Abstract
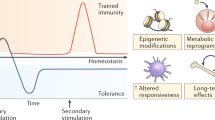
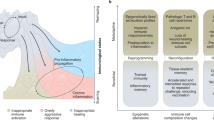
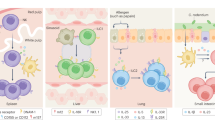
Adaptation is the ability of cells, tissues and organisms to rapidly and reversibly modify their properties to maximize fitness in a changing environment. The activity of immune-system components unfolds in the remarkably heterogeneous milieus to which they are exposed in different tissues, during homeostasis or during various acute or chronic pathological states. Therefore, adaptation is essential for immune cells to tune their responses to a large variety of contexts and conditions. The adaptation of immune cells reflects the integration of multiple inputs acting simultaneously or in a temporal sequence, which eventually leads to transcriptional reprogramming and to various functional consequences, some of which extend beyond the duration of the stimulus. A range of adaptive responses have been observed in both adaptive immune cells and innate immune cells; these are referred to with terms such as ‘plasticity’, ‘priming’, ‘training’, ‘exhaustion’ and ‘tolerance’, among others, all of which can be useful for defining a certain immunological process or outcome but whose underlying molecular frameworks are often incompletely understood. Here we review and analyze mechanisms of adaptation and memory in immunity with the aim of providing basic concepts that rationalize the properties and molecular bases of these essential processes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wilson, C. B., Rowell, E. & Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 9, 91–105 (2009).
Monticelli, S. DNA (hydroxy)methylation in T helper lymphocytes. Trends Biochem. Sci. https://doi.org/10.1016/j.tibs.2019.01.009 (2019).
Netea, M. G. et al. Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098 (2016).
Monticelli, S. & Natoli, G. Short-term memory of danger signals and environmental stimuli in immune cells. Nat. Immunol. 14, 777–784 (2013).
Seeley, J. J. & Ghosh, S. Molecular mechanisms of innate memory and tolerance to LPS. J. Leukoc. Biol. 101, 107–119 (2017).
Sun, J. C., Ugolini, S. & Vivier, E. Immunological memory within the innate immune system. EMBO J. 33, 1295–1303 (2014).
O’Sullivan, T. E., Sun, J. C. & Lanier, L. L. Natural killer cell memory. Immunity 43, 634–645 (2015).
Sun, J. C., Beilke, J. N. & Lanier, L. L. Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009).
DuPage, M. & Bluestone, J. A. Harnessing the plasticity of CD4+ T cells to treat immune-mediated disease. Nat. Rev. Immunol. 16, 149–163 (2016).
Sen, D. R. et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Matzinger, P. Friendly and dangerous signals: is the tissue in control? Nat. Immunol. 8, 11–13 (2007).
Varol, C., Mildner, A. & Jung, S. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 33, 643–675 (2015).
Monticelli, S. & Natoli, G. Transcriptional determination and functional specificity of myeloid cells: making sense of diversity. Nat. Rev. Immunol. 17, 595–607 (2017).
Glass, C. K. & Natoli, G. Molecular control of activation and priming in macrophages. Nat. Immunol. 17, 26–33 (2016).
Ghisletti, S. et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 (2010).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Mancino, A. et al. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev. 29, 394–408 (2015).
Garber, M. et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822 (2012).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Kaikkonen, M. U. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325 (2013).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844 (2014).
Gautier, E. L. et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J. Exp. Med. 211, 1525–1531 (2014).
Rosas, M. et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648 (2014).
Haldar, M. et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156, 1223–1234 (2014).
Scott, C. L. et al. The transcription factor ZEB2 is required to maintain the tissue-specific identities of macrophages. Immunity 49, 312–325.e315 (2018).
Mass, E. et al. Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238 (2016).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Deczkowska, A. et al. Mef2C restrains microglial inflammatory response and is lost in brain ageing in an IFN-I-dependent manner. Nat. Commun. 8, 717 (2017).
