Abstract
MicroRNA miR-138, which is highly expressed in neurons, represses herpes simplex virus 1 (HSV-1) lytic cycle genes by targeting viral ICP0 messenger RNA, thereby promoting viral latency in mice. We found that overexpressed miR-138 also represses lytic processes independently of ICP0 in murine and human neuronal cells; therefore, we investigated whether miR-138 has targets besides ICP0. Using genome-wide RNA sequencing/photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation followed by short interfering RNA knockdown of candidate targets, we identified the host Oct-1 and Foxc1 messenger mRNAs as miR-138’s targets, whose gene products are transcription factors important for HSV-1 replication in neuronal cells. OCT-1 has a known role in the initiation of HSV transcription. Overexpression of FOXC1, which was not known to affect HSV-1, promoted HSV-1 replication in murine neurons and ganglia. CRISPR–Cas9 knockout of FOXC1 reduced viral replication, lytic gene expression and miR-138 repression in murine neuronal cells. FOXC1 also collaborated with ICP0 to decrease heterochromatin on viral genes and compensated for the defect of an ICP0-null virus. In summary, miR-138 targets ICP0, Oct-1 and Foxc1 to repress HSV-1 lytic cycle genes and promote epigenetic gene silencing, which together enable favourable conditions for latent infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw high-throughput sequencing data for RNA-seq (Figs. 1g and 3 and Extended Data Figs. 2 and 8b) and PAR-CLIP (Fig. 3 and Extended Data Fig. 5) experiments have been deposited with the Gene Expression Omnibus under accession no. GSE127504. The HSV-1 KOS strain genome sequence and annotation were obtained from GenBank with accession no. JQ673480.1. Human (hg19) and mouse (mm10) genome sequences and annotations were downloaded from the Harvard Medical School Research Computing server (https://rc.hms.harvard.edu). Source data are provided with this paper.
References
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Ebert, M. S. & Sharp, P. A. Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012).
Guo, Y. E. & Steitz, J. A. Virus meets host microRNA: the destroyer, the booster, the hijacker. Mol. Cell. Biol. 34, 3780–3787 (2014).
Bruscella, P. et al. Viruses and miRNAs: more friends than foes. Front. Microbiol. 8, 824 (2017).
Girardi, E., López, P. & Pfeffer, S. On the importance of host microRNAs during viral infection. Front. Genet. 9, 439 (2018).
Skalsky, R. L. & Cullen, B. R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64, 123–141 (2010).
Roizman, B. et al. in Fields Virology 6th edn (eds Knipe, D.M. et al.) 1823–1897 (Lippincott Williams & Wilkins, 2013).
Wysocka, J. & Herr, W. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28, 294–304 (2003).
Nogueira, M. L., Wang, V. E. H., Tantin, D., Sharp, P. A. & Kristie, T. M. Herpes simplex virus infections are arrested in Oct-1-deficient cells. Proc. Natl Acad. Sci. USA 101, 1473–1478 (2004).
Cai, W. & Schaffer, P. A. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66, 2904–2915 (1992).
Lee, J. S., Raja, P. & Knipe, D. M. Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. mBio 7, e02007-15 (2016).
Oh, J. & Fraser, N. W. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82, 3530–3537 (2008).
Cliffe, A. R. & Knipe, D. M. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82, 12030–12038 (2008).
Herrera, F. J. & Triezenberg, S. J. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 78, 9689–9696 (2004).
Kwiatkowski, D. L., Thompson, H. W. & Bloom, D. C. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83, 8173–8181 (2009).
Cliffe, A. R., Coen, D. M. & Knipe, D. M. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 4, e00590-12 (2013).
Raja, P. et al. A herpesviral lytic protein regulates the structure of latent viral chromatin. mBio 7, e00633-16 (2016).
Cliffe, A. R., Garber, D. A. & Knipe, D. M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83, 8182–8190 (2009).
Wang, Q.-Y. et al. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl Acad. Sci. USA 102, 16055–16059 (2005).
Umbach, J. L. et al. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783 (2008).
