications (2023)
-
CREB1-driven CXCR4hi neutrophils promote skin inflammation in mouse models and human patients
Nature Communications (2023)