Matcovitch-Natan, O. et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016).
Saijo, K. et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 137, 47–59 (2009).
Kayama, H. et al. Heme ameliorates dextran sodium sulfate-induced colitis through providing intestinal macrophages with noninflammatory profiles. Proc. Natl Acad. Sci. USA 115, 8418–8423 (2018).
Casanova-Acebes, M. et al. Neutrophils instruct homeostatic and pathological states in naive tissues. J. Exp. Med. 215, 2778–2795 (2018).
Ng, L. G., Ostuni, R. & Hidalgo, A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 19, 255–265 (2019).
Casanova-Acebes, M. et al. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 (2013).
Puga, I. et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat. Immunol. 13, 170–180 (2011).
Mackay, L. K. & Kallies, A. Transcriptional regulation of tissue-resident lymphocytes. Trends Immunol. 38, 94–103 (2017).
Ditadi, A., Sturgeon, C. M. & Keller, G. A view of human haematopoietic development from the Petri dish. Nat. Rev. Mol. Cell Biol. 18, 56–67 (2017).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Gentek, R. et al. Hemogenic endothelial fate mapping reveals dual developmental origin of mast cells. Immunity 48, 1160–1171.e1165 (2018).
Li, Z. et al. Adult connective tissue-resident mast cells originate from late erythro-myeloid progenitors. Immunity 49, 640–653.e645 (2018).
Smith, F. L. & Baumgarth, N. B-1 cell responses to infections. Curr. Opin. Immunol. 57, 23–31 (2019).
Gentek, R. et al. Epidermal γ δ T cells originate from yolk sac hematopoiesis and clonally self-renew in the adult. J. Exp. Med. 215, 2994–3005 (2018).
Smith, N. L. et al. Developmental origin governs CD8+ T cell fate decisions during infection. Cell 174, 117–130.e114 (2018).
Shemer, A. et al. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun. 9, 5206 (2018).
Cronk, J. C. et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med. 215, 1627–1647 (2018).
Zhu, Y. et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity 47, 597 (2017).
Bowman, R. L. et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 17, 2445–2459 (2016).
Pope, S. D. & Medzhitov, R. Emerging Principles of Gene Expression Programs and Their Regulation. Mol. Cell 71, 389–397 (2018).
Smale, S. T., Tarakhovsky, A. & Natoli, G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 32, 489–511 (2014).
Foster, S. L., Hargreaves, D. C. & Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 447, 972–978 (2007).
Kamada, R. et al. Interferon stimulation creates chromatin marks and establishes transcriptional memory. Proc. Natl Acad. Sci. USA 115, E9162–E9171 (2018).
Daniel, B. et al. The nuclear receptor PPARγ controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity 49, 615–626.e616 (2018).
Qiao, Y. et al. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and toll-like receptor signaling. Immunity 39, 454–469 (2013).
Cheng, S. C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Wendeln, A. C. et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338 (2018).
Naik, S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017).
Ordovas-Montanes, J. et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature 560, 649–654 (2018).
McLane, L.M., Abdel-Hakeem, M.S. & Wherry, E.J. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019).
Hotchkiss, R. S., Monneret, G. & Payen, D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 13, 862–874 (2013).
Chen, J. & Ivashkiv, L. B. IFN-γ abrogates endotoxin tolerance by facilitating Toll-like receptor-induced chromatin remodeling. Proc. Natl Acad. Sci. USA 107, 19438–19443 (2010).
Novakovic, B. et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e1314 (2016).
Park, S. H. et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat. Immunol. 18, 1104–1116 (2017).
Burrill, D. R. & Silver, P. A. Making cellular memories. Cell 140, 13–18 (2010).
Ptashne, M. Regulation of transcription: from lambda to eukaryotes. Trends Biochem. Sci. 30, 275–279 (2005).
Seeley, J. J. et al. Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature 559, 114–119 (2018).
Ramirez-Carrozzi, V. R. et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138, 114–128 (2009).