Jurak, I. et al. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. J. Virol. 84, 4659–4672 (2010).
Kim, J. Y., Mandarino, A., Chao, M. V., Mohr, I. & Wilson, A. C. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog. 8, e1002540 (2012).
Linderman, J. A. et al. Immune escape via a transient gene expression program enables productive replication of a latent pathogen. Cell Rep. 18, 1312–1323 (2017).
Du, T., Zhou, G. & Roizman, B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc. Natl Acad. Sci. USA 108, 18820–18824 (2011).
Thompson, R. L., Preston, C. M. & Sawtell, N. M. De novo synthesis of VP16 coordinates the exit from HSV latency in vivo. PLoS Pathog. 5, e1000352 (2009).
Pan, D. et al. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe 15, 446–456 (2014).
Bogerd, H. P. et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 88, 8065–8076 (2014).
Pan, D. & Coen, D. M. Quantification and analysis of thymidine kinase expression from acyclovir-resistant G-string insertion and deletion mutants in herpes simplex virus-infected cells. J. Virol. 86, 4518–4526 (2012).
Cai, W. Z. & Schaffer, P. A. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 63, 4579–4589 (1989).
Pan, D. et al. Herpes simplex virus 1 lytic infection blocks microRNA (miRNA) biogenesis at the stage of nuclear export of pre-miRNAs. mBio 10, e02856-18 (2019).
Haraguchi, T., Ozaki, Y. & Iba, H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37, e43 (2009).
Gilding, L. N. & Somervaille, T. C. P. The diverse consequences of FOXC1 deregulation in cancer. Cancers (Basel) 11, 184 (2019).
Berry, F. B., Saleem, R. A. & Walter, M. A. FOXC1 transcriptional regulation is mediated by N- and C-terminal activation domains and contains a phosphorylated transcriptional inhibitory domain. J. Biol. Chem. 277, 10292–10297 (2002).
Wang, W., Zhao, L.-J., Tan, Y.-X., Ren, H. & Qi, Z.-T. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis 33, 1113–1120 (2012).
Sossey-Alaoui, K. & Plow, E. F. miR-138-mediated regulation of KINDLIN-2 expression modulates sensitivity to chemotherapeutics. Mol. Cancer Res. 14, 228–238 (2016).
Huang, H. et al. MIR-138-5P inhibits the progression of prostate cancer by targeting FOXC1. Mol. Genet. Genomic Med. 8, e1193 (2020).
Yu, C. et al. MicroRNA-138-5p regulates pancreatic cancer cell growth through targeting FOXC1. Cell. Oncol. (Dordr.) 38, 173–181 (2015).
Bai, X. et al. Inhibition of lung cancer growth and metastasis by DHA and its metabolite, RvD1, through miR-138-5p/FOXC1 pathway. J. Exp. Clin. Cancer Res. 38, 479 (2019).
Siegel, G. et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716 (2009).
Wang, X. et al. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 589, 726–729 (2015).
Yeh, Y.-M., Chuang, C.-M., Chao, K.-C. & Wang, L.-H. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1α. Int. J. Cancer 133, 867–878 (2013).
Acharya, M., Huang, L., Fleisch, V. C., Allison, W. T. & Walter, M. A. A complex regulatory network of transcription factors critical for ocular development and disease. Hum. Mol. Genet. 20, 1610–1624 (2011).
Iwafuchi-Doi, M. et al. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell 62, 79–91 (2016).
O’Connor, C. M., Vanicek, J. & Murphy, E. A. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J. Virol. 88, 5524–5532 (2014).
Trobaugh, D. W. et al. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506, 245–248 (2014).
Ruelas, D. S. et al. MicroRNA-155 reinforces HIV latency. J. Biol. Chem. 290, 13736–13748 (2015).
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581 (2005).
Mulik, S. et al. Role of miR-132 in angiogenesis after ocular infection with herpes simplex virus. Am. J. Pathol. 181, 525–534 (2012).
Bhela, S. et al. Critical role of microRNA-155 in herpes simplex encephalitis. J. Immunol. 192, 2734–2743 (2014).
Ingle, H. et al. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 8, ra126 (2015).
Sawtell, N. M. & Thompson, R. L. De novo herpes simplex virus VP16 expression gates a dynamic programmatic transition and sets the latent/lytic balance during acute infection in trigeminal ganglia. PLoS Pathog. 12, e1005877 (2016).
Tal-Singer, R. et al. The transcriptional activation domain of VP16 is required for efficient infection and establishment of latency by HSV-1 in the murine peripheral and central nervous systems. Virology 259, 20–33 (1999).
Stern, S., Tanaka, M. & Herr, W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341, 624–630 (1989).
Whitlow, Z. & Kristie, T. M. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J. Virol. 83, 9591–9595 (2009).
Kolb, G. & Kristie, T. M. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J. Virol. 82, 9555–9563 (2008).
Elliott, G. & O’Hare, P. Equine herpesvirus 1 gene 12, the functional homologue of herpes simplex virus VP16, transactivates via octamer sequences in the equine herpesvirus IE gene promoter. Virology 213, 258–262 (1995).
Katzenell, S., Cabrera, J. R., North, B. J. & Leib, D. A. Isolation, purification, and culture of primary murine sensory neurons. Methods Mol. Biol. 1656, 229–251 (2017).
Ng, A. H. M. et al. A comprehensive library of human transcription factors for cell fate engineering. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0742-6 (2020).
Tischer, B. K., von Einem, J., Kaufer, B. & Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40, 191–197 (2006).
Sen, J., Liu, X., Roller, R. & Knipe, D. M. Herpes simplex virus US3 tegument protein inhibits Toll-like receptor 2 signaling at or before TRAF6 ubiquitination. Virology 439, 65–73 (2013).
Heckman, K. L. & Pease, L. R. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 (2007).
Sinani, D., Cordes, E., Workman, A., Thunuguntia, P. & Jones, C. Stress-induced cellular transcription factors expressed in trigeminal ganglionic neurons stimulate the herpes simplex virus 1 ICP0 promoter. J. Virol. 87, 13042–13047 (2013).
Chen, S.-H. et al. Suppression of transcription factor early growth response 1 reduces herpes simplex virus lethality in mice. J. Clin. Invest. 118, 3470–3477 (2008).
Pan, D., Pesola, J. M., Li, G., McCarron, S. & Coen, D. M. Mutations inactivating herpes simplex virus 1 microRNA miR-H2 do not detectably increase ICP0 gene expression in infected cultured cells or mouse trigeminal ganglia. J. Virol. 91, e02001-16 (2017).
Gottwein, E. & Cullen, B. R. A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J. Virol. 84, 5229–5237 (2010).
Danan, C., Manickavel, S. & Hafner, M. PAR-CLIP: a method for transcriptome-wide identification of RNA binding protein interaction sites. Methods Mol. Biol. 1358, 153–173 (2016).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Acknowledgements
We thank B. Cullen, C. Jones and S.-H. Chen for the generous provision of plasmids, K. Holton from Harvard Medical School Research Computing for help with the sequencing data analysis, and the Core Facility of Zhejiang University School of Medicine and the Biopolymers Facility at Harvard Medical School for expertise and instrument availability. This work was supported by the National Key R & D Program of China (no. 2017YFC1200204 to D.P.), National Natural Science Foundation of China (no. 81671993 to D.P.), Natural Science Foundation of Zhejiang Province, China (no. LR18H190001 to D.P.), National Institutes of Health (no. P01 AI098681 to D.M.C. and D.M.K.), a Harvard Medical School Dean’s Initiative for Innovation Grant (D.M.K. and D.M.C.), a Natural Sciences and Engineering Research Council of Canada Postgraduate Fellowship and a Peter and Carolyn Lynch Foundation Fellowship (to A.H.M.N.), the National Human Genome Research Institute (no. RM1 HG008525 to G.M.C.) and the Blavatnik Biomedical Accelerator at Harvard University (to G.M.C.).