Toyama, B. H. et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982 (2013).
Ghoneim, H. E. et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell 170, 142–157.e119 (2017).
Mitroulis, I. et al. Modulation of myelopoiesis progenitors is an integral component of trained immunity. Cell 172, 147–161.e112 (2018).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Deal, R. B., Henikoff, J. G. & Henikoff, S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 (2010).
Dion, M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Kraushaar, D. C. et al. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 14, R121 (2013).
Margueron, R. et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Quintin, J. et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232 (2012).
Stone, B.A. Chemistry of β-glucans. in Chemistry, Biochemistry, and Biology of 1→3-β-Glucans and Related Polysaccharides (eds. Bacic, A. et al.) Ch. 2.1 (Academic Press, 2009).
Williams, D. L. Overview of (1→3)-β-D-glucan immunobiology. Mediators Inflamm. 6, 247–250 (1997).
Ariizumi, K. et al. Identification of a novel, dendritic cell-associated molecule, dectin-1, by subtractive cDNA cloning. J. Biol. Chem. 275, 20157–20167 (2000).
Brown, G. D. & Gordon, S. Immune recognition. A new receptor for β-glucans. Nature 413, 36–37 (2001).
Palma, A. S. et al. Ligands for the β-glucan receptor, Dectin-1, assigned using “designer” microarrays of oligosaccharide probes (neoglycolipids) generated from glucan polysaccharides. J. Biol. Chem. 281, 5771–5779 (2006).
Nono, I., Ohno, N., Masuda, A., Oikawa, S. & Yadomae, T. Oxidative degradation of an antitumor (1→3)-β-D-glucan, grifolan. J. Pharmacobiodyn. 14, 9–19 (1991).
Ozment, T. R., Goldman, M. P., Kalbfleisch, J. H. & Williams, D. L. Soluble glucan is internalized and trafficked to the Golgi apparatus in macrophages via a clathrin-mediated, lipid raft-regulated mechanism. J. Pharmacol. Exp. Ther. 342, 808–815 (2012).
Miura, T., Ohno, N., Miura, N. N., Shimada, S. & Yadomae, T. Inactivation of a particle β-glucan by proteins in plasma and serum. Biol. Pharm. Bull. 20, 1103–1107 (1997).
Miura, T. et al. Inactivation of (1→3)-β-D-glucan in mice. Biol. Pharm. Bull. 18, 1797–1801 (1995).
Chen, K. et al. Super-low dose endotoxin pre-conditioning exacerbates sepsis mortality. EBioMedicine 2, 324–333 (2015).
Lira, F. S. et al. Endotoxin levels correlate positively with a sedentary lifestyle and negatively with highly trained subjects. Lipids Health Dis. 9, 82 (2010).
Szeto, C. C. et al. Endotoxemia is related to systemic inflammation and atherosclerosis in peritoneal dialysis patients. Clin. J. Am. Soc. Nephrol. 3, 431–436 (2008).
Wiedermann, C. J. et al. Association of endotoxemia with carotid atherosclerosis and cardiovascular disease: prospective results from the Bruneck Study. J. Am. Coll. Cardiol. 34, 1975–1981 (1999).
Olsen, A. W., Brandt, L., Agger, E. M., van Pinxteren, L. A. & Andersen, P. The influence of remaining live BCG organisms in vaccinated mice on the maintenance of immunity to tuberculosis. Scand. J. Immunol. 60, 273–277 (2004).
Kaveh, D. A., Garcia-Pelayo, M. C. & Hogarth, P. J. Persistent BCG bacilli perpetuate CD4 T effector memory and optimal protection against tuberculosis. Vaccine 32, 6911–6918 (2014).
Queiroz, A. & Riley, L. W. Bacterial immunostat: Mycobacterium tuberculosis lipids and their role in the host immune response. Rev. Soc. Bras. Med. Trop. 50, 9–18 (2017).