Author information
Authors and Affiliations
Contributions
D.P. conceived the study. B.S., X. Yang, Q.W. and D.P. performed molecular cloning and the experiments in cell culture. X. Yang, J.M.-L., F.H. and D.P. generated recombinant viruses. X. Yang, F.H., X. Yu, J.M.P. and S.M.C. contributed to the animal studies. F.H. and E.A.H.V. performed the immunofluorescence assays. F.H. conducted the experiments using mouse primary neurons. P.R., Q.W. and D.P. performed the ChIP experiments. A.H.M.N. and G.M.C. developed the methods of deriving mixed neurons from iPSCs. H.S.O. differentiated the iPSCs into sensory neurons with a separate protocol and performed the experiments using the iPSC-derived neurons. J.M.P. provided statistical advice. D.P. and D.M.C. prepared the manuscript. D.P., D.M.K. and D.M.C. provided supervision. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.H.M.N. and G.M.C. are inventors on patent nos. WO2018049382 and WO2018204262 filed by the Presidents and Fellows of Harvard College. Full disclosure for G.M.C. is available on http://arep.med.harvard.edu/gmc/tech.html. A.H.M.N. and G.M.C. are co-founders and have equity in GC Therapeutics. Other authors declare no conflict of interest.
Additional information
Peer review information Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
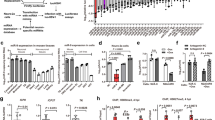
Extended Data Fig. 1 Additional data about effects of miR-138 on HSV-1 replication.
a, Neuro-2a cells were mock-infected or infected with the indicated virus (MOI = 5) for 16 h before qRT-PCR analysis of miR-138 and let-7a levels. b, In 4 independent experiments performed as in Fig. 1b, the ratios of the average titers from scrambled transfected cells over those from miR-138 transfected cells were calculated separately for WT and M138 viruses and plotted. c, 293 T cells were transfected with 40 nM miRNA mimic for 16 h, and then infected with WT or M138 virus (left graph), or 7134 (ICP0-null) virus (right graph) for 48 h (MOI = 0.1) before viral titer measurements. d, Same as c, except that Vero cells were infected (MOI = 0.01) and no significant difference was detected between miR-138 and either of the two controls. For b, n = 4 independent experiments. For other panels, n = 3 (a, c right), 4 (d) or 8 (c left) biologically independent samples per condition. For all panels, data are presented as mean values ± S.D. and were analyzed by unpaired, two tailed t tests (a, b), two-way (c left, d) or one-way (c right) ANOVA with Bonferroni’s multiple comparisons tests.
Extended Data Fig. 2 Global effects of miR-138 on viral gene expression.
Same experiment as Fig. 1g, but effects on individual viral transcripts are plotted. Blue and red bars represent data for WT and M138 virus, respectively. n = 3 biologically independent samples per condition. Each bar represents the read count mean value for the indicated transcript from scrambled mimic transfected cells (after being normalized to total reads of the sample) divided by that from miR-138 mimic transfected cells.
Extended Data Fig. 3 Expression of miR-138, US11 and US12 from recombinant viruses.
a, 293 T cells were infected with M138miR138 or M138nomiR138 virus (MOI = 5). At 8 hpi, the cells were harvested for RNA purification and Northern blot hybridization. RNA from M138miR138 infected cells was run alongside a dilution series of synthetic miR-138 in the gel and hybridized with a miR-138 probe (left panel). RNA from M138nomiR138 infected cells was run alongside a dilution series of synthetic miR-M138b in the gel and hybridized with a miR-M138b probe (right panel). The integrity of RNA from both samples was verified by ethidium bromide staining, which is shown below the Northern blot images for a set of bands ~80 bases. This experiment was performed once. b, Vero cells were infected with the viruses indicated at the bottom at an MOI of 5 and harvested at 8 h post-infection for qRT-PCR analysis of US11 and US12 mRNA levels normalized to ICP27 mRNA levels. c, miR-138 expression from 293 T cells infected by the indicated viruses (MOI = 5, 8 h post-infection) as measured by qRT-PCR. For b and c, n = 3 biologically independent samples and data are presented as mean values ± s.d. Data were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons tests.
Extended Data Fig. 4 Additional in vivo data about miR-138 expressing and control recombinant viruses.
a, Viral DNA and RNA levels in TG infected with WTmiR138 and WTnomiR138 at 5 dpi. The DNA or RNA molecules measured are labeled at the top of each graph. Viruses are indicated at the bottom. Each point represents a value from one trigeminal ganglion, and the horizontal lines represent the geometric means. The displayed n numbers represent the numbers of mice used per condition. Data were analyzed by two-tailed, unpaired t tests. b, Same as a, but WTmiR138R and WTnomiR138 are compared. c, Same as a, but M138miR138R and M138nomiR138 are compared.
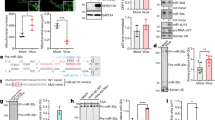
Extended Data Fig. 5 PAR-CLIP experiments identified viral and host targets of miR-138.
a, miR-138 expression in 293 Tcontrol and 293T138 cells. b, PAR-CLIP procedure for detecting viral targets of miR-138 with, from left to right, a schematic showing the samples, a cartoon showing the crosslinking procedure, a representative autoradiograph of an SDS polyacrylamide gel showing a band corresponding to the Ago-RNA complex, the subsequent steps, and an agarose gel showing PCR amplification of the cDNA library. The experiment was performed once. c, Read counts of the indicated sequences from the indicated samples. d, 293T cells were co-transfected with 20 nM miRNA mimic and 100 ng/ml plasmid for 48 h before Western blot analysis of FLAG-tagged UL39 using an anti-FLAG antibody and analysis of ICP0 using an ICP0 antibody. This experiment was repeated once with similar results. e, miR-138 levels in Neuro-2a, N2A138 and N2Aanti138 cells. f, Diagram showing the “anti138” sequence expressed in N2Aanti138 cells. The sequence has a “tough decoy” secondary structure. Red and black horizontal lines represent anti138 and miR-138, respectively. Curved lines above and below the main structure represent bulges (extra nucleotides not bound to miR-138) designed to prevent cleavage. g, miR-138 and total read counts in the N2A138 and N2Aanti138 cells in a PAR-CLIP experiment. h, Fraction of 5’ UTR, CDS or 3’ UTR sites in total sites identified by the single PAR-CLIP approach (panels 1 and 2) or the combined PAR-CLIP/RNAseq approach (panels 3 and 4) in 293 T (panels 1 and 3) and Neuro-2a cells (panels 2 and 4). Relative to the single PAR-CLIP approach, the combined approach identified significantly higher fractions of targets with 3’ UTR sites (P = 0.034 and 0.0006 for 293 T and Neuro-2a cells, respectively by Fisher’s exact tests). For a and e, n = 3 biologically independent samples and data are presented as mean values ± s.d.
Extended Data Fig. 6 Low expression of OCT-1 and FOXC1 in mouse TG.
a, Fixed trigeminal ganglion cryo-sections were stained using an anti-FOXC1 or anti-OCT-1 rabbit primary antibody (red) or without a rabbit primary antibody (control), stained with a mouse anti-Tuj1 antibody (green), and stained with DAPI (blue). Similar results were obtained for OCT-1 using a different OCT-1 antibody (data not shown). This experiment was repeated twice with similar results. b, Neuro-2a cells were mock-transfected or transfected with 200 ng/ml OCT-1 or FOXC1 expressing plasmid for 48 h. The cells were then fixed and stained with an anti-OCT-1 or anti-FOXC1 antibody (green) or without a primary antibody, and stained with DAPI (blue). This experiment was repeated once with similar results.