Xue, J. et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40, 274–288 (2014).
Murray, P. J. et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Zanoni, I. et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236 (2016).
Saccani, S., Pantano, S. & Natoli, G. Two waves of nuclear factor kB recruitment to target promoters. J. Exp. Med. 193, 1351–1359 (2001).
Czimmerer, Z. et al. The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48, 75–90.e76 (2018).
Kang, K. et al. Interferon-γ represses m2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity 47, 235–250.e234 (2017).
Piccolo, V. et al. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 18, 530–540 (2017).
Schleicher, U. et al. TNF-mediated restriction of arginase 1 expression in myeloid cells triggers type 2 NO synthase activity at the site of infection. Cell Rep. 15, 1062–1075 (2016).
Nelson, V. L. et al. PPARγ is a nexus controlling alternative activation of macrophages via glutamine metabolism. Genes Dev. 32, 1035–1044 (2018).
Glass, C. K. & Saijo, K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat. Rev. Immunol. 10, 365–376 (2010).
Qiao, Y., Kang, K., Giannopoulou, E., Fang, C. & Ivashkiv, L. B. IFN-γ induces histone 3 lysine 27 trimethylation in a small subset of promoters to stably silence gene expression in human macrophages. Cell Rep. 16, 3121–3129 (2016).
Ezenwa, V. O. & Jolles, A. E. Epidemiology. Opposite effects of anthelmintic treatment on microbial infection at individual versus population scales. Science 347, 175–177 (2015).
Osborne, L. C. et al. Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 345, 578–582 (2014).
Rückerl, D. et al. Macrophage origin limits functional plasticity in helminth-bacterial co-infection. PLoS Pathog. 13, e1006233 (2017).
Kratochvill, F. et al. TNF counterbalances the emergence of M2 tumor macrophages. Cell Rep. 12, 1902–1914 (2015).
Bosurgi, L. et al. Macrophage function in tissue repair and remodeling requires IL-4 or IL-13 with apoptotic cells. Science 356, 1072–1076 (2017).
Harrison, O.J. et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science 363, eaat6280 (2019).
Blander, J. M., Longman, R. S., Iliev, I. D., Sonnenberg, G. F. & Artis, D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 18, 851–860 (2017).
Acknowledgements
We are grateful to members of the G.N. and R.O. laboratories for scientific discussions and to S. Monticelli for critical reading of the manuscript. Research on related topics in the G.N. laboratory is supported by the European Research Council (ERC Advanced Grant 692789, MEDICI), the European Commission (Consortium grant SYSCYD), the Cariplo Foundation and the Italian Ministry of University and Research (MIUR, grant METRIC (METabolic Regulation of Inflammatory Cells). Research in the R.O. laboratory is supported by grants from the European Research Council (ERC Starting Grant 759532, X-TAM), the Italian Telethon Foundation (SR-Tiget grant award F04), the Italian Ministry of Health (GR-2016-02362156), the Associazione Italiana per la Ricerca sul Cancro (AIRC MFAG, 20247), the Cariplo Foundation (2015–0990) and the European Union (Infect-ERA 126).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Natoli, G., Ostuni, R. Adaptation and memory in immune responses. Nat Immunol 20, 783–792 (2019). https://doi.org/10.1038/s41590-019-0399-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0399-9
This article is cited by
-
LFA-1 nanoclusters integrate TCR stimulation strength to tune T-cell cytotoxic activity
Nature Communications (2024)
-
Trained innate immunity modulates osteoblast and osteoclast differentiation
Stem Cell Reviews and Reports (2024)
-
Injury-experienced satellite cells retain long-term enhanced regenerative capacity
Stem Cell Research & Therapy (2023)
-
Remembering foods and foes: emerging principles of transcriptional memory
Cell Death & Differentiation (2023)
-
Nanoparticle-mediated TRPV1 channel blockade amplifies cancer thermo-immunotherapy via heat shock factor 1 modulation
Nature Communications (2023)