Extended Data Fig. 7 Additional results with HSV1FOXC1 and HSV1FOXC1delAD viruses.
a, Mice were infected on the cornea with 2 × 105 PFU per eye of the indicated viruses. Viral titers in eye swabs collected at 1 d post-infection were determined. b, Following infection as in a, TG were harvested at 31 d post-infection and analyzed for viral genome levels normalized to mouse adipsin gene levels by qPCR. c, Following infection as in a, TG were harvested at 5 d post-infection and viral titers in TG were determined. d, Following mock infection (left) or infection of mice at the cornea with 2 × 105 PFU per eye of HSV1FOXC1delAD (middle) or HSV1FOXC1 (right), fixed TG cryo-sections were stained by DAPI (blue) and an anti-FLAG antibody (green) that can detect FLAG-tagged FOXC11 and FOXC1delAD proteins. This experiment was performed once. e, After corneal inoculation with 2 × 105 pfu/eye of the indicated viruses, mouse TG were harvested at 29 d post-infection and analyzed for viral genome levels normalized to mouse adipsin gene levels by qPCR. No significant difference was detected in e. For all panels, the n numbers represent the numbers of mice used per condition. The horizontal lines represent geometrical means. Data were analyzed by two-tailed, unpaired t tests (a, b, c) or one-way ANOVA with Bonferroni’s multiple comparisons tests (e) with the P values indicated.
Extended Data Fig. 8 Global effects of FOXC1 on viral gene expression.
a, Additional data for Fig. 6b. Neuro-2a cells were transfected with 200 ng/ml plasmid for 40 h. KOS was then added (MOI = 2). The cells were incubated at 4 °C for 1 h to allow attachment, washed by PBS and incubated at 37 °C for 2 h before qRT-PCR analysis for the transcript indicated at the top normalized to host GAPDH levels. n = 6 biologically independent samples. The horizontal lines represent mean values. Data were analyzed by two-tailed, unpaired t tests. b, Neuro-2a cells were transfected with 200 ng/ml pcDNA or pFOXC1human for 40 h and infected with KOS for 5 h at an MOI of 1 before RNA-seq analysis. n = 3 biologically independent samples. Each bar represents the mean read count for the indicated transcript from pFOXC1human transfected cells (after being normalized by total reads from that sample) divided by that from pcDNA transfected cells.
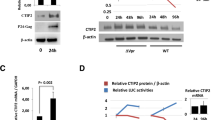
Extended Data Fig. 9 FOXC1 is important for HSV-1 replication and gene expression.
a, Left, Foxc1 CDS is depicted as a blue box with the DBD in green. Expanded below is the region of deletion in N2AFOXC1 knockout cells, showing sequences in WT and KO cell lines. The PAM sequence required for guide RNA target recognition is shown in green and the target sequence in red. Right, expression of FOXC1 relative to β-tubulin (loading control) in Neuro-2a and N2AFOXC1 knockout cells was analyzed by Western blots. This experiment was repeated twice with similar results. b, Neuro-2a and N2AFOXC1 knockout cells were infected with KOS (MOI = 0.5) for the indicated times. Mean viral titers ± s.d. are shown. c, Additional data for Fig. 6e. N2AFOXC1 knockout or Neuro-2a cells were transfected with 400 ng/ml pcDNA or pFOXC1human. 40 h later, the cells were infected with KOS for 5 h at an MOI of 1 before qRT-PCR analyses of the transcript indicated at the top. Horizontal lines represent geometrical means. Data were analyzed by two-way ANOVA with Bonferroni’s multiple comparisons tests. For b and c, n = 3 biological independent samples per condition.
Extended Data Fig. 10 Foxc1 reduced heterochromatin associated with viral genes.
Neuro-2a cells were transfected with 500 ng/ml pcDNA or pFoxc1human, as indicated, for 40 h and infected with KOS for 2 h (MOI = 2) before ChIP-qPCR analysis for association of H3K9me3 with the genes indicated at the bottom. n = 3 biologically independent samples. Mean values ± s.d. are shown. Data were analyzed by two-way ANOVA with Bonferroni’s multiple comparisons tests.
Supplementary information
Supplementary Information
Supplementary Tables 1–3.
Source data
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed northern blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots and gels.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Unprocessed western blots.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Sun, B., Yang, X., Hou, F. et al. Regulation of host and virus genes by neuronal miR-138 favours herpes simplex virus 1 latency. Nat Microbiol 6, 682–696 (2021). https://doi.org/10.1038/s41564-020-00860-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-020-00860